Abstract
The Glu-134–Arg-135 residues in rhodopsin, located near the cytoplasmic end of the C helix, are involved in G protein binding, or activation, or both. Furthermore, the charge-neutralizing mutation Glu-134 to Gln-134 produces hyperactivity in the activated state and produces constitutive activity in opsin. The Glu/Asp-Arg charge pair is highly conserved in equivalent positions in other G protein-coupled receptors. To investigate the structural consequences of charge-neutralizing mutations at Glu-134 and Arg-135 in rhodopsin, single spin-labeled side chains were introduced at sites in the cytoplasmic domains of helices C (140), E (227), F (250), or G (316) to serve as “molecular sensors” of the local helix bundle conformation. In each of the spin-labeled rhodopsins, a Gln substitution was introduced at either Glu-134 or Arg-135, and the electron paramagnetic resonance spectrum of the spin label was used to monitor the structural response of the helix bundle. The results indicate that a Gln substitution at Glu-134 induces a photoactivated conformation around helices C and G even in the dark state, an observation of potential relevance to the hyperactivity and constitutive activity of the mutant. In contrast, little change is induced in helix F, which has been shown to undergo a dominant motion upon photoactivation. This result implies that the multiple helix motions accompanying photoactivation are not strongly coupled and can be induced to take place independently. Gln substitution at Arg-135 produces only minor structural changes in the dark- or light-activated conformation, suggesting that this residue is not a determinant of structure in the regions investigated, although it may be functionally important.
Keywords: G protein-coupled receptors, signal transduction, constitutive activation, EPR, site-directed spin-labeling
The structure and the molecular mechanism of activation of G protein-coupled receptors have been the focus of numerous biochemical and biophysical studies (2, 3). Rhodopsin has often served as the prototypic system, and low resolution structural models of bovine (4), frog (5), and squid (6) rhodopsins have been constructed based on cryoelectron microscopy of two-dimensional crystals. Recently, tertiary structures for the cytoplasmic loops of bovine rhodopsin also have been proposed based on the NMR studies of individual peptide segments corresponding to the cytoplasmic interhelical loops, C-terminal peptide sequence, and the total cytoplasmic domain (7). Additional experiments that use systematic mutagenesis of rhodopsin to introduce double cysteine replacements followed by crosslinking (8, 9) and double histidine replacements followed by metal ion binding (10) have provided clues to the protein topology in solution. Site-directed spin-labeling studies of rhodopsin have provided insights into both the dark structure and the apparent rigid-body movement of the helices relative to one another upon photoactivation (9, 11–13).
The charge-pair amino acids, Glu(Asp)-134 and Arg-135, near the cytoplasmic end of helix C, is highly conserved in G protein-coupled receptors (14), and replacements of the charge pair by neutral amino acids in rhodopsin and other G protein-coupled receptors affect G protein activation (15, 16). Previous work with rhodopsin has implicated the charge pair in the binding of transducin (15) or the release of GDP from the activated rhodopsin–transducin complex (17), or both. In addition, Glu-134 may be the acceptor of the proton from the protonated Schiff base upon light activation (18). Recently, the charge-neutralizing mutation E134Q has been found to cause constitutive activation of opsin (19) and, probably, rhodopsin (20) suggesting that the mutation may directly affect the conformational state of the protein. If this is the case, it should be possible to detect the perturbation by using spin labels introduced at the proper sites in the protein.
Previous site-directed spin-labeling studies have shown that spin-labeled side chains located at either site 227 or 250 in the cytoplasmic domains of the E and F helices, respectively (Fig. 1), undergo changes in mobility upon light activation. These changes were interpreted to reflect a rigid-body outward motion of the F helix in the cytoplasmic domain of the molecule (12). Increases in distance between the C and F helices upon photoactivation, determined from the interaction of spin labels in the two helices, supported this conclusion (9). Spin-labeled side chains at site 140 or 316, near the cytoplasmic ends of helices C and G, respectively (Fig. 1), also report photo-induced structural changes (8, 11, 21). The changes at site 140 were interpreted to reflect motion of the C helix upon photoactivation (11, 22). The specific origin of the photoactivated structural change detected by a spin label at C316 has not yet been identified, but it provides a “signature” for the photoactivated structure of rhodopsin in the vicinity of the G helix.
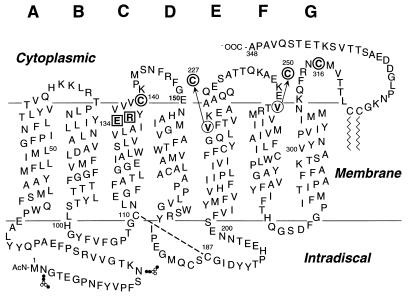
Figure 1.
