Abstract
Carbamoyl-phosphate synthetases (CPSs) utilize two molecules of ATP at two internally duplicated domains, B and C. Domains B and C have recently been shown to be structurally [Thoden, J. B., Holden, H. M., Wesenberg, G., Raushel, F. M. & Rayment, I. (1997) Biochemistry 36, 6305–6316] and functionally [Guy, H. I. & Evans, D. R. (1996) J. Biol. Chem. 271, 13762–13769] equivalent. We have carried out a site-directed mutagenic analysis that is consistent with ATP binding to a palmate motif rather than to a Walker A/B motif in domains B and C. To accommodate our present findings, as well as the other recent findings of structural and functional equivalence, we are proposing a novel mechanism for CPS. In this mechanism utilization of ATP bound to domain C is coupled to carbamoyl-phosphate synthesis at domain B via a nucleotide switch, with the energy of ATP hydrolysis at domain C allowing domain B to cycle between two alternative conformations.
Carbamoyl-phosphate (CP) is a high-energy biological compound that plays a key role in the introduction of both ammonia and single carbon units into the metabolic pool. CP may serve either as the precursor for synthesis of pyrimidine nucleotides or, in an alternative pathway, as the precursor for arginine. In addition to serving as a required component of almost all proteins, arginine is used in the liver for ammonia detoxification via the urea cycle and, under appropriate physiological conditions, it is also used in a variety of tissues for nitric oxide formation. Depending on the physiological role, CP synthetases (CPSs) vary in mode of regulation and subunit composition (1). However, all of the CPSs thus far sequenced display marked primary sequence identity (2, 3), and all catalyze the formation of CP, Pi, and two ADP from HCO3−, NH3, and two ATP.
Sequence analysis demonstrated regions of internal duplication within CPS and suggested, on the basis of similarities to the ATP-binding sequence motifs of Walker A/B proteins (i.e., proteins involving a βαβ nucleotide binding fold; ref. 4), that each region of internal duplication contains one ATP site (5, 6). Subsequent studies (7–10) demonstrated that the synthetase duplications correspond to independently folded domains (B and C) and that binding of one ATP molecule is localized to each of the two synthetase domains (6, 11–13). A great deal of experimental evidence (14–23) has established that the role of one ATP is to form the enzyme-bound intermediate carboxy-phosphate (CxP) and that domain B binds this ATP (ATPB) whereas domain C binds the other ATP molecule (ATPC).
When we carried out the presently described studies, further definition of the utilization of ATPB and ATPC by CPS could not be based on a solved three-dimensional structure because there was none available for any CPS at that time. However, we found that domains B and C of CPS could each be fit to the structural coordinates for the biotin carboxylase (BN) component of Escherichia coli acetyl Co-A carboxylase (24). This BN homology modeling seemed appropriate for the following reasons. First, there is significant sequence identity shared by portions of BN (as well as other biotin-dependent enzymes) and each of the CPS domains B and C (2). Also, both CPS and BN couple cleavage of ATP to ADP with the activation of HCO3− to yield CxP, and both catalyze reaction of the activated carboxyl group with an amino group (NH3 for CPS and the N1′ group of biotin for BN; ref. 25). A final rationale for the homology modeling was that the BN structure appears to represent a general fold for enzymes that catalyze formation of an amide bond accompanied by cleavage of ATP to ADP and formation of an acyl-phosphate intermediate (26). The ATP binding site within this structure is comprised of a “palmate” motif (27) rather than the Walker A/B motif (4). Other members of this N-ligase structural group (enzymes catalyzing amide ligation and sharing structural folding patterns) are glutathione synthetase (GS; ref. 27), which catalyzes phosphorylation of a γ-glutamyl-cysteinyl carboxyl group and ligation to the amino group of glycine, and d-alanine:d-alanine ligase (AA; ref. 28), which catalyzes phosphorylation of one d-alanyl carboxyl group and ligation to the amino group of the other molecule of d-alanine. Although our homology modeling served as the basis for the other studies described here, reliance solely on it has been rendered unnecessary by the recent report of the x-ray crystal structure for E. coli CPS (29). Domains B and C were found to superimpose with an rmsd of 1.1 Å and superposition of domain B on the BN coordinates yielded a rms value of 1.7 Å.
