Abstract
There is considerable concern that bovine prions from cattle with bovine spongiform encephalopathy (BSE) may have been passed to humans (Hu), resulting in a new form of Creutzfeldt–Jakob disease (CJD). We report here the transmission of bovine (Bo) prions to transgenic (Tg) mice expressing BoPrP; one Tg line exhibited incubation times of ≈200 days. Like most cattle with BSE, vacuolation and astrocytic gliosis were confined in the brainstems of these Tg mice. Unexpectedly, mice expressing a chimeric Bo/Mo PrP transgene were resistant to BSE prions whereas mice expressing Hu or Hu/Mo PrP transgenes were susceptible to Hu prions. A comparison of differences in Mo, Bo, and Hu residues within the C terminus of PrP defines an epitope that modulates conversion of PrPC into PrPSc and, as such, controls prion transmission across species. Development of susceptible Tg(BoPrP) mice provides a means of measuring bovine prions that may prove critical in minimizing future human exposure.
Keywords: Creutzfeldt–Jakob disease, transgenics, bioassays
Investigations of the prion diseases have taken on new significance with the reports of more than 20 cases of an atypical, variant Creutzfeldt–Jakob disease (vCJD) in teenagers and young adults (1–5). To date, all of these cases have been reported from Great Britain and France. It now seems possible that bovine prions from “mad cows” passed to humans through the consumption of tainted beef products. It is generally thought that prion-contaminated offal initially from sheep and later from cattle was used in the manufacture of meat and bone meal, and that this is the source of prions responsible for bovine spongiform encephalopathy (BSE) (6, 7).
Understanding the species barrier is paramount in our efforts to evaluate the impact of the BSE epidemic in Britain on human health (8). It has been estimated that almost one million cattle were infected with BSE prions with an incubation time of about 5 years. This may be an underestimation of the disease incidence, as most cattle were slaughtered between 2 and 3 years of age (9). Nevertheless, more than 160,000 cattle, primarily dairy cows, have died of BSE over the past decade (8). In the late 1970s, the hydrocarbon-solvent extraction method used in the rendering of offal began to be abandoned resulting in meat and bone meal with a much higher fat content (6). It is now thought that this change in the rendering process allowed scrapie prions from sheep to survive rendering and to be passed into cattle (10, 11).
Although many plans have been offered for the culling of older cattle to minimize the spread of BSE (8), it seems more important to monitor the frequency of prion disease in cattle as they are slaughtered for human consumption. No reliable, specific test for prion disease in live animals is available (12), but immunoblotting of the brainstems of cattle for PrPSc might provide a reasonable approach to establish the incidence of subclinical BSE in cattle entering the human food chain (13–17). Determining how early in the incubation period PrPSc can be detected by immunological methods is complicated by the lack of a reliable, sensitive, and relatively rapid bioassay.
We report here on the production of Tg mice expressing bovine PrP (TgBoPrP) that are susceptible to BSE prions and exhibit incubation times of less than 250 days. These mice faithfully reproduce the hallmarks of the bovine disease, with a similar localization of pathology and PrPSc accumulation in the brainstem. Furthermore, PrP fragments identified after limited proteolytic digestion of PrPSc in the brains of Tg(BoPrP) mice were indistinguishable from those found in brains of cattle afflicted with BSE. Our studies also provide insight into the nature of the “species barrier” for transmission of prions (18, 19) and suggest a method for producing Tg mice that are highly susceptible to foreign prions from humans and ungulates.
MATERIALS AND METHODS
Sources of BSE Inocula, Reagents, and Laboratory Animals.
BSE isolates used in this study were described previously (16). Normal CD-1 and FVB mice were obtained from Charles River Laboratories. Tg(MoPrP-A)4053 mice were described previously (20). BoPrP or MBo2M PrP ORF cassettes (16) were ligated into the cosTet vector for microinjection (21). MBo2M PrP was constructed as described previously for similar chimeric PrP transgenes (22) resulting in eight bovine substitutions in MoPrP corresponding to HuPrP residues: 97, 109, 138, 143, 145, 155, 184, and 186. Fertilized oocytes from FVB/Prnp0/0 mice were produced by repeated backcrosses of Prnp0/0 mice (23) with FVB mice. Founder Tg(BoPrP)Prnp0/0 and Tg(MBo2M)Prnp0/0 mice were identified by PCR screening for transgene integration by using a Beckman robotic workstation. Tg mice from the F2 generation were sacrificed, and the level of BoPrPC expression in the brain was determined by dot blot by using 2-fold dilutions of the homogenate that were compared with BoPrPC in bovine brain.
Determination of BSE Incubation Periods.
Tg(BoPrP)Prnp0/0 mice from lines chosen for transmission studies and control mice were inoculated intracerebrally with 30 μl of a 10% homogenate prepared with PBS. The well being of the mice was monitored daily and the neurologic status was assessed semiweekly. Mice were scored positive for prion disease when two of three signs of neurologic dysfunction were present and progressive deterioration of the animals was apparent. Most reliable signs of neurologic dysfunction for monitoring prion disease in mice are (i) truncal ataxia, (ii) increased tone of the tail, and (iii) lack of forelimb extensor response when lifted by the tail (19, 22, 24).
Immunological Reagents.
The anti-PrP rabbit polyclonal antiserum designated #9095 was raised against a synthetic peptide corresponding to residues 90–145 of BoPrP. Three rabbits were immunized with 0.25 mg of the peptide dispersed into adjuvant, and RIBI/three booster immunizations were performed with the same amount of the peptide in RIBI adjuvant. The antiserum was used at a dilution of 1:2,000 for Western immunoblotting and at a dilution of 1:1,000 for histoblotting. The antiserum reacted strongly with PrP from cattle, mice, and Syrian hamsters.
Preparation of Brain Homogenates and Western Blotting.
Brain tissue was homogenized in PBS by repeated extrusion through syringe needles of decreasing diameter. Samples that were digested with proteinase K were diluted to approximately 2 mg/ml of total protein as determined by BCA assay (Bio-Rad) in PBS containing 0.5% Nonidet P-40 and 0.5% sodium deoxycholate immediately before digestion. Proteinase K (50 μg/ml; Boehringer Mannheim) was added and samples were incubated at 37°C for 1 h. The reaction was terminated by addition of Pefabloc (Boehringer Mannheim) to 1 mM. Control aliquots were added immediately to sample buffer. An equal volume of 2× SDS sample buffer containing 125 mM Tris⋅HCl at pH 7.4, 4% SDS, and 20% glycerol was added to all samples and each was boiled for 5 min before loading on an 11% SDS polyacrylamide gel. Procedures for immunoblotting and development by using the ECL system have been described (22).