A secondary structure model of bovine rhodopsin shows the sites of point mutations used in the present study. E134 and R135 (□) were replaced one at a time by Q, and this mutation was coupled to either C140 or V227C or V250C or C316 (○) for spin labeling. The dashed line between C110 and C187 in the intradiscal domain shows the conserved disulfide bond. The glycosylation sites at N-2 and N-15 are also shown.
In the present work, a single spin-labeled side chain at position 140, 227, 250, or 316 was used to monitor changes in the conformation of the helix bundle because of charge-neutralizing mutations in the pair E134/R135 of rhodopsin. The results indicate that the constitutively active mutation E134Q causes the dark rhodopsin structure to assume a photoactivated conformation around helices C and G but not around helix F. This result suggests that the multiple helix movements that apparently accompany photoactivation may be induced to occur independently and are thus not strongly coupled. The mutation R135Q results in only small localized structural perturbations and does not induce a photoactivated-like state in the dark.
MATERIALS AND METHODS
Materials.
Restriction endonucleases, T4 DNA ligase, and calf intestine alkaline phosphatase were purchased from New England Biolabs and GIBCO/BRL. Deoxyadenosine 5′-[α-35S]triphosphate was obtained from DuPont/NEN. The DNA sequenase version 2.0 sequencing kit used was from United States Biochemical. Dodecyl maltoside (DM) was obtained from Anatrace, Maumee, OH. 11-cis-Retinal was a gift from R. Crouch (Medical University of South Carolina and the National Eye Institute, National Institutes of Health). Anti-rhodopsin mAb, rho-1D4 (23), was purified from a myeloma cell line provided by R. S. Molday (University of British Columbia) and was coupled to cyanogen bromide-activated Sepharose™ 4B (Pharmacia LKB). The sulfhydryl-specific spin label (1-oxyl-2, 2, 5, 5-tetramethyl pyrroline-3-methyl)methanethiosulfonate was a gift from Kalman Hideg (University of Pecs, Hungary). The 4,4′-dithiodipyridine (PDS) was obtained from Aldrich Chemicals (Milwaukee, WI). The nonapeptide corresponding to the C-terminal sequence of rhodopsin was synthesized at the Biopolymer Laboratories, Center for Cancer Research, Massachusetts Institute of Technology.
Construction of Mutant Opsins.
Mutagenesis was carried out by restriction fragment replacement in the synthetic opsin gene as cloned in the vector pMT4 (24). Four mutant opsin genes that contained replacements at positions E134, or R135, or both with intact native cysteine residues at positions 140 and 316 were previously described (15). A set of mutants that contained replacements at position 134 or 135 including C316S was constructed by ligation of 4,301-bp XbaI fragments of the above mutants with 1,881-bp XbaI fragments of plasmids that contained C316S mutation (8, 21). Another set of mutants that contained the above replacements (position 134 or 135) and C140S mutation was prepared by ligation of 908-bp BsaAI fragments of the above mutants with 5,274-bp BsaAI fragments of plasmids that contained the rhodopsin gene with C140S (8). For the construction of mutants that contained single cytoplasmic cysteines at position 227 or 250, 1,812-bp XbaI-NdeI fragments of plasmids that contained mutations at positions 134, or 135, or both, together with C140S, were ligated with 328-bp XbaI-ApaI fragments that contained either V227C or V250C (13) and a 4,042-bp ApaI-NdeI fragment of rhodospin genes that contained a C316S replacement. Digestions with restriction endonucleases and analyses were carried out as recommended by the suppliers, and the restriction fragments were purified by extraction of DNA bands from agarose gels. Ligated DNAs were transformed into Escherichia coli DH5α (25). All plasmid preparations were carried out by using the alkaline lysis procedure (26).
Expression, Purification, and Characterization of Mutant Opsins.
Wild-type (wt) and mutant-opsin genes were transiently expressed in COS-1 cells. Cells were harvested 56 h after transfection and were incubated with 11-cis-retinal as described (24). Mutant rhodopsins were solubilized by the addition of 1/10 vol of 10% DM and were purified by using the immunoaffinity method as described (24). Mutant rhodopsins were eluted with 1 ml of the buffer containing 2 mM sodium phosphate (pH 6.0), 0.05% DM, and 50 μM C-terminal nonapeptide.