We have utilized site-directed mutagenesis to determine that appropriate CPS residues display the functional characteristics expected for ATP binding to a palmate structural motif (27) but not those expected for ATP binding to potential Walker A/B structural motifs (4). The fact that domains B and C are structurally equivalent suggested to us that they should display at least quasi-equivalent utilization of the two ATP molecules. This hypothesis is also consistent with a recent report demonstrating potential functional equivalence of domains B and C (30); in surprising contrast to the distinct functional behavior of domains B and C within the intact CPS, Guy and Evans (30) found that CPS from which either domain B or C has been deleted can dimerize to display full functional ability.
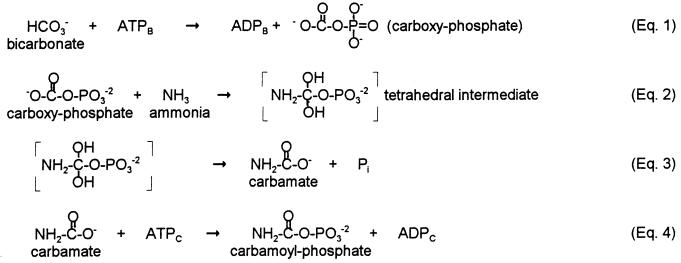
However, neither this study nor our own present findings are consistent with the sequential reaction mechanism (Fig. 1) for CPS first proposed by Jones and Lipmann in 1960 (31), and still generally utilized as the working mechanistic model for CPS (1, 30, 32, 33). As noted above, CxP has been demonstrated to be an intermediate (14–21), establishing the validity of Eq. 1. Although the remainder of the proposed pathway has not been established experimentally, it has been generally assumed that the CPS reaction proceeds sequentially from Eq. 1: CxP undergoes nucleophilic attack by NH3 to yield a tetrahedral intermediate (Eq. 2); the tetrahedral intermediate breaks down to yield carbamate plus Pi (Eq. 3); and finally carbamate interacts with the second molecule of ATP in a carbamate kinase reaction to yield CP (Eq. 4).
Figure 1.
Sequential mechanism for CPS (31).
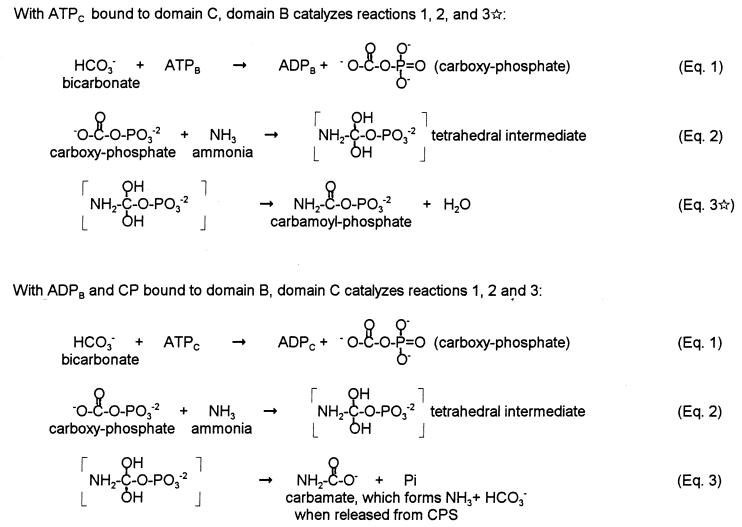
We are proposing an alternative mechanism for CPS (Fig. 2, discussed in detail below) in which usage of the two molecules of ATP is coupled, with the first ATP bound (ATPC in intact CPS) acting as a molecular switch to trigger conformational changes in domain B. Domain B can then catalyze the formation of CP via Eqs. 1, 2, and 3⋆, and finally catalysis of Eqs. 1, 2, and 3 on domain C allows return to the initial CPS conformation and the release of products. Although many of the features of the original scheme, including its pioneering proposal of energy transfer via the γ-phosphate group of ATP, are retained in Fig. 2, this revised scheme also incorporates the more recent knowledge of the role of ATP in effecting protein conformational switches (34).