Neuropathology.
Brain tissue was immersion-fixed in 10% buffered formalin solution after sacrificing the animals. The brains were embedded in paraffin and histological sections were prepared and stained with hematoxylin and eosin for evaluation of spongiform degeneration. Peroxidase immunohistochemistry with antibodies to glial fibrillary acidic protein was used to evaluate the degree of reactive astrocytic gliosis.
The hydrolytic autoclaving technique was performed as described previously (25).
Histoblots for PrPSc.
Histoblots were performed as described (15). Animals were sacrificed by asphyxiation with CO2. The brain was removed rapidly and frozen in powdered dry ice. Ten-micron thick cryostat sections were cut, mounted on glass, thawed, and pressed to nitrocellulose membrane wetted in lysis buffer containing 0.5% Nonidet P-40/0.5% Na deoxycholate,100 mM NaCl,10 mM EDTA, and 10 mM Tris⋅HCl, pH 7.8 (15). The slide was pressed to the nitrocellulose for 25 s and checked for complete transfer. To eliminate PrPC from the section, the membranes were air-dried, rehydrated for 1 h in TBST buffer containing 0.05% Tween 20, 100 mM NaCl, and 10 mM Tris⋅HCl, pH 7.8, and exposed for 18 h at 37°C to proteinase K (400 μg/ml) in a buffer containing 0.5% Brij 35, 100 mM NaCl, and 10 mM Tris⋅HCl, pH 7.8. To terminate the reaction, the blots were rinsed three times in TBST and immersed for 30 min in TBST containing 3 mM phenylmethylsulfonyl fluoride. To enhance the immunostaining of PrPSc, the histoblots were exposed to 3 M guanidinium isothiocyanate for 10 min at room temperature in 20 mM Tris⋅HCl, pH 7.8, and rinsed three times with TBST before immunostaining.
RESULTS AND DISCUSSION
Native and Chimeric Bovine PrP Transgenes.
Based on our prior experience in creating Tg mice susceptible to Hu prions (26–28), we initially utilized two different ORFs to create Tg mice susceptible to BSE prions. The complete bovine PrP(BoPrP) ORF used in this study was derived from the brain of a Hereford bull afflicted with BSE, PG31/90. Sequencing of the ORF (16) revealed that this version contains six copies of the octarepeat sequence. We employed an alternative strategy based on transgenetic studies that had shown that a chimeric Hu/Mo PrP ORF, termed MHu2M, conferred susceptibility to Hu prions (20, 26). In the analogous Bo/Mo PrP chimeric ORF, termed MBo2M PrP, DNA sequences including the central region of Mo PrP between amino acid residues 97 and 186 were replaced with a corresponding fragment from the PG31/90 BoPrP ORF (20, 21, 26). To minimize confusion, we reference the positions of amino acid residues in the PrP ORF relative to the sequences of Syrian hamster (SHa) and Hu PrP, except where explicitly stated. The BoPrP and MBo2M ORF cassettes were inserted into a cosTet vector and used to produce Tg mice (21).
Construction of Transgenic Mice Expressing BoPrP and MBo2M PrP.
Initially, we attempted to construct Tg(BoPrP) mice by using FVB mice that express MoPrP-A. Two lines, designated Tg(BoPrP)883 and Tg(BoPrP)333, were obtained, but the transgene used to construct these lines was eventually found to be defective because neither line expressed BoPrP at levels detectable by Western blotting when tested by using antisera specific for BoPrP (29). Because both Tg lines expressed MoPrP-A (Prnpa) genes, we used the mice as Prnpa/a controls; moreover, they allowed us to assess the remote possibility that sequences other than PrP within the cosTet vector could influence the specificity for passaging of BSE prions.
Our success in transmitting Hu prions to Tg(MHu2M) mice (26) prompted us to construct similar mice expressing MBo2M PrP. Mice deficient for PrP (Prnp0/0) were crossed onto the FVB background and used as recipients for microinjection (23). Three Tg(MBo2M)Prnp0/0 founders were obtained. Only two were fertile and could be bred for preliminary screening for expression of MBo2M PrP. Of these, one expressed only low levels of MBo2M PrP, and the other line, Tg(MBo2M)14586/Prnp0/0, was found to express approximately 8–16 times the amount of MBo2M PrP per gram of total protein when compared with bovine brain (G.T., data not shown). The latter line was selected for transmission studies.
In a parallel study, we constructed a new BoPrP transgene to create lines of Tg(BoPrP) mice that efficiently express BoPrP. To ensure that any interfering effects of endogenous PrPs were eliminated (27, 30), FVB/Prnp0/0 mice described above were used as recipients for microinjection. Four lines of Tg(BoPrP)/Prnp0/0 mice were established; two of these, Tg(BoPrP)4092/Prnp0/0 and Tg(BoPrP)4125/Prnp0/0, expressed 4–8 and 8–16 times more BoPrP per gram of protein than that found in bovine brain, respectively (M.R.S. and O.N., data not shown).
Resistance of Tg(MBo2M) Mice to Infection with BSE Prions.
Extracts prepared from the brainstems of several cows with histologically positive BSE and PrPSc by Western blotting (16) were inoculated into Tg(MBo2MPrP) mice. None of the mice developed clinical signs of neurologic dysfunction, more than 600 days after inoculation (Table 1). This was surprising because the MBo2M ORF was composed of BoPrP and MoPrP-A sequences, and previous studies had shown that Prnpa mice are susceptible to BSE, with incubation times exceeding 400 days (31). That the inocula used for intracerebral injection of the Tg(MBo2M)PrP mice contained infectious prions was shown by transmission of prion disease to FVB, Tg(BoPrP)883, and Tg(BoPrP)333 mice expressing MoPrP-A. These mice exhibited incubation times of 400–650 days (Table 1), similar to that reported by others (31, 32). Tg(MoPrP-A)4053 mice inoculated with GJ248/85 BSE prions exhibited signs of CNS dysfunction at ≈320 days (Table 1); in contrast, these mice inoculated with RML Mo prions developed disease in ≈50 days (33). Thus, Tg(MBo2MPrP) mice were highly resistant to BSE prions whereas mice expressing wild-type Mo PrP-A sequence were susceptible to BSE prions but required prolonged incubation times.