UV/Visible Spectroscopy and the Rates of Metarhodopsin II (Meta II) Decay.
The UV/Vis spectra of purified rhodopsin mutants were taken with a Perkin–Elmer λ7 UV/Vis spectrophotometer. For bleaching, the samples were illuminated for 30 sec at room temperature with light (λ > 495 nm) from a fiber optic light source.
The rates of Meta II decay were measured by monitoring the fluorescence increase due to retinal release (27). The assay was carried out with 2 μg of rhodopsin in 200 μl of 2 mM sodium phosphate (pH 6.0) and 0.05% DM at 20°C. After the bleaching for 30 sec, the fluorescence was measured by using excitation and emission wavelengths at 295 nm (slit = 0.25 nm) and 330 nm (slit = 12 nm).
Titration and Spin Labeling of Sulfhydryl Groups.
The quantitation of reactive sulfhydryls in the folded proteins was carried out by reaction with PDS as described (1, 28). The reactive sulfhydryl content was determined from the absorbance at 324 nm (29).
The procedure for spin labeling of the sulfhydryl groups with methanethiosulfonate and EPR spectroscopy were as described (21). The EPR spectra to be compared were normalized to equal the number of spins.
RESULTS
Characterization of the Rhodopsin Mutants.
Mutant opsins were transiently expressed in COS-1 cells to levels comparable to the wt (data not shown) and were purified to homogeneity by immunoaffinity procedures previously described (24). Each mutant rhodopsin showed chromophore formation with an absorption maximum at 500 nm (Table 1) and wt-like behavior after bleaching and acidification (data not shown). Most of the mutants showed Meta II decay rates comparable to that of the wt (Table 1).
Table 1.
Characterization of mutant rhodopsins
| Rhodopsin Mutants | λmax (nm) | A280/A500 | Meta II Decay (T½, min) |
|---|---|---|---|
| wt | 500 | 1.7 | 12.2 |
| C140/C316S | 500 | 1.7 | 10.7 |
| E134Q/C140/C316S | 500 | 1.8 | 10.8 |
| R135Q/C140/C316S | 500 | 1.8 | 13.0 |
| C140S/V227C/C316S | 500 | 1.8 | 8.3 |
| E134Q/C140S/V227C/C316S | 500 | 1.7 | 11.5 |
| R135Q/C140S/V227C/C316S | 500 | 1.7 | 11.9 |
| C140S/V250C/C316S | 500 | 1.8 | 9.9 |
| E134Q/C140S/V250C/C316S | 500 | 1.8 | 14.1 |
| R135Q/C140S/V250C/C316S | 501 | 1.7 | 14.0 |
| C140S/C316 | 500 | 1.7 | 12.1 |
| E134Q/C140S/C316 | 500 | 1.7 | 12.1 |
| R135Q/C140S/C316 | 500 | 1.7 | 13.2 |
Approximately 2 mol of sulfhydryl groups per mol rhodopsin were found by using the PDS titration in the wt protein, consistent with earlier results (28). These reactive sulfhydryl groups are apparently those in wt cysteines C140 and C316 (21, 28). In the work presented here, rhodopsin mutants were prepared that contained a single cysteine substituted at site 140, 227, 250, or 316 in a C140S/C316S mutant background for the purpose of attachment of spin label sensors. Each of these mutants had ≈1 mol of reactive cysteine per mol of rhodopsin as determined by the PDS titration. These mutants, after reaction of the substituted cysteine residue with methanethiosulfonate to generate the nitroxide side chain designated R1 (Fig. 2), will be referred to as “sensor mutants” (SM) and denoted as SM140, SM227, SM250, and SM316, depending on the position of cysteine available for the labeling. Because the EPR spectrum of the R1 side chain is sensitive to the protein conformation, the various SMs were used to monitor conformational changes in the state of the helix bundle due to the additional mutations E134Q or R135Q.
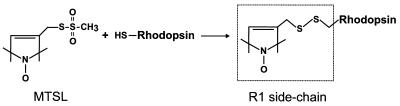
Figure 2.
The reaction of the methanethiosulfonate reagent produces the spin-labeled side chain R1.
Structural Changes in Rhodopsin Due to Mutations E134Q and R135Q.
The dynamics of the R1 side chain in a folded protein, reflected by the EPR spectral lineshape, is determined by interactions of the side chain with the local environment (30). In the present work, we use changes in spectral lineshape in a qualitative way to infer the changes in R1 dynamics and hence local protein structure. The term “mobility” will be used to refer to the general dynamic state of an R1 side chain population and will encompass both frequency and amplitude effects. Many of the spectra reflect multiple conformations of the side chain, each with a distinct mobility. A shift in the relative population of such states also will be described in terms of a change in general mobility.