Figure 2.
Coupled mechanism for CPS.
The sequential mechanism (Fig. 1) was assumed in interpreting the recent x-ray structure for CPS (29), and the authors did not consider any mechanistic implications of the finding of structural equivalence for domains B and C. Strikingly, the apparent ATP sites of domains B and C were found to be far removed (with a linear distance of about 35 Å) and it was proposed that, in analogy to tryptophan synthase (35), the presumed carbamate intermediate is channeled through the interior of CPS. Detailed evaluation of this channeling proposal will not be possible until the x-ray coordinates are available (June 1998 is listed by the Protein Data Bank as the estimated date of release for the E. coli CPS coordinates). However, the passage of the hydrophobic (and chemically stable) indole group through the well-defined 25-Å hydrophobic tunnel of tryptophan synthase may not be an appropriate model for possible movement of the labile and polar carbamate intermediate. And, as discussed below, the spatially distinct active sites would be consistent with our proposed coupled reaction mechanism (Fig. 2).
MATERIALS AND METHODS
Molecular biological procedures were performed essentially as described in Sambrook et al. (36). Standard yeast genetic and molecular biological procedures were performed as described (37). Site-directed mutagenesis was performed by the recombinant PCR method (38) or the unique site-elimination method (39) as described for yeast arginine-specific CPS (33, 40). The presence of desired mutations and the absence of any unexpected mutations in each mutagenesis cassette was confirmed by sequencing. To determine whether mutant constructions could produce CP, their ability to functionally complement CPS-disrupted yeast strain LPL26 was assessed (41). Generation time determination, crude extract preparation, enzymatic activity assay, protein determination, SDS/PAGE, and Western blot analysis were as previously utilized for CPS mutational analysis (33, 40).
RESULTS AND DISCUSSION
Homology Modeling of Yeast CPS Domains B and C with a BN Template.
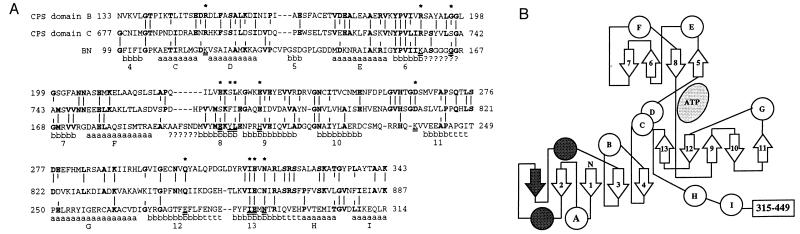
As discussed above, sole reliance on our homology modeling has been eliminated by the recent report of the x-ray crystal structure for E. coli CPS (29). However, because the CPS structural coordinates are not yet available, the homology modeling is still necessary to provide a three-dimensional structure for CPS upon which to base interpretation of the site-directed mutagenesis data. The template BN, as determined by x-ray crystallography (24, 26), is composed of four structural regions: residues 1–105, 106–203, 204–318, and 319–449. As shown in Fig. 3A, portions of the yeast CPS domains B and C (residues 133–343 and 677–887) share sequence identity (20% and 17%, respectively) with residues 99–314 of BN, the region comprising the two internal structural regions of BN. Also shown in Fig. 3A are the secondary structural elements observed in the BN solved structure. Fig. 3B shows the topological pattern of 9 α-helices and 13 β-strands shared by BN and the other members of the N-ligase structural group (ref. 26).