Table 1.
Susceptibility and resistance of transgenic mice to BSE prions
| Inoculum | Recipient | Transgene expression | Incubation time, days ± SE | n/no |
|---|---|---|---|---|
| Mice deficient for PrP (Prnp0/0) | ||||
| BSE(PG31/90) | Tg(BoPrP)4125 | 8–16 × | 234 ± 8 | 10/10 |
| BSE(PG31/90) | Tg(BoPrP)4092 | 4–8 × | 319 ± 15 | 8/8 |
| BSE(GJ248/85) | Tg(BoPrP)4125 | 8–16 × | 281 ± 19 | 10/10 |
| BSE(GJ248/85) | Tg(BoPrP)4092 | 4–8 × | 343 ± 18 | 8/8 |
| BSE(GJ248/85) | Tg(MBo2M)14586 | 8–16 × | >600 | 0/15 |
| BSE(PG31/90) | Tg(MBo2M)14586 | 8–16 × | >600 | 0/13 |
| BSE(574C) | Tg(MBo2M)14586 | 8–16 × | >600 | 0/13 |
| Mice expressing MoPrP-A | ||||
| BSE(GJ248/85) | FVB | 0 | 628 ± 47 | 2/3 |
| BSE(PG31/90) | FVB | 0 | 448 ± 29 | 2/2 |
| BSE(574C) | FVB | 0 | 525 ± 34 | 4/4 |
| BSE(PG31/90) | Tg(MoPrP-A)4053 | 8–16 × | >310 | 4/10 |
| BSE(GJ248/85) | Tg(MoPrP-A)4053 | 8–16 × | 322 ± 6 | 8/9 |
| BSE(PG31/90) | Tg(BoPrP)333 | 0* | 426 ± 11 | 8/8 |
| BSE(PG31/90) | Tg(BoPrP)833 | 0* | 395 ± 22 | 9/9 |
| BSE → Tg(BoPrP)333 | CD-1 | 0 | 155 ± 3 | 16/16 |
No BoPrP was detected by Western immunoblotting.
Susceptibility of Tg(BoPrP) Mice to BSE Prions.
Two lines of Tg(BoPrP)Prnp0/0 mice expressing BoPrP were inoculated with a 10% homogenate derived from the medulla of a Hereford bull (case PG31/90) clinically ill with BSE. The Tg(BoPrP)4125/Prnp0/0 mice with the highest level of BoPrP expression were found to be highly susceptible to BSE prions, with 100% of animals exhibiting clinical signs within 250 days after inoculation. Tg(BoPrP)4092/Prnp0/0 mice with an intermediate level of BoPrP expression exhibited a longer incubation period with the same inoculum; they had a mean incubation period of ≈320 days. Similar incubation times were obtained when these Tg lines were inoculated with another BSE isolate (Table 1).
Neuropathology of BSE in Tg(BoPrP) Mice.
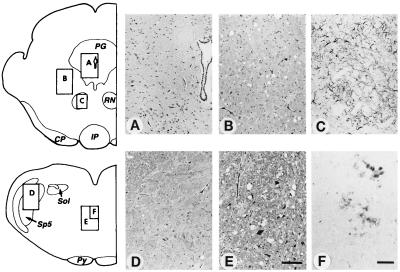
We examined the neuropathology of both lines of Tg(BoPrP) mice infected with either of two BSE isolates: PG31/90 or GJ248/85 (Fig. 1). The distribution of spongiform degeneration was similar to that previously reported in cattle afflicted with BSE (34). It was most intense in the thalamus, hypothalamus, and tegmentum of the midbrain, pons, and medulla (Fig. 1). Like BSE in cattle, there was little or no vacuolation in the cerebral cortex or basal ganglia (Fig. 1). It should be noted that this contrasts with virtually all Mo-passaged scrapie strains where we observe vacuolation of the cerebral cortex (35, 36). The pattern of neuropathology in Tg(BoPrP) mice differed from that of BSE in cattle by the absence of vacuolation in the nucleus of the spinal trigeminal tract, the nucleus of tractus solitarius, and the periaqueductal grey of the midbrain. The midbrain is where the most intense vacuolation in cattle was found. PrPSc colocalized with vacuolation in the Tg(BoPrP) mice when visualized by using either PrP immunohistochemistry or hydrolytic autoclaving (Fig. 1). No amyloid plaques were identified by hematoxylin and eosin staining or by PrP immunohistochemistry.
Figure 1.
Neuropathology of Tg(BoPrP)Prnp0/0 mice inoculated with BSE prions. (A) No pathological changes were found in the periaqueductal grey of the midbrain. (B) Mild to moderate vacuolar degeneration was found in the reticular formation of the midbrain tegmentum. (C) Reactive astrocytic gliosis colocalized with sites of vacuolar degeneration: astrogliosis in the red nucleus is shown here. (D) Little or no vacuolar degeneration was found in the tract or the nucleus of the spinal tract of the trigeminal nerve in the medulla. (E) Moderate to severe vacuolar degeneration occurred in the medial tegmentum of the medullary reticular formation. (F) Small PrP-immunopositive primitive plaque-like deposits colocalized with sites of the most severe vacuolar degeneration. Diagrams show locations of photomicrographs. Hematoxylin and eosin stain was used in A, B, D, and E. Glial fibrillary acidic protein immunohistochemistry was used in C. PrP immunohistochemistry was used in F. Bar in E = 100 μm and also applies to A, B, and D. Bar in F = 50 μm and also applies to C. Italicized letters identify selected brainstem structures: CP, cerebral peduncle; IP, interpeduncular nucleus; PG, periaqueductal grey; Py, pyramidal tract; RN, red nucleus; Sol, nucleus and tractus solitarius; Sp5, nucleus of the spinal tract of the trigeminal nerve.
Localization of PrPSc in the Brains of Tg(BoPrP) Mice.