Because of the fundamental difference in effects detected by R1 side chains at sites 316 and 140 from those at sites 250 and 227, we discuss these groups separately below.
Structural Changes Sensed by R1 at C316 and C140.
Residues 316 and 140 lie near the cytoplasmic termination of the transmembrane G and C helices, respectively (Fig. 1). The relatively broad, multicomponent lineshapes of the EPR spectra of SM316 and SM140 indicate tertiary contacts of the nitroxides in the structure (Fig. 3 A and B, red traces; ref. 21). As a result, the spectral lineshapes are sensitive indicators of local tertiary structure.
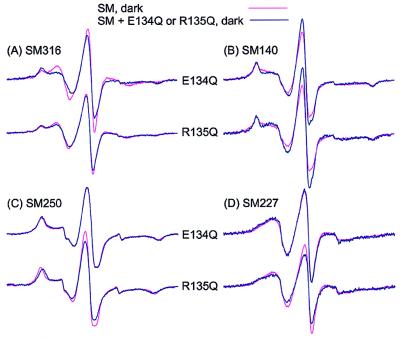
Figure 3.
The EPR spectra of the spin-labeled rhodopsin SMs show the effect of mutation E134Q or R135Q in the dark state: (A) SM316, (B) SM140, (C) SM250, and (D) SM227. Each superimposed spectral pair compares the spin-labeled rhodopsin SM alone (red trace) with the indicated mutant (blue trace).
Fig. 3A compares the EPR spectrum of SM316 in the dark state (red traces) with those from SM316/E134Q and SM316/R135Q (blue traces), also in the dark state. The data reveal a difference between the spectra of SM316 and SM316/E134Q, although those of SM316 and SM316/R135 are rather similar.
Fig. 4A compares the spectra of SM316, SM316/E134Q, and SM316/R135Q in the dark and after photoactivation to the Meta II state. The change in the SM316 spectrum due to photoactivation was previously reported (21) and is similar to that shown here. As shown, the difference between the spectra of SM316 and SM316/E134Q in the dark (Fig. 3A, top trace) is the same as the difference between the spectra of SM316 in the dark and photoactivated state (Fig. 4A, top trace). Thus, we surmise that the E134Q mutation induces the photoactivated structure of the protein in the neighborhood of C316, near the end of the G helix. Consistent with this conclusion is the result that photoactivation of the SM316/E134Q mutation produces little spectral change (Fig. 4A, middle trace); i.e., the spectra of the photoactivated SM316 and dark SM316/E134Q rhodopsins are similar. Photoactivation of the SM316/R135Q, on the other hand, results in a spectral change comparable to the SM316 (Fig. 4A, bottom trace) consistent with the weak perturbation of the dark structure noted above.
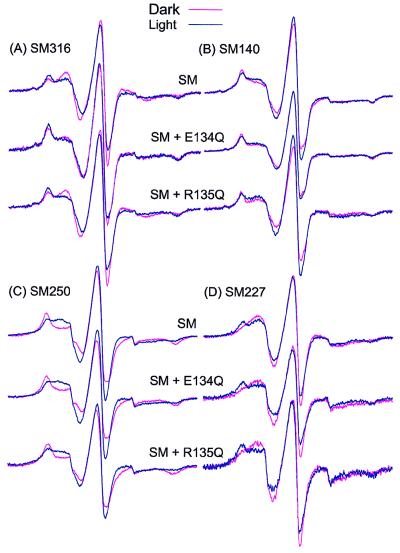
Figure 4.
The EPR spectra of the spin-labeled rhodopsin SMs with and without E134Q or R135Q mutations shows the effects of photoactivation to the Meta II state: (A) SM316, (B) SM140, (C) SM250, and (D) SM227. Each superposed spectral pair compares the dark (red trace) and photoactivated (blue trace) states of the indicated mutants.
Fig. 3B compares the corresponding EPR spectra for SM140 in the dark state. The spectra of SM140 and SM140/E134Q show a small but significant difference, apparently because of an increase in population of a more mobile state, as reflected by the increase in intensity of the central resonance line. The spectra of SM140 and SM140/R135Q also show a similar difference in the dark state that can be attributed to a shift in the spin population to a more mobile state. The underlying structural perturbation because of R135Q is apparently localized to the neighborhood of SM140, and is sensed only weakly in SM316, as noted above.