Figure 3.
(A) Alignment of yeast arginine-specific CPS domain B (residues 133–343) and domain C (residues 677–887) with BN (residues 99–314; ref. 42). The alignment is essentially that of Simmer et al. (2) and is based on alignment of five different CPSs. Vertical lines indicate sequence identity between the CPS domains or between BN and one or both of the CPS domains; the identical residues are shown in bold. a, b, and t indicate α-helix, β-sheet, and turn structural elements, respectively, in the solved structure of BN, and ? indicates residues too disordered to be included in the solved structure (24). The 12 BN residues conserved or conservatively substituted in the ATP binding sites of BN, GS, and AA (26) are indicated by underlining of the BN residues and by stars above their positions. (B) Topological diagram of residues 1–314 of the BN structure, simplified from Artymiuk et al. (26). α-Helix elements are shown as circles and β-sheet elements as arrows. The unshaded elements (helices A–I and sheets 1–13; also indicated on the bottom line of A) are those that are found in all members of the N-ligase family and the shaded elements are those unique to BN (26). The topology of residues 315–449 (data not shown) is also unique to BN.
The ATP binding sites have been determined to be in equivalent positions for GS (27) and AA (28), and are suggested to be in corresponding positions for BN (24, 26; Fig. 3B) and E. coli CPS (29). This ATP site is not comprised of the frequently observed βαβ nucleotide binding fold, where (i) a glycine-rich Walker A sequence forms a loop (the P-loop) about the nucleotide triphosphate group, with the loop connecting a β strand with an α helix, and (ii) a Walker B sequence corresponds to a hydrophobic strand of parallel β sheet, terminated by a carboxylate, that flanks the triphosphate binding site (4). Instead, interaction with ATP involves a “palmate” motif (27) comprised of two structural components: (i) a 4-stranded antiparallel β-sheet (β-strands 5–8 in Fig. 3B) plus the loop connecting strands 6 and 7, and (ii) a 5-stranded antiparallel β-sheet (β-strands 9–13 in Fig. 3B) plus the loop connecting strands 10 and 11. The ATP is sandwiched between these two β-sheets, binding at the face of the sheets. Both loops of this motif are flexible, presumably to permit entry of ATP and substrates and then to swing inward to cover and stabilize the substrates and intermediates during catalysis. The loops also serve to exclude water from the large catalytic cavity. In addition to the loop mobility, the second region of BN (β5–β8) appears to serve as a “lid” that closes down on the active site when the substrates are positioned for catalysis.
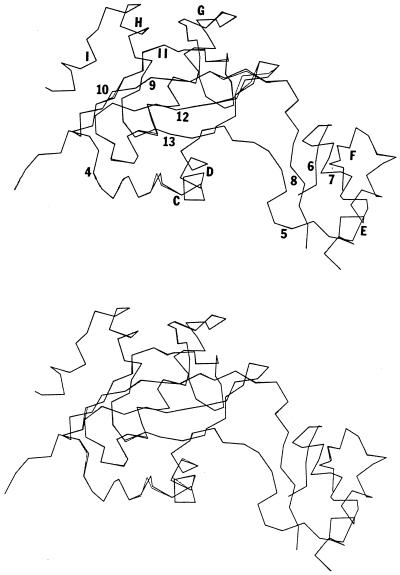
We utilized the x-ray coordinates for residues 99–314 of BN (24) as a template for modeling the homologous portions of CPS domains B and C (residues 133–343 and 677–887, respectively), using Molecular Simulations (Waltham, MA) software, including quanta and x-plor, on a Silicon Graphics 4D/35 workstation. The quanta spin program was used to optimize the van der Waals interactions of the substituted side chain atoms. Energy minimization was carried out using the charmm program (43) to yield the final CPS model structures. The modeled structures for CPS domains B and C, as well as a comparison to the BN backbone, are shown in Fig. 4. The backbones of the three structures are generally superimposable, with only small dislocations occurring in a few regions of the molecule. Twelve residues are conserved (or conservatively substituted) in the ATP binding sites for GS and AA. These 12 residues are also generally conserved in BN and in domains B and C of CPS (Fig. 3A) and occur at positions of near-superposition in the models. These homology models should serve as effective working models for the folded structure of domains B and C, which will of course be supplanted by the actual crystal structure of E. coli CPS (29) as soon as possible.