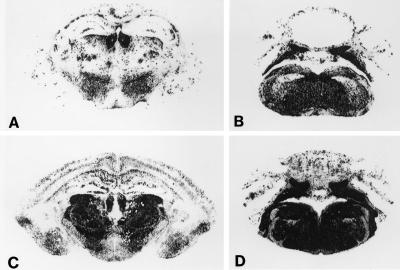
Histoblotting demonstrated PrPSc deposition highly localized to the brainstem of Tg(BoPrP)4125/Prnp0/0 mice (Fig. 2). For these studies, we used an anti-PrP rabbit polyclonal antiserum designated #9095 that was raised against a synthetic peptide corresponding to residues 90–145 of BoPrP. This antiserum shows a broad specificity, with a high affinity for bovine, mouse, hamster, and sheep PrPs. We compared the pattern of PrPSc accumulation in Tg(BoPrP) mice inoculated with BSE prions with the pattern in congenic B6.I-1 (Prnpb/b) mice (33) inoculated with 301V prions. The 301V strain was derived after transmission of BSE prions to Prnpb/b mice (31, 37). Although the patterns of PrPSc accumulation were virtually identical in the brainstem (Fig. 2 B and D), there were marked differences in the accumulation patterns in the diencephalon and cerebral cortex (Fig. 2 A and C). In Tg(BoPrP) mice, there was diffusely scattered, patchy accumulation of PrPSc in the thalamus, whereas the entire thalamus was strongly immunoreactive for PrPSc in B6.I-1 mice. In Tg(BoPrP) mice, there were a few punctate deposits of PrPSc in the cerebral cortex, whereas there was significantly more PrPSc in the cerebral cortex of B6.I-1 mice that deposited in a laminar fashion. The accumulation of PrPSc in the brainstems of Tg(BoPrP) mice inoculated with BSE prions is reminiscent of the pattern of PrPSc accumulation in cow brains infected with BSE (16). The distribution of PrPSc in Tg(BoPrP) brain was more extensive than the distribution of vacuolar degeneration.
Figure 2.
Regional distribution of PrPSc in the brains of Tg(BoPrP) mice inoculated with BSE prions. Sections A and B were from Tg(BoPrP)Prnp0/0 mice inoculated with BSE prions, and sections C and D were taken from congenic B6.I-1 (Prnpb/b) mice inoculated with 301V prions. A and C were through hippocampus and thalamus, and B and D were through the brainstem.
The ability of the Tg(BoPrP)Prnp0/0 mice to mimic the CNS distribution of PrPSc found in cattle with BSE is reminiscent of the ability of Tg(MHu2M) mice to mimic the distribution of PrPSc deposition in two of the inherited prion diseases. Fatal familial insomnia (FFI) is caused by the D178N mutation whereas one form of familial CJD (fCJD) is caused by the E200K mutation (38–40). Transmission of FFI prions from human brain to Tg(MHu2M) mice produced marked accumulation of PrPSc confined to the thalamus similar to that found in humans with FFI; in contrast, fCJD(E200K) prions resulted in PrPSc deposition throughout the cortex, thalamus, and hypothalamus as seen in humans with fCJD(E200K) (28). The mechanism of selective neuronal targeting in the prion diseases is unclear, but recent studies have documented the ability of Asn-linked glycosylation to modify the distribution of both PrPC and PrPSc in concert with the particular strain of prion (41–44).
Characteristics of PrPSc in the Brains of Tg(BoPrP) Mice.
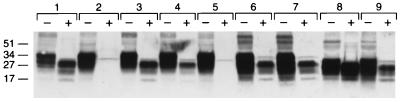
We identified PrPSc in brains of Tg(BoPrP) mice afflicted with BSE by Western blotting by using the anti-PrP rabbit polyclonal 9095 antiserum described above. Both the Tg(BoPrP)4125/Prnp0/0 and Tg(BoPrP)4092/Prnp0/0 lines were found to contain large amounts of a protein of ≈34 kDa, indistinguishable in size from that found in the bovine PG31/90 brain from which the BoPrP ORF used to construct the transgene was derived (Fig. 3). The increased size of BoPrP compared with MoPrP is predominantly because of the presence of six octarepeats rather than five. In addition to the extra octarepeat, the N-terminal region contains two single base insertions relative to SHa/Hu PrP; however, the region corresponding to residues 90–231 is identical in size to SHa/Hu PrP.
Figure 3.
Western blots of PrP in brains from Tg mice inoculated with BSE prions and cattle with BSE. Each undigested control aliquot (− lanes) contained approximately 5 μg of total protein; aliquots digested with proteinase K (+ lanes) consisted of the equivalent of 25 μg of undigested, total protein. Samples in lane pairs: 1, bovine medulla from case PG31/90; 2, Tg(BoPrP)4092/Prnp0/0, uninoculated; 3, Tg(BoPrP)4092/Prnp0/0 clinically ill after inoculation with case PG31/90; 4, Tg(BoPrP)4092/Prnp0/0 clinically ill after inoculation with case GJ248/85; 5, Tg(BoPrP)4125/Prnp0/0, uninoculated; 6, Tg(BoPrP)4125/Prnp0/0 clinically ill after inoculation with case PG31/90; 7, Tg(BoPrP)4125/Prnp0/0 clinically ill after inoculation with case GJ248/85; 8, Tg(BoPrP)333 clinically ill after inoculation with PG31/90 prions; 9, Tg(BoPrP)333 clinically ill at approximately 500 days after inoculation with the sheep scrapie SSBP/1 isolate.
After digestion with proteinase K, a series of truncated polypeptides of ≈28 kDa, ≈23 kDa, and ≈17 kDa were revealed in BSE-infected PG31/90 brain corresponding to the diglycosylated, monoglycosylated, and unglycosylated form, respectively (Fig. 3). Indistinguishable patterns of protease-resistant BoPrP fragments were identified in Tg(BoPrP)4092/Prnp0/0 brains infected with either PG31/90 or GJ248/85 brain inocula and, in slightly lower amounts, in Tg(BoPrP)4125/Prnp0/0. Interestingly, the ratios of the three glycoforms were similar in bovine and Tg(BoPrP) mouse brains and in those reported for vCJD patients (45), with a predominance of the fully glycosylated fragment. BSE prions passaged into normal Prnpa/a mice showed a similar pattern of protease-resistant fragments, but these displayed a slightly lower molecular size compared with BoPrPSc found in the brains of BSE-infected Tg(BoPrP) mice and cattle or with the MoPrPSc fragments observed after hydrolysis of brain extracts of Prnpa/a mice that had been inoculated with a sheep scrapie isolate.