Fig. 4B shows the effect of photoactivation to Meta II on the SM140 spectra. The change in the SM140 spectrum (top trace) is similar to that previously reported (21) and reflects a small increase in the general mobility of the R1 side chain upon photoactivation. As for SM316/E134Q, photoactivation of the SM140/E134Q produces little additional change in the spectrum (middle trace). This finding suggests that the E134Q mutation has already produced a photoactivated-like structure in the vicinity of C140, just as it did at C316. However, in this case the spectra of photoactivated SM140 and dark SM140/E134Q are not identical. Photoactivation of SM140/R135Q produces small spectral changes indicating an increase in label mobility (bottom trace), similar to that of the SM140 alone (top trace).
The conclusion from these experiments is that the constitutively active mutation E134Q produces structural changes in the neighborhood of sites 316 and 140 in the dark state that resemble those produced by photoactivation. The mutation R135Q results in a localized structural change but does not produce a photoactivated conformation, because a change similar to that in the SMs is observed after photoactivation.
Structural Changes Sensed by R1 at 250 and 227.
Based on previous work, the R1 nitroxide side chain at position 250 in helix F faces the interior of the helix bundle and is strongly immobilized by extensive tertiary contacts. The R1 nitroxide at site 227 is located on the outer surface of helix E, facing the lipid bilayer, and has a high mobility. Upon Meta II formation, R1 side chains at 250 and 227 become more mobile and less mobile, respectively (12). Apparently, an outward movement of helix F reduces tertiary interactions on its buried surface and increases tertiary contact with the outer surface of adjacent helix E (12). Thus R1 side chains at sites 250 and 227 are equivalent monitors of helix F motion but have opposite changes in mobility.
Fig. 3 C and D shows the EPR spectra for SM250 and SM227 in the dark state with and without the mutations E134Q and R135Q. In contrast to the results with SM316 and SM140, the presence of the E134Q mutation has essentially no effect on the EPR spectral lineshapes. Thus, this mutation does not perturb the position of the F helix in the dark state. The mutation R135Q results in a small but distinct decrease in the splitting of the outer hyperfine extrema in SM250/R135Q relative to SM250 (Fig. 3C, bottom trace), indicating a slight increase in R1 mobility. In SM227, R135Q produces a slight decrease in R1 mobility, seen most clearly in the decrease in the central resonance intensity. Together, these effects suggest a small outward displacement of helix F.
Fig. 4 C and D presents the changes in EPR spectra observed for SM250 and SM227 upon photoactivation with and without mutations E134Q and R135Q. As is evident, the spectral changes for proteins carrying the charge-neutralizing mutations are strikingly similar to those in the SMs alone, indicating little perturbation of the F helix in either the dark or the photoactivated structure by these mutations.
The general conclusion from these experiments is that neither the constitutively active mutation E134Q nor the mutation R135Q produces a significant effect on the tertiary structure of the protein in the vicinity of the F helix. The small change in mobility of R1 at site 250 due to R135Q mutation is consistent with a slight outward displacement of the F helix but much smaller than that caused by photoactivation.
DISCUSSION
Structural Changes Induced by E134Q and R135Q.
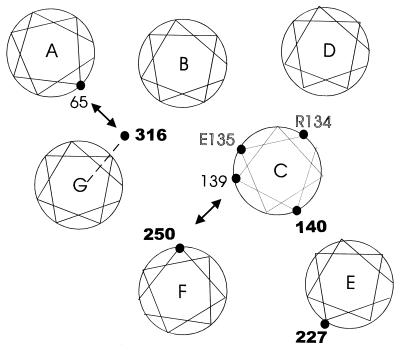
In the experiments reported here, spin-labeled side chains have been used to monitor structural perturbations in rhodopsin due to charge-neutralizing mutations in the G protein-coupled receptor conserved sequence Glu/Asp–Arg, occurring at positions 134 and 135 in rhodopsin. In Fig. 5, the approximate topological locations of the sites of interest are mapped onto a two-dimensional projection representing a section at the membrane–aqueous boundary of the cytoplasmic domain, based on the recent model for the helix packing in rhodopsin derived from cryoelectron microscopy (5). The mapping of the residues on the helical surfaces is based on previous site-directed spin-labeling studies (8, 9, 11, 12) and based on disulfide crosslinking (8, 9). The location of residue 65 is based on site-directed spin-labeling data (J. Klein, C.A., W.H. & H.G.K., unpublished work) and is consistent with that predicted by Baldwin based on statistical analysis of sequence data (14). The position of the C helix in this section is changed from earlier models based on cryoelectronmicroscopy but remains consistent with the previous spin labeling data. In Fig. 5, boldface numbers indicate the location of spin-label sensor sites used in this study. Photoactivation of rhodopsin to the Meta II state produces distinctive “signature” changes in the EPR spectra of spin-label sensors at each of the sites studied.