Figure 4.
Superimposed Cα traces of BN residues 99–314 (dark) with residues 133–343 of CPS domain B (Upper) or residues 677–887 of CPS domain C (Lower).
Site-Directed Mutagenic Analysis of CPS.
We have carried out site-directed mutagenesis studies to determine whether appropriate CPS residues display the functional characteristics expected for ATP utilization via the Walker A/B structural motif and/or for ATP utilization via a palmate structural motif. For domain B, CPS peptide 227–242 (KSLKGWKEVEYEVVRD) conforms most closely (6) to the primary sequence expected for the Walker B motif (K/RXXXGXXXLOOO-space-D, where X can be any amino acid residue and O is a generally nonpolar residue; ref. 4). In addition, CPS 227–242 is the counterpart of the rat liver CPS peptide identified as near the ATPB site by affinity analog labeling (12). We have previously shown that Y237 and R241 are not critical residues, but that E234, E236, and E238 are critical for function and that D242 appears to occupy a critical structural position (33). These findings were not consistent with the expectations for conventional functioning of a Walker B motif that D242 would be critical for function and that hydrophobic interaction would constitute the sole functional role of residues 236–241. In the present study, we have created mutations targeting four additional residues within the CPS peptide 227–242 that are conserved in all or most CPSs: K227A, G231V, W232A, and K233A. We were especially interested in the behavior of mutants involving K227 and G231 because these residues would be expected to be critical if CPS utilizes the standard interactions with ATP previously observed for the Walker B sequence motif. The functional effect of each mutation was determined by in vivo screening with the CPS-disrupted yeast strain LPL26 (41). All four mutants displayed wild-type functional behavior, indicating that none of the targeted residues plays a critical role in CPS structure and/or function.
We have also carried out site-directed mutagenic screening of the domain C equivalent of the putative Walker B motif: peptide 773–784 (KFIEGAQEIDVD). We have targeted the residues corresponding to those that were found to be critical within domain B: E780, D782, and D784. Mutations were designed to (i) determine if any feature of the side chain is essential (E780A, D782A, D784A); (ii) determine whether the spatial arrangement of the carboxyl group is critical (E780D, D782E, D784E); (iii) determine if hydrogen-bonding potential is sufficient for functionality or if the carboxyl groups are critical (E780Q, D782N, D784N); and (iv) determine if a substitution of the corresponding residue in the BN sequence can allow functioning (E780H, D782E, D784N). The analysis of these mutants (Table 1) revealed that the presence of a carboxyl group and its exact spatial arrangement appears to be critical only for E780, although functional groups with hydrogen bonding potential are required at positions 782 and 784. We have also constructed and analyzed the K773A mutant (Table 1). Although the terminal lysyl residue is a critical feature of the Walker B motif, its replacement by alanine resulted in an apparently fully functional CPS.
Table 1.