Measuring Bovine Prions.
Mice inoculated intracerebrally with BSE brain extracts require more than a year to develop disease (31, 32, 46). Depending on the titer of the inoculum, the structures of PrPC and PrPSc, and other host factors, such as protein X, the number of inoculated animals developing disease can vary over a wide range. Some investigators have stated that transmission of BSE to mice is quite variable with incubation periods exceeding one year (32), whereas others report low prion titers of BSE brain homogenates (31, 46) compared with rodent brain (47–50). Moreover, endpoint titrations of BSE prions in cattle suggest much higher titers of prions in bovine brain than those determined by mouse bioassays (J. Wilesmith, personal communication). Such problems with the measurement of bovine prions emphasize the urgent need for the Tg mice described in this report.
Other attempts at assaying BSE prions have used animals from various species. Brain extracts from BSE cattle caused disease in cattle, sheep, mice, pigs, and mink after intracerebral inoculation (51–54), but prions in brain extracts from sheep with scrapie fed to cattle produced illness substantially different from BSE (55). All of the previously available bioassay systems suffer from severe limitations that limit their usefulness. Apart from the cost involved, the long incubation periods and low efficiency of transmission of prions, heightened in some cases by the species barrier caused by lack of PrP sequence identity, have conspired to severely impede progress in performing routine measurements of titers of BSE prions.
An Epitope Modulating Prion Transmission.
Because of our earlier success with the transmission of Hu prions to Tg(MHu2M) mice (26), the resistance of Tg(MBo2M) mice to BSE prions was puzzling (Table 1). A comparison of the MoPrP-A, MBo2M PrP, and MHu2M PrP translated sequences shows that Hu residue substitutions in MHu2M extended from 97 to 168 whereas Bo substitutions in MBo2M extended from 97 to 186. This finding raised the possibility that residues 184 and 186, which are not homologous in Bo and Mo PrP and lie at the C-terminal end of the chimeric region, might account for the differences in susceptibility of Tg(MHu2M) and Tg(MBo2M) mice to prion infection. Alternatively, residue 203, which is a Val in Mo and Hu PrP and is an Ile in BoPrP, might be responsible for this difference in susceptibility to prions. In Tg(MBo2M) mice, residue 203 is a Val and thus might prevent conversion of MBo2M PrPC into PrPSc.
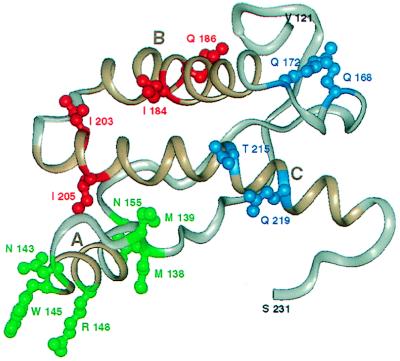
The availability of three-dimensional structures for the Mo PrP(121–231) and SHa PrP(90–231) prion protein fragments (56, 57) and more recently the full-length SHa(29–231) (58) has allowed us to view the impact of changes in the PrP sequence in a spatial context. This led us to understand the sequentially disparate but spatially proximal epitope that provides the PrPC/protein X binding interface (59). With this in mind, we studied the spatial juxtaposition of the residues that contribute to the Hu/Bo species barrier that we believe form a subset of the PrPC/PrPSc binding interface (59). Residues 184, 186, 203, and 205 were identified within the three-dimensional structure of SHa PrP(90–231) derived by solution NMR (57). These residues are seen to cluster on one side of the PrPC structure and are spatially distinct from the discontinuous epitope consisting of residues 168, 172, 215, and 219 that binds to protein X (27, 59) (Fig. 4). To this collection of residues, we added those known to be polymorphic from an extensive analysis of the PrP genes from more than 40 species (60). These residues are candidates for contributing to the species barrier. As can be seen in Fig. 4, almost all of these residues cluster and enlarge the epitope formed by residues 184, 186, 203, and 205. Certainly, other residues (e.g., 204) that are spatial neighbors of the residues highlighted in Fig. 4 are candidates for this epitope; we expect that their mutation would create a species barrier, but this has yet to be tested. A systematic study of residues that should contribute to this epitope will allow us to define the extent of the PrPC/PrPSc interface and thus reach an understanding of the species barrier at a molecular level. This epitope includes regions that are known to remain structurally constant as well as a portion that is known to undergo a substantial conformational reorganization (61). Perhaps this explains why PrPSc can homodimerize (62) as well as bind to PrPC (30, 63) and simultaneously act as a template for assisting the conversion of PrPC into PrPSc (28). Transgenic studies have shown that some mutations associated with inherited human disease create transmissible disease (178, 200, 210) (ref. 28; J. A. Mastrianni and S.B.P., unpublished data) whereas others create disease in the founder’s lineage that are not transmissible to murine hosts expressing wild-type PrP (e.g., 102) (27, 64). Seven point mutations (178, 180, 183, 198, 200, 208, 210) known to cause inherited prion diseases, including those that are known to create a transmissible encephalopathy, map to this region of the structure as well (for review see ref. 65).
Figure 4.
PrP residues governing the transmission of prions. NMR structure of recombinant SHaPrP (90–231) (57) shown with the putative epitope formed by residues 184, 186, 203, and 205 highlighted in red. Residue numbers correspond to SHaPrP. Additional residues (138, 139, 143, 145, 148, and 155) that might participate in controlling the transmission of prions across species are depicted in green. Residues 168, 172, 215, and 219 that form the epitope for the binding of protein X are shown in blue. The three helices (A, B, and C) are highlighted in brown. The illustration was generated with biosym/insight ii.
Designing Transgenes.
The identification of a species-specific epitope that modulates the conversion of PrPC into PrPSc has important implications for the design of PrP transgenes. Whether optimal Bo/Mo chimeric transgenes should contain Mo or Bo residues at positions 184, 186, 203, and 205 remains to be established. Similarly it is unknown whether improved Hu/Mo chimeric transgenes can be constructed by simultaneously mutating Hu residues at these same positions to Mo residues. Mutagenesis at any or all of these positions may overcome the paradoxically long incubation times found in Tg(MHu2M)Prnp0/0 mice expressing high levels of the transgene product (G.T. and S.B.P., unpublished data) as well as the resistance of Tg(MBo2M) mice to Bo prions (Table 1).