Figure 5.
A structural model of the rhodopsin helix packing in a projection near the cytoplasmic membrane–aqueous boundary based on cryoelectron microscopy data (5). The mapping of specific residues on each helix is based on spin-labeling and disulfide crosslinking data (see Discussion). The double arrows indicate sites where substituted cysteines can be crosslinked by disulfide formation and where attached nitroxides show spin-spin interaction indicating proximity. The dashed line to residue 316 indicates uncertainty regarding the mapping of this residue (see Discussion). The residues in bold type are those to which spin label sensors are attached to monitor local structure. The residues in outline identify the location of the charged residues mutated in the present study.
The EPR spectrum of R1 at site SM140 shows changes upon photoactivation that have been attributed to a probable movement of helix C (11, 22). The constitutively active mutation E134Q produces a spectral change in SM140, in the dark, that is of a similar type and magnitude but not identical to that produced by photoactivation of SM140 in the absence of the mutation (Figs. 3B and 4B, top trace). Detailed interpretation of the spectral difference between SM140 and SM140/E134Q may be problematic, because the presence of two mutations (C140R1 and E134Q) in the same region of helix C may lead to enhanced structural perturbation. However, it is significant that photoactivation of the SM140/E134Q produces no further changes, implying that the mutation has already produced a protein conformation equivalent to that of the photoactivated state in this region.
Residue 316 lies near the termination of the G transmembrane helix (Fig. 1), although it is not yet clear whether this residue lies in a helical structure. This uncertainty is indicated by the dashed line in Fig. 5. However, residue 316 is known to lay in proximity to residue 65 in helix A (8), indicated by the double arrow. The presence of an immobilized component in the EPR spectrum of SM316 may arise from a tertiary interaction of the nitroxide with helices A, or B, or both helices or the C-terminal domain that extends from helix G. Photoactivation of rhodopsin produces changes in the SM316 spectrum (Fig. 4A, top trace) that may arise from relative motion of any or all of helices A, B, or G, or the C-terminal domain.
It is remarkable that the difference between the EPR spectra of SM316 and the SM316/E134Q mutant in the dark is essentially identical to that between the dark and photoactivated SM316. This implies that the mutation produces a photoactivated-like conformation around 316 in the dark state. The mechanism of coupling of helix C perturbations because of E134Q to the distant site 316 is unknown. One intriguing possibility is that favorable helix packing in the A-B-C group (Fig. 5) results in a concerted motion of these helices that would produce concomitant changes in SM140 and SM316.
R1 side chains at sites 227 and 250 undergo opposite changes in mobility upon photoactivation of rhodopsin that have been interpreted in terms of an outward motion of helix F relative to helix C (8, 9). In accord with this interpretation, R1 side chains placed at both sites 139 and 250 show spin-spin interactions that indicate close proximity in the dark state (indicated by the double arrow in Fig. 5), with an increase in distance upon photoactivation (9). Introduction of the E134Q mutation in helix C leaves the EPR spectral lineshapes in SM227 and SM250 relatively unchanged in both the dark and the photoactivated states. Thus the photoactivated-like perturbations observed around 140 and 316 due to this mutation are not coupled to a movement of the F helix. The observation raises the possibility that the helical movement detected by R1 at sites 140 and 316 may occur independently of those associated with the motion of helix F in the normal excitation pathway of rhodopsin, perhaps corresponding to the formation of the two Meta II states suggested by Hofmann and Arnis (18).
The mutation R135Q produces only small changes in structure of the helix bundle in the dark state detected by the spin labels. That a small difference in EPR spectra is detected between SM140 and SM140/R135Q is perhaps not surprising, because the spin label and R135Q mutation are in close proximity in helix C (Fig. 5). The effect of R135Q mutation is apparently local, because it produces only a small fraction of the photoactivated change detected in SM316. The slight changes in structure detected by spin labels at sites 250 and 227 because of R135Q are consistent with an outward displacement of helix F but much smaller than that resulting from photoactivation. Again, these are likely to be local perturbations resulting from the direct contact of helix C, containing R135, with helix F in the structural model (Fig. 5). Under any condition, the structural perturbation induced by the R135Q mutation is not globally equivalent to that produced by photoactivation, and photoactivation of the R135Q mutant leads to spectral changes similar to those in the spin-labeled proteins with the wt residue at 135. The above arguments suggest that R135 is not a critical structural residue. Cohen et al.(19) found that the R135Q mutation showed normal transducin activation in membranes, although it was from 10- to 20-fold less active than wt rhodopsin in detergent solution.