Effect of CPS mutations on the ability of transformants to support the growth of the CPS-disrupted strain LPL26 and on CP synthesis activity in crude extracts prepared from transformants.
| Mutant | Generation time, (h)
|
Specific activity
|
||
|---|---|---|---|---|
| 30°C | 37°C | − Gly | + Gly | |
| Wild-type CPS | 4.9 | 5.7 | 1.05 | 3.35 |
| K773A | 5.5 | 4.7 | 1.23 | 5.12 |
| E780A | u | u | u | u |
| E780D | u | u | u | u |
| E780Q | u | u | u | u |
| E780H | u | u | u | u |
| D782A | u | u | u | u |
| D782E | 4.7 | 6.0 | 0.62 | 2.12 |
| D782N | 33.5 | 96.3 | 0.16 | 0.26 |
| D784A | u | u | u | u |
| D784E | 4.8 | 4.8 | 0.27 | 0.57 |
| D784N | u* | u* | 0.19 | 0.46 |
| D784Q | 43.3 | 91.5 | u | u |
| E317A | u | u | u | u |
| N319A | u | u | u | u |
Glycine activation indicates conformational coupling between the synthetase and glutamine amidotransferase subunits of CPS, with glycine binding to the latter as a glutamine analog (44); the presence of such coupling serves as one indication that the mutation in CPS has not resulted in a gross conformational change. Specific activity is nmol CP synthesized/min per unit CPS protein, with amount of CPS protein determined by densitometry of Western blots. u, Below the assay detection limit; u*, D784N failed to grow in liquid media but showed essentially wild-type growth on solid medium.
Site-directed mutagenic screening has also been carried out on peptide 131–139 (KYNVKVLGT), which contains elements of the Walker A sequence (K/R-space-GXXXXGKT/S) and occurs at the expected distance from the putative Walker B sequence (6). When we substituted G138, which should be essential if it is involved in forming a P loop, with the bulky amino acid valine, the construction displayed wild type functional behavior in the in vivo screen. The T139A mutant, with elimination of the terminal threonine residue that is also critical for the Walker A motif, supported growth at ≈30% of the wild-type rate. These findings, together with those on the potential Walker B motifs of domains B and C, very strongly suggest that CPS is not a member of the Walker A/B motif structural family. It should be noted that another region of CPS (including G196 and G199 of domain B and G741 of domain C) has been proposed as the Walker A motif, and that these corresponding three glycyl residues were critical for activity of E. coli CPS (22). However, this domain C region of CPS contains only one glycyl residue and neither the domain B nor the domain C region contains the expected terminal K or S/T residues; also, there is no correctly spaced Walker B motif for this other potential Walker A motif.
Our site-directed mutagenic analysis provides support for utilization of a “palmate” ATP binding motif by CPS and argues against its utilization of a Walker A/B ATP binding motif. The glycine-containing loop in the palmate motif (between β6 and β7) differs from that of the Walker A motif by not containing the terminal conserved residues KS/T (27). The CPS region containing G196 and G199 (domain B) or G741 (domain C) form this loop in our modeled structure, whereas the other CPS region considered as a potential Walker A motif (131–139 of domain B or 675–683 of domain C) forms a loop that connects the ATP-binding region of CPS to another region of the protein. In the palmate structure, the homologous CPS peptides 227–242 (domain B) and 773–788 (domain C) form β-strand 9, which contains 1 of the 12 conserved active site residues: the critical residues E234 and E780 are thus predicted to form hydrogen bonds with the ribose hydroxyl group of ATP.
To further confirm that the palmate motif modeling is correct, we carried out mutagenic screening of CPS residues E317 and N319, which would correspond to 2 more of the 12 residues conserved at the ATP binding sites of known palmate motif enzymes (26). Both E317 and N319 are expected to be near the ATP phosphate groups and to be coordinated to the Mg2+. In keeping with these proposed critical roles, neither E317A nor N319A displayed activity in the in vivo screen. The domain C counterpart to E317 is E861, which is conserved in all CPSs. Previous studies have shown that the E. coli CPS analog of E861 is an essential residue (45). Thus, from the present and previous studies, 4 of the 12 conserved palmate motif active site residues are known to also be critical for CPS functioning. During the present studies, additional site-directed mutagenic analysis of E. coli CPS was reported (32, 46). Significant functional changes resulted (32, 46) from the site-directed mutation of 3 more of the 12 conserved palmate motif residues within both domains B and C (counterparts to R150/R694, R190/R734, and Q303/Q848) but not from the mutation of a fourth conserved residue in either domain (counterparts to E226/S772). Consistent with our present findings, the E. coli CPS counterparts of E234, E317, N319, and E780 were found to be critical (32); the domain C counterparts of E317 and N319 were also found to be critical (46). In summary, both our own mutagenic analysis and the recently reported one on E. coli CPS are consistent with the proposal that domains B and C utilize a palmate ATP-binding motif.