Implications for Public Health and Monitoring the Food Supply.
The Tg(BoPrP) mice make possible, for the first time, an accurate determination of BSE prion titers in brain and other tissues. Determining the titers of BSE prions in muscle, pancreas, liver, and intestine that are commonly consumed by humans may be of utmost importance. Because the bioassay for bovine prions in non-Tg mice is so insensitive (31, 46), the levels of prions in these bovine tissues remain unknown. If the distribution of bovine prions proves to be different from that presumed for sheep (66), then assumptions about the efficacy of the specified bovine offals ban of November 1989 that prohibited human consumption of CNS and lymphoid tissues from cattle older than 6 months of age may need to be reassessed.
These Tg(BoPrP) mice also make possible for the first time the evaluation of drugs and other medicinal products derived from cattle for prion contamination. For example, collagen from cattle is used widely in plastic and reconstructive surgery, and gelatin is used in foods and in the production of a wide variety of drug capsules. The availability of Tg(BoPrP) mice will also make possible epidemiologic studies on the frequency of BSE in countries such as the United States and Canada that have been thought to be spared (67). With these Tg(BoPrP) mice, it is now possible to determine the frequency of sporadic BSE, particularly in older cattle (68). Such studies will determine whether or not it will be important to produce Tg cattle that are resistant to prions either through PrP transgenes that bind tightly to protein X and act as dominant negatives or by genetic ablation of the PrP gene (23, 65, 69).
Acknowledgments
We thank G. Wells, J. Wilesmith, and R. Bradley for the BSE brain tissue; R. Kimberlin for the SSBP/1 isolate; and J. Cayetano and V. Itri for excellent technical assistance. This work was supported by grants from the National Institutes of Health (NS14069, AG08967, AG02132, and NS22786) and the American Health Assistance Foundation, as well as by gifts from the G. Harold and Leila Y. Mathers Foundation, the Sherman Fairchild Foundation, the Bernard Osher Foundation, and Centeon. D.W. was supported by a Deutsche Forschungsgemeinschaft Fellowship.
ABBREVIATIONS
- BSE
bovine spongiform encephalopathy
- Hu
human
- CJD
Creutzfeldt–Jakob disease
- Bo
bovine
- Tg
transgenic
- PrP
prion protein
- PrPC
cellular isoform of PrP
- PrPSc
scrapie isoform of PrP
- vCJD
variant CJD
- SHa
Syrian hamster
- Prnp0/0
PrP-deficient mice
- FFI
fatal familial insomnia
References
- 1.Bateman D, Hilton D, Love S, Zeidler M, Beck J, Collinge J. Lancet. 1995;346:1155–1156. doi: 10.1016/s0140-6736(95)91828-0. [DOI] [PubMed] [Google Scholar]
- 2.Britton T C, Al-Sarraj S, Shaw C, Campbell T, Collinge J. Lancet. 1995;346:1155. doi: 10.1016/s0140-6736(95)91827-2. [DOI] [PubMed] [Google Scholar]
- 3.Chazot G, Broussolle E, Lapras C I, Blättler T, Aguzzi A, Kopp N. Lancet. 1996;347:1181. doi: 10.1016/s0140-6736(96)90638-8. [DOI] [PubMed] [Google Scholar]
- 4.Will R G, Ironside J W, Zeidler M, Cousens S N, Estibeiro K, Alperovitch A, Poser S, Pocchiari M, Hofman A, Smith P G. Lancet. 1996;347:921–925. doi: 10.1016/s0140-6736(96)91412-9. [DOI] [PubMed] [Google Scholar]
- 5.Cousens S N, Vynnycky E, Zeidler M, Will R G, Smith P G. Nature (London) 1997;385:197–198. doi: 10.1038/385197a0. [DOI] [PubMed] [Google Scholar]
- 6.Wilesmith J W, Ryan J B M, Atkinson M J. Vet Rec. 1991;128:199–203. doi: 10.1136/vr.128.9.199. [DOI] [PubMed] [Google Scholar]
- 7.Nathanson N, Wilesmith J, Griot C. Am J Epidemiol. 1997;145:959–969. doi: 10.1093/oxfordjournals.aje.a009064. [DOI] [PubMed] [Google Scholar]
- 8.Anderson R M, Donnelly C A, Ferguson N M, Woolhouse M E J, Watt C J, Udy H J, MaWhinney S, Dunstan S P, Southwood T R E, Wilesmith J W, Ryan J B M, Hoinville L J, Hillerton J E, Austin A R, Wells G A H. Nature (London) 1996;382:779–788. doi: 10.1038/382779a0. [DOI] [PubMed] [Google Scholar]
- 9.Stekel D J, Nowak M A, Southwood T R E. Nature (London) 1996;381:119. doi: 10.1038/381119a0. [DOI] [PubMed] [Google Scholar]
- 10.Wilesmith J W. Semin Virol. 1991;2:239–245. [Google Scholar]
- 11.Kimberlin R H. In: Bovine Spongiform Encephalopathy: The BSE Dilemma. Gibbs C J Jr, editor. New York: Springer; 1996. pp. 155–175. [Google Scholar]
- 12.Hsich G, Kenney K, Gibbs C J, Lee K H, Harrington M G. N Engl J Med. 1996;335:924–930. doi: 10.1056/NEJM199609263351303. [DOI] [PubMed] [Google Scholar]
- 13.Hope J, Reekie L J D, Hunter N, Multhaup G, Beyreuther K, White H, Scott A C, Stack M J, Dawson M, Wells G A H. Nature (London) 1988;336:390–392. doi: 10.1038/336390a0. [DOI] [PubMed] [Google Scholar]
- 14.Serban D, Taraboulos A, DeArmond S J, Prusiner S B. Neurology. 1990;40:110–117. doi: 10.1212/wnl.40.1.110. [DOI] [PubMed] [Google Scholar]