Possible Structure/Activity Relationships in the E134Q Constitutively Active Mutant.
The E134Q mutant produces a Meta II species that has hyperactivity with regard to transducin activation and shows constitutive activity in the opsin state (19, 31). The above results provide a possible relationship between protein structural changes and these activities. Interestingly, we have shown previously that the charge neutralization leading to hyperactivity does not appear to affect the phosphorylation of the receptor. The mutations in which both of the residues are changed to Ala have little effect on phosphorylation (32). This result implies that there are different determinants for the binding of the G protein and the kinase. Although no structural perturbations were caused by mutations at site 135, the neutralization of this charge was shown essentially to abolish G protein activity in DM solutions. This observation suggests that this residue may be important for direct contact with the G protein either in binding or in activation, at least in DM solutions.
When rhodopsin passes from the dark state to the photoactivated Meta II state, two protons are taken up. Proton uptake is blocked in the E134Q mutation, suggesting that protonation of E134 may account for one of the adsorbed protons (18), although other possibilities exist. If this is the case, the activated form of the receptor requires protonation of E134. Electrostatically, the mutation E134Q is equivalent to protonation of E134. If electrostatic interactions of E134 are essential to maintaining the conformation of helix C in the dark state, for example through a salt bridge, the mutation should “preset” the conformation around this site in the photoactivated state. This would account for the hyperactivity and constitutive activity because the conformational equilibria in the photoexcited protein would be shifted in favor of the activated state.
The data presented here show that the E134Q mutation does in fact produce a photoactivated-like state in the dark around helices C and G, presetting the structure in this region to the activated form, lending support to the simple picture outlined above. However, only part of the protein structure is preset, and one does not expect significant activity in the dark state without concomitant movement of the F helix, which is the dominant movement upon photoactivation (12).
A number of mutants in rhodopsin have been found that have constitutive activity higher than that of E134Q (33), and it is anticipated that some of these mutants may involve perturbation of the F helix structure. Future studies will explore structural changes in these mutations by using the methods presented above.
Acknowledgments
We thank U. L. RajBhandary for the helpful discussions during the experiment. We also thank Ms. Judith Carlin for assisting in the preparation of the manuscript. This work was supported by National Institutes of Health Grants EY05216 (W.L.H.) and GM 28289 (H.G.K.) and the Jules Stein Professorship Endowment (W.L.H.).
ABBREVIATIONS
- Opsin mutations are designated by using the single letter code for the original amino acid
the amino acid number and the single letter code for the substituted amino acid, in that order. DM, dodecyl maltoside
- Meta II
Metarhodopsin II
- R1
the designation for the spin-label side chain
- SM
sensor mutant
- wt
wild type
- PDS
4,4′-dithiodipyridine
Footnotes
This is paper No. 27 in the series Structure and Function in Rhodopsin. The previous paper is ref. 1.