Novel Mechanism for Coupled ATP Use by CPS: ATP Binding at One Domain Triggers a Conformational Switch That Allows CP Synthesis at the Other Domain.
The finding that domains B and C are structurally equivalent and that they conform to the structure expected for N-ligases (our present work and ref. 29), as well as the recent finding that domains B and C are functionally equivalent (30), have prompted us to re-evaluate the sequential reaction pathway for CPS (Fig. 1) that was proposed in 1960 by Jones and Lipmann (31). Although domain B catalysis of the carbamate synthesis reaction described by Eqs. 1-3 is consistent with the known N-ligase functionality of the palmate structural group, domain C catalysis of the carbamate kinase reaction of Eq. 4 is not. Nor is the catalysis of distinct reaction types consistent with the structural equivalence and the apparent functional equivalence of domains B and C. Finally, utilization of the sequential pathway by independent active sites of homologous domains B and C would require movement of the labile carbamate intermediate from domain B to domain C, an apparent distance of 35 Å (29).
It is very well established (14–23) that one molecule of ATP binds to domain B and is used to form CxP (Eq. 1), but the validity of Eqs. 2-4 has only been assumed by CPS researchers (1, 30, 32, 33, 45). There have been no reported attempts to directly identify carbamate as an intermediate (Eq. 3), presumably because it is in equilibrium with the CPS substrates NH3 and HCO3− (31) and would thus be very difficult to definitively identify. Two lines of indirect evidence have suggested that ATPC functions in a carbamate kinase reaction (Eq. 4). The ability of domain C to catalyze the formation of ATP from CP and ADP has been interpreted as reflecting the reverse of Eq. 4 (47). However, as previously noted (16), this reaction more likely reflects the reversal of Eq. 1, CxP formation, with CP acting as a CxP analog because several biotin-dependent enzymes (for which CxP is an intermediate whereas neither carbamate nor CP is involved in the reaction) can also catalyze the formation of ATP from CP plus ADP (25). Furthermore, the synthesis of ATP from CP plus ADP can also be catalyzed by glutamine synthetase (48) and formyltetrahydrofolate synthetase (49), where presumably CP is serving, respectively, as an analog of glutamyl-phosphate or of formyl-phosphate. Reported sequence identity (13.5%) between domain C of E. coli CPS and Pseudomonas aerouginosa carbamate kinase (50) has also been interpreted as supporting the idea of a functional relationship between the two enzymes. However, the sequence comparison (50) failed to consider the requirement for 22 gaps in the alignment to produce the observed 42 identities. When re-analyzed with appropriate gap penalties (51), there is no significant identity between carbamate kinase and either domain B or C of CPS.
We are proposing an alternative mechanism (Fig. 2) for CPS in which the energy derived from the 2 ATP molecules is coupled. In Fig. 2, ATPC binds to domain C and acts as a nucleotide switch for domain B, similar to ATP usage by nitrogenase, myosin, and signal transduction proteins (34). The “switched-on” conformation of domain B then sequentially binds ATPB, HCO3−, and NH3 and carries out the portion of the N-ligase reaction described by Eqs. 1 and 2. However, rather than proceeding through Eq. 3, the “switched-on” conformation of domain B allows the direct collapse of the tetrahedral intermediate to CP, as shown in Eq. 3⋆. This route of collapse is possibly facilