Abstract
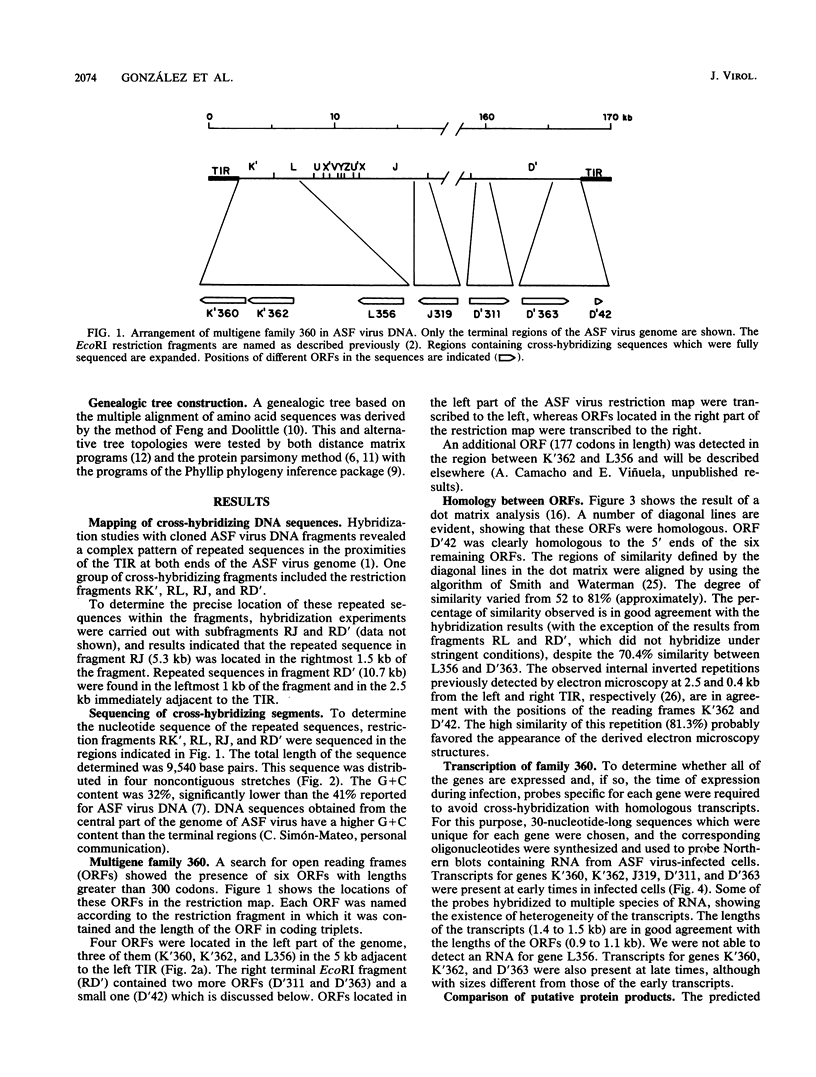
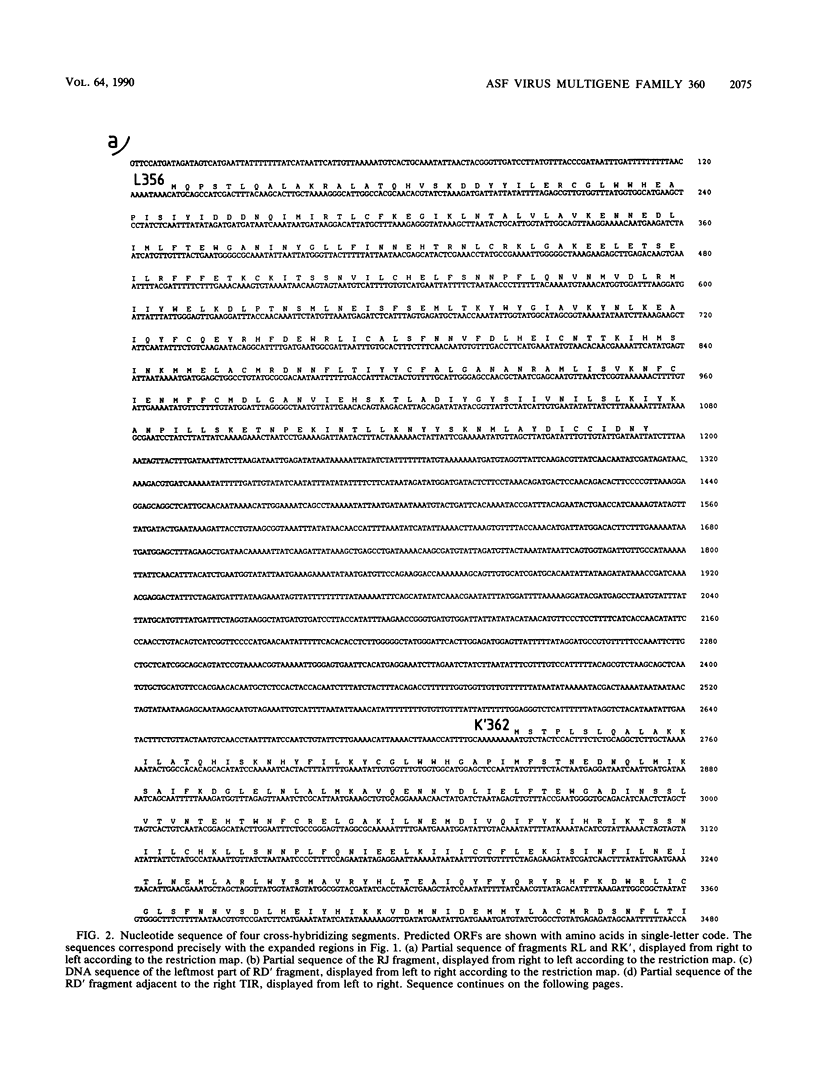
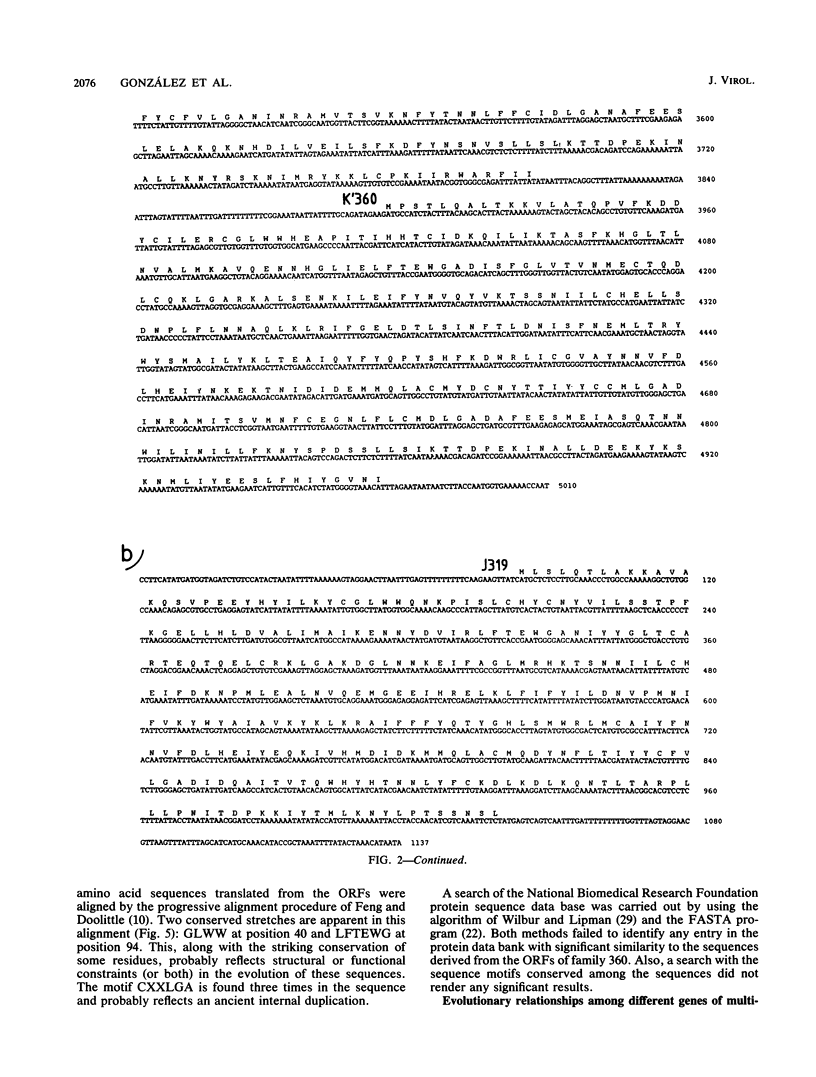
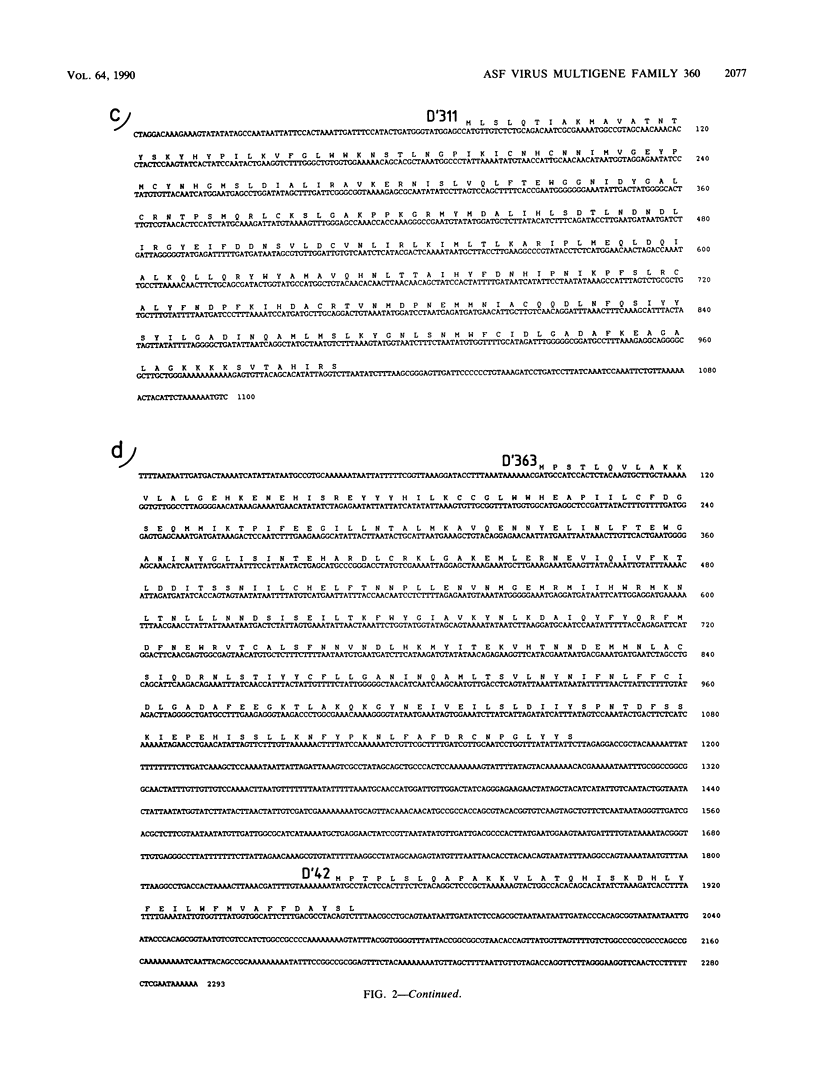
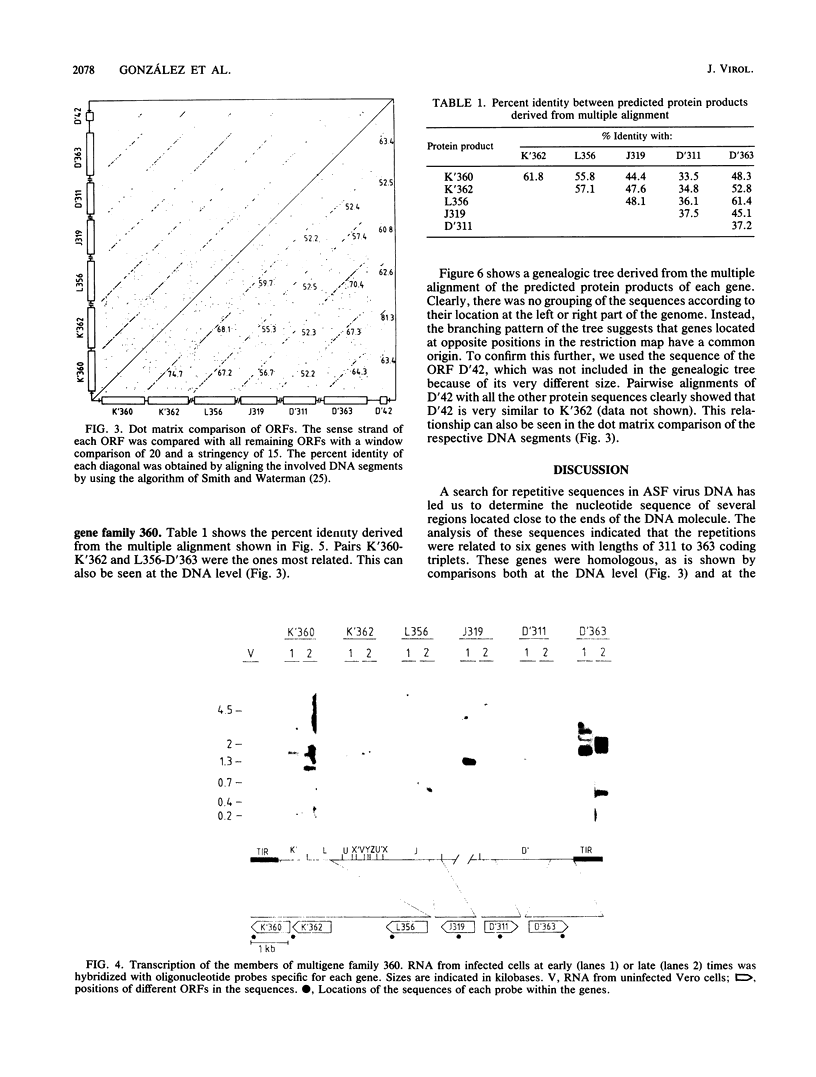
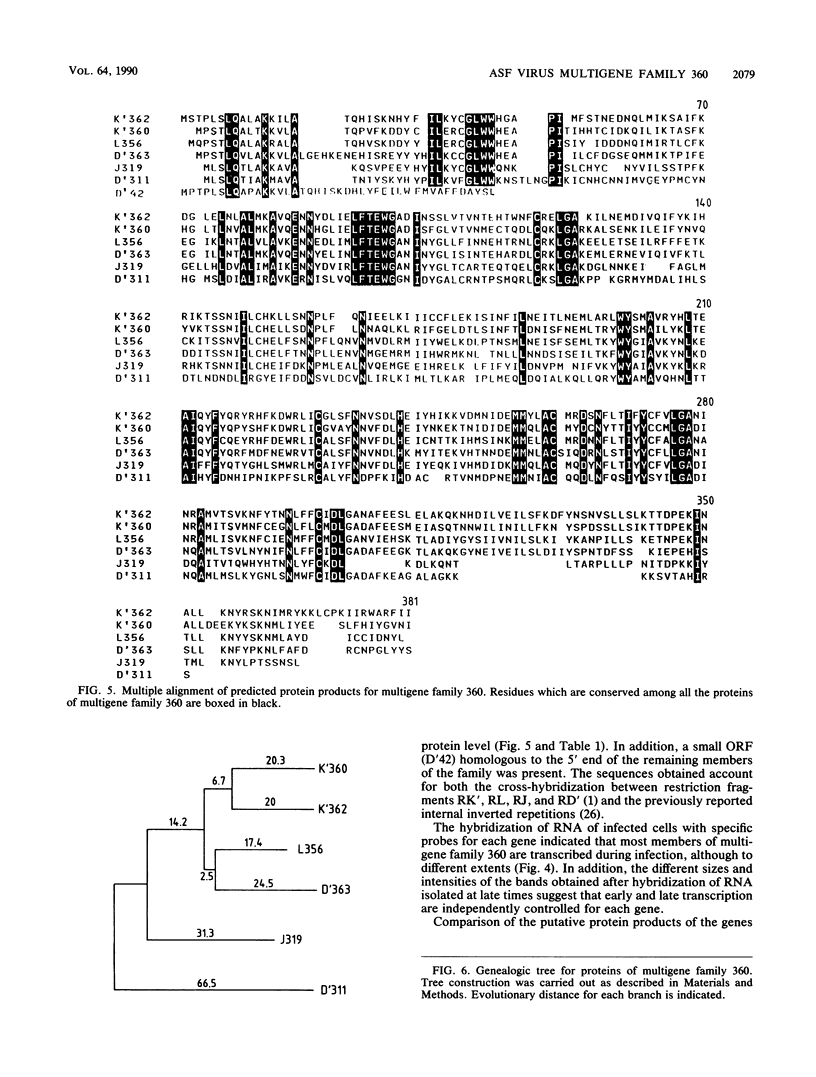
A group of cross-hybridizing DNA segments contained within the restriction fragments RK', RL, RJ, and RD' of African swine fever virus DNA were mapped and sequenced. Analysis of these sequences revealed the presence of a family of homologous open reading frames in regions close to the DNA ends. The whole family is composed of six open reading frames with an average length of 360 coding triplets (multigene family 360), four of which are located in the left part of the genome and two of which are in the right terminal EcoRI fragment. In close proximity to the right terminal inverted repeat, we found an additional small open reading frame which was homologous to the 5'-terminal portion of the other open reading frames, suggesting that most of that open reading frame has been deleted. These repeated sequences account for the previously described inverted internal repetitions (J.M. Sogo, J.M. Almendral, A. Talavera, and E. Viñuela, Virology 133:271-275, 1984). Most of the genes of multigene family 360 are transcribed in African swine fever virus-infected cells. A comparison of the predicted protein sequences of family 360 indicated that several residues are conserved, suggesting that an overall structure is maintained for every member of the family. The transcription direction of each open reading frame, as well as the evolutionary relationships among the genes, suggests that the family originated by gene duplication and translocation of sequences between the DNA ends.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almendral J. M., Almazán F., Blasco R., Viñuela E. Multigene families in African swine fever virus: family 110. J Virol. 1990 May;64(5):2064–2072. doi: 10.1128/jvi.64.5.2064-2072.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almendral J. M., Blasco R., Ley V., Beloso A., Talavera A., Viñuela E. Restriction site map of African swine fever virus DNA. Virology. 1984 Mar;133(2):258–270. doi: 10.1016/0042-6822(84)90393-3. [DOI] [PubMed] [Google Scholar]
- Archard L. C., Mackett M., Barnes D. E., Dumbell K. R. The genome structure of cowpox virus white pock variants. J Gen Virol. 1984 May;65(Pt 5):875–886. doi: 10.1099/0022-1317-65-5-875. [DOI] [PubMed] [Google Scholar]
- Blasco R., Agüero M., Almendral J. M., Viñuela E. Variable and constant regions in African swine fever virus DNA. Virology. 1989 Feb;168(2):330–338. doi: 10.1016/0042-6822(89)90273-0. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes L., Carrascosa A. L., Moreno M. A., Viñuela E. Titration of African swine fever (ASF) virus. J Gen Virol. 1976 Sep;32(3):471–477. doi: 10.1099/0022-1317-32-3-471. [DOI] [PubMed] [Google Scholar]
- Esposito J. J., Cabradilla C. D., Nakano J. H., Obijeski J. F. Intragenomic sequence transposition in monkeypox virus. Virology. 1981 Mar;109(2):231–243. doi: 10.1016/0042-6822(81)90495-5. [DOI] [PubMed] [Google Scholar]
- Feng D. F., Doolittle R. F. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol. 1987;25(4):351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- Fitch W. M., Margoliash E. Construction of phylogenetic trees. Science. 1967 Jan 20;155(3760):279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- González A., Talavera A., Almendral J. M., Viñuela E. Hairpin loop structure of African swine fever virus DNA. Nucleic Acids Res. 1986 Sep 11;14(17):6835–6844. doi: 10.1093/nar/14.17.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwal G. J., Moss B. Analysis of a large cluster of nonessential genes deleted from a vaccinia virus terminal transposition mutant. Virology. 1988 Dec;167(2):524–537. [PubMed] [Google Scholar]
- Ley V., Almendral J. M., Carbonero P., Beloso A., Viñuela E., Talavera A. Molecular cloning of African swine fever virus DNA. Virology. 1984 Mar;133(2):249–257. doi: 10.1016/0042-6822(84)90392-1. [DOI] [PubMed] [Google Scholar]
- Maizel J. V., Jr, Lenk R. P. Enhanced graphic matrix analysis of nucleic acid and protein sequences. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7665–7669. doi: 10.1073/pnas.78.12.7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Moyer R. W., Graves R. L., Rothe C. T. The white pock (mu) mutants of rabbit poxvirus. III. Terminal DNA sequence duplication and transposition in rabbit poxvirus. Cell. 1980 Nov;22(2 Pt 2):545–553. doi: 10.1016/0092-8674(80)90364-5. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. F., Waterman M. S. Identification of common molecular subsequences. J Mol Biol. 1981 Mar 25;147(1):195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Sogo J. M., Almendral J. M., Talavera A., Viñuela E. Terminal and internal inverted repetitions in African swine fever virus DNA. Virology. 1984 Mar;133(2):271–275. doi: 10.1016/0042-6822(84)90394-5. [DOI] [PubMed] [Google Scholar]
- Upton C., McFadden G. Tumorigenic poxviruses: analysis of viral DNA sequences implicated in the tumorigenicity of Shope fibroma virus and malignant rabbit virus. Virology. 1986 Jul 30;152(2):308–321. doi: 10.1016/0042-6822(86)90134-0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Weston K., Barrell B. G. Sequence of the short unique region, short repeats, and part of the long repeats of human cytomegalovirus. J Mol Biol. 1986 Nov 20;192(2):177–208. doi: 10.1016/0022-2836(86)90359-1. [DOI] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]