- 15.Taraboulos A, Jendroska K, Serban D, Yang S-L, DeArmond S J, Prusiner S B. Proc Natl Acad Sci USA. 1992;89:7620–7624. doi: 10.1073/pnas.89.16.7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prusiner S B, Fuzi M, Scott M, Serban D, Serban H, Taraboulos A, Gabriel J-M, Wells G, Wilesmith J, Bradley R, DeArmond S J, Kristensson K. J Infect Dis. 1993;167:602–613. doi: 10.1093/infdis/167.3.602. [DOI] [PubMed] [Google Scholar]
- 17.Grathwohl K-U D, Horiuchi M, Ishiguro N, Shinagawa M. J Virol Methods. 1997;64:205–216. doi: 10.1016/s0166-0934(97)02197-6. [DOI] [PubMed] [Google Scholar]
- 18.Pattison I H. In: Slow, Latent and Temperate Virus Infections, NINDB Monograph 2. Gajdusek D C, Gibbs C J Jr, Alpers M P, editors. Washington, DC: U.S. Government Printing Office; 1965. pp. 249–257. [Google Scholar]
- 19.Scott M, Foster D, Mirenda C, Serban D, Coufal F, Wälchli M, Torchia M, Groth D, Carlson G, DeArmond S J, Westaway D, Prusiner S B. Cell. 1989;59:847–857. doi: 10.1016/0092-8674(89)90608-9. [DOI] [PubMed] [Google Scholar]
- 20.Telling G C, Haga T, Torchia M, Tremblay P, DeArmond S J, Prusiner S B. Genes Dev. 1996;10:1736–1750. doi: 10.1101/gad.10.14.1736. [DOI] [PubMed] [Google Scholar]
- 21.Scott M R, Köhler R, Foster D, Prusiner S B. Protein Sci. 1992;1:986–997. doi: 10.1002/pro.5560010804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott M, Groth D, Foster D, Torchia M, Yang S-L, DeArmond S J, Prusiner S B. Cell. 1993;73:979–988. doi: 10.1016/0092-8674(93)90275-u. [DOI] [PubMed] [Google Scholar]
- 23.Büeler H, Fischer M, Lang Y, Bluethmann H, Lipp H-P, DeArmond S J, Prusiner S B, Aguet M, Weissmann C. Nature (London) 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 24.Carlson G A, Goodman P A, Lovett M, Taylor B A, Marshall S T, Peterson-Torchia M, Westaway D, Prusiner S B. Mol Cell Biol. 1988;8:5528–5540. doi: 10.1128/mcb.8.12.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muramoto T, Kitamoto T, Tateishi J, Goto I. Am J Pathol. 1992;140:1411–1420. [PMC free article] [PubMed] [Google Scholar]
- 26.Telling G C, Scott M, Hsiao K K, Foster D, Yang S-L, Torchia M, Sidle K C L, Collinge J, DeArmond S J, Prusiner S B. Proc Natl Acad Sci USA. 1994;91:9936–9940. doi: 10.1073/pnas.91.21.9936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Telling G C, Scott M, Mastrianni J, Gabizon R, Torchia M, Cohen F E, DeArmond S J, Prusiner S B. Cell. 1995;83:79–90. doi: 10.1016/0092-8674(95)90236-8. [DOI] [PubMed] [Google Scholar]
- 28.Telling G C, Parchi P, DeArmond S J, Cortelli P, Montagna P, Gabizon R, Mastrianni J, Lugaresi E, Gambetti P, Prusiner S B. Science. 1996;274:2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 29.Oberdieck U, Xi Y-G, Pocchiari M, Diringer H. Arch Virol. 1994;136:99–110. doi: 10.1007/BF01538820. [DOI] [PubMed] [Google Scholar]
- 30.Prusiner S B, Scott M, Foster D, Pan K-M, Groth D, Mirenda C, Torchia M, Yang S-L, Serban D, Carlson G A, Hoppe P C, Westaway D, DeArmond S J. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 31.Fraser H, Bruce M E, Chree A, McConnell I, Wells G A H. J Gen Virol. 1992;73:1891–1897. doi: 10.1099/0022-1317-73-8-1891. [DOI] [PubMed] [Google Scholar]
- 32.Lasmézas C I, Deslys J-P, Robain O, Jaegly A, Beringue V, Peyrin J-M, Fournier J-G, Hauw J-J, Rossier J, Dormont D. Science. 1997;275:402–405. doi: 10.1126/science.275.5298.402. [DOI] [PubMed] [Google Scholar]
- 33.Carlson G A, Ebeling C, Yang S-L, Telling G, Torchia M, Groth D, Westaway D, DeArmond S J, Prusiner S B. Proc Natl Acad Sci USA. 1994;91:5690–5694. doi: 10.1073/pnas.91.12.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wells G A H, Wilesmith J W. Brain Pathol. 1995;5:91–103. doi: 10.1111/j.1750-3639.1995.tb00580.x. [DOI] [PubMed] [Google Scholar]
- 35.Fraser H, Dickinson A G. J Comp Pathol. 1973;83:29–40. doi: 10.1016/0021-9975(73)90024-8. [DOI] [PubMed] [Google Scholar]
- 36.DeArmond S J, Prusiner S B. Am J Pathol. 1995;146:785–811. [PMC free article] [PubMed] [Google Scholar]
- 37.Bruce M, Chree A, McConnell I, Foster J, Pearson G, Fraser H. Philos Trans R Soc London B. 1994;343:405–411. doi: 10.1098/rstb.1994.0036. [DOI] [PubMed] [Google Scholar]
- 38.Medori R, Tritschler H-J, LeBlanc A, Villare F, Manetto V, Chen H Y, Xue R, Leal S, Montagna P, Cortelli P, Tinuper P, Avoni P, Mochi M, Baruzzi A, Hauw J J, Ott J, Lugaresi E, Autilio-Gambetti L, Gambetti P. N Engl J Med. 1992;326:444–449. doi: 10.1056/NEJM199202133260704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldfarb L, Korczyn A, Brown P, Chapman J, Gajdusek D C. Lancet. 1990;336:637–638. doi: 10.1016/0140-6736(90)93443-s. [DOI] [PubMed] [Google Scholar]