References
- 1.Cai K, Langen R, Hubbell W L, Khorana H G. Proc Natl Acad Sci USA. 1997;94:14267–14272. doi: 10.1073/pnas.94.26.14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helmreich E J M, Hofmann K-P. Biochim Biophys Acta. 1996;1286:285–322. doi: 10.1016/s0304-4157(96)00013-5. [DOI] [PubMed] [Google Scholar]
- 3.Wess J. FASEB J. 1997;11:346–354. [PubMed] [Google Scholar]
- 4.Unger V M, Schertler G F X. Biophys J. 1995;68:1776–1786. doi: 10.1016/S0006-3495(95)80354-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unger V M, Hargrave P A, Baldwin J M, Schertler G F X. Nature (London) 1997;389:203–206. doi: 10.1038/38316. [DOI] [PubMed] [Google Scholar]
- 6.Davies A, Schertler G F X, Gowen B E, Saibil H R. J Struct Biol. 1996;117:36–44. doi: 10.1006/jsbi.1996.0067. [DOI] [PubMed] [Google Scholar]
- 7.Yeagle P L, Alderfer J L, Albert A D. Biochemistry. 1997;36:9649–9654. doi: 10.1021/bi970908a. [DOI] [PubMed] [Google Scholar]
- 8.Yang K, Farrens D L, Altenbach C, Hubbell W L, Khorana H G. Biochemistry. 1996;35:14040–14046. doi: 10.1021/bi962113u. [DOI] [PubMed] [Google Scholar]
- 9.Farrens D L, Altenbach C, Yang K, Hubbell W L, Khorana H G. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 10.Sheikh S P, Zvyaga T A, Lichtarge O, Sakmar T P, Bourne H R. Nature (London) 1996;383:347–350. doi: 10.1038/383347a0. [DOI] [PubMed] [Google Scholar]
- 11.Farahbakhsh Z T, Ridge K D, Khorana H G, Hubbell W L. Biochemistry. 1995;34:8812–8819. doi: 10.1021/bi00027a033. [DOI] [PubMed] [Google Scholar]
- 12.Altenbach C, Yang K, Farrens D L, Farahbakhsh Z T, Khorana H G, Hubbell W L. Biochemistry. 1996;35:12470–12478. doi: 10.1021/bi960849l. [DOI] [PubMed] [Google Scholar]
- 13.Yang K, Farrens D L, Hubbell W L, Khorana H G. Biochemistry. 1996;35:12464–12469. doi: 10.1021/bi960848t. [DOI] [PubMed] [Google Scholar]
- 14.Baldwin J M, Schertler G F X, Unger V M. J Mol Biol. 1997;272:144–164. doi: 10.1006/jmbi.1997.1240. [DOI] [PubMed] [Google Scholar]
- 15.Franke R R, Sakmar T P, Graham R M, Khorana H G. J Biol Chem. 1992;267:14767–14774. [PubMed] [Google Scholar]
- 16.Scheer A, Fanelli F, Costa T, DeBenedetti P G, Cotecchia S. Proc Natl Acad Sci USA. 1997;94:808–813. doi: 10.1073/pnas.94.3.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnis S, Fahmy K, Hofmann K P, Sakmar T P. J Biol Chem. 1994;269:23879–23881. [PubMed] [Google Scholar]
- 18.Arnis S, Hofmann K-P. Proc Natl Acad Sci USA. 1993;90:7849–7853. doi: 10.1073/pnas.90.16.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen G B, Yang T, Robinson P R, Oprian D D. Biochemistry. 1993;32:6111–6115. doi: 10.1021/bi00074a024. [DOI] [PubMed] [Google Scholar]
- 20.Acharya S, Karnik S S. J Biol Chem. 1996;271:25406–25411. doi: 10.1074/jbc.271.41.25406. [DOI] [PubMed] [Google Scholar]
- 21.Resek J F, Farahbakhsh Z T, Hubbell W L, Khorana H G. Biochemistry. 1993;32:12025–12032. doi: 10.1021/bi00096a012. [DOI] [PubMed] [Google Scholar]
- 22.Farahbakhsh Z T, Hideg K, Hubbell W L. Science. 1993;262:1416–1419. doi: 10.1126/science.8248781. [DOI] [PubMed] [Google Scholar]
- 23.Molday R S, Mackenzie D. Biochemistry. 1983;22:653–660. doi: 10.1021/bi00272a020. [DOI] [PubMed] [Google Scholar]
- 24.Oprian D D, Molday R S, Kaufman R J, Khorana H G. Proc Natl Acad Sci USA. 1987;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoue H, Nojima H, Okayama H. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 27.Farrens D L, Khorana H G. J Biol Chem. 1995;270:5073–5076. doi: 10.1074/jbc.270.10.5073. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y S, Hubbell W L. Memb Biochem. 1978;1:107–130. doi: 10.3109/09687687809064162. [DOI] [PubMed] [Google Scholar]
- 29.Grassetti D R, Murray Jr J F. Arch Biochem Bipohys. 1967;119:41–49. doi: 10.1016/0003-9861(67)90426-2. [DOI] [PubMed] [Google Scholar]
- 30.Mchaourab H S, Lietzow M A, Hideg K, Hubbell W L. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 31.Fahmy K, Sakmar T P. Biochemistry. 1993;32:7229–7236. doi: 10.1021/bi00079a020. [DOI] [PubMed] [Google Scholar]
- 32.Thurmond R L, Cruzenet C, Reeves P J, Khorana H G. Proc Natl Acad Sci USA. 1997;94:1715–1720. doi: 10.1073/pnas.94.5.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao V R, Oprian D D. Ann Rev Biophys Biomol Struct. 1996;25:287–314. doi: 10.1146/annurev.bb.25.060196.001443. [DOI] [PubMed] [Google Scholar]