- 40.Hsiao K, Meiner Z, Kahana E, Cass C, Kahana I, Avrahami D, Scarlato G, Abramsky O, Prusiner S B, Gabizon R. N Engl J Med. 1991;324:1091–1097. doi: 10.1056/NEJM199104183241604. [DOI] [PubMed] [Google Scholar]
- 41.Bruce M E, McBride P A, Farquhar C F. Neurosci Lett. 1989;102:1–6. doi: 10.1016/0304-3940(89)90298-x. [DOI] [PubMed] [Google Scholar]
- 42.Hecker R, Taraboulos A, Scott M, Pan K-M, Torchia M, Jendroska K, DeArmond S J, Prusiner S B. Genes Dev. 1992;6:1213–1228. doi: 10.1101/gad.6.7.1213. [DOI] [PubMed] [Google Scholar]
- 43.DeArmond S J, Yang S-L, Lee A, Bowler R, Taraboulos A, Groth D, Prusiner S B. Proc Natl Acad Sci USA. 1993;90:6449–6453. doi: 10.1073/pnas.90.14.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeArmond, S. J., Sánchez, H., Yehiely, F., Qiu, Y., Ninchak-Casey, A., Daggett, V., Camerino, A. P., Cayetano, J., Rogers, M., Groth, D., Torchia, M., Tremblay, P., Scott, M. R., Cohen, F. E. & Prusiner, S. B. (1997) Neuron, in press. [DOI] [PubMed]
- 45.Collinge J, Sidle K C L, Meads J, Ironside J, Hill A F. Nature (London) 1996;383:685–690. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- 46.Taylor K C. Vet Rec. 1991;129:522–526. [PubMed] [Google Scholar]
- 47.Hunter G D, Millson G C, Chandler R L. Res Vet Sci. 1963;4:543–549. [Google Scholar]
- 48.Eklund C M, Kennedy R C, Hadlow W J. J Infect Dis. 1967;117:15–22. doi: 10.1093/infdis/117.1.15. [DOI] [PubMed] [Google Scholar]
- 49.Kimberlin R, Walker C. J Gen Virol. 1977;34:295–304. doi: 10.1099/0022-1317-34-2-295. [DOI] [PubMed] [Google Scholar]
- 50.Prusiner S B, Cochran S P, Groth D F, Downey D E, Bowman K A, Martinez H M. Ann Neurol. 1982;11:353–358. doi: 10.1002/ana.410110406. [DOI] [PubMed] [Google Scholar]
- 51.Fraser H, McConnell I, Wells G A H, Dawson M. Vet Rec. 1988;123:472. doi: 10.1136/vr.123.18.472. [DOI] [PubMed] [Google Scholar]
- 52.Dawson M, Wells G A H, Parker B N J. Vet Rec. 1990;126:112–113. [PubMed] [Google Scholar]
- 53.Dawson M, Wells G A H, Parker B N J, Scott A C. Vet Rec. 1990;127:338. [PubMed] [Google Scholar]
- 54.Bruce M E, Will R G, Ironside J W, McConnell I, Drummond D, Suttie A, McCardle L, Chree A, Hope J, Birkett C, Cousens S, Fraser H, Bostock C J. Nature (London) 1997;389:498–501. doi: 10.1038/39057. [DOI] [PubMed] [Google Scholar]
- 55.Robinson M M, Hadlow W J, Knowles D P, Huff T P, Lacy P A, Marsh R F, Gorham J R. J Comp Path. 1995;113:241–251. doi: 10.1016/s0021-9975(05)80039-8. [DOI] [PubMed] [Google Scholar]
- 56.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. Nature (London) 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 57.James T L, Liu H, Ulyanov N B, Farr-Jones S, Zhang H, Donne D G, Kaneko K, Groth D, Mehlhorn I, Prusiner S B, Cohen F E. Proc Natl Acad Sci USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Donne D G, Viles J H, Groth D, Mehlhorn I, James T L, Cohen F E, Prusiner S B, Wright P E. Proc Natl Acad Sci USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaneko K, Zulianello L, Scott M, Cooper C M, Wallace A C, James T L, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1997;94:10069–10074. doi: 10.1073/pnas.94.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bamborough P, Wille H, Telling G C, Yehiely F, Prusiner S B, Cohen F E. Cold Spring Harbor Symp Quant Biol. 1996;61:495–509. [PubMed] [Google Scholar]
- 61.Peretz D, Williamson R A, Matsunaga Y, Serban H, Pinilla C, Bastidas R, Rozenshteyn R, James T L, Houghten R A, Cohen F E, Prusiner S B, Burton D R. J Mol Biol. 1997;273:614–622. doi: 10.1006/jmbi.1997.1328. [DOI] [PubMed] [Google Scholar]
- 62.Bellinger-Kawahara C G, Kempner E, Groth D F, Gabizon R, Prusiner S B. Virology. 1988;164:537–541. doi: 10.1016/0042-6822(88)90569-7. [DOI] [PubMed] [Google Scholar]
- 63.Kocisko D A, Come J H, Priola S A, Chesebro B, Raymond G J, Lansbury P T, Jr, Caughey B. Nature (London) 1994;370:471–474. doi: 10.1038/370471a0. [DOI] [PubMed] [Google Scholar]
- 64.Hsiao K K, Groth D, Scott M, Yang S-L, Serban H, Rapp D, Foster D, Torchia M, DeArmond S J, Prusiner S B. Proc Natl Acad Sci USA. 1994;91:9126–9130. doi: 10.1073/pnas.91.19.9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prusiner S B. Science. 1997;278:245–251. doi: 10.1126/science.278.5336.245. [DOI] [PubMed] [Google Scholar]
- 66.Hadlow W J, Kennedy R C, Race R E. J Infect Dis. 1982;146:657–664. doi: 10.1093/infdis/146.5.657. [DOI] [PubMed] [Google Scholar]
- 67.Wilesmith J W, Ryan J B M, Hueston W D. Res Vet Sci. 1992;52:323–331. doi: 10.1016/0034-5288(92)90032-w. [DOI] [PubMed] [Google Scholar]
- 68.Marsh R F, Bessen R A, Lehmann S, Hartsough G R. J Gen Virol. 1991;72:589–594. doi: 10.1099/0022-1317-72-3-589. [DOI] [PubMed] [Google Scholar]
- 69.Kaneko K, Wille H, Mehlhorn I, Zhang H, Ball H, Cohen F E, Baldwin M A, Prusiner S B. J Mol Biol. 1997;270:574–586. doi: 10.1006/jmbi.1997.1135. [DOI] [PubMed] [Google Scholar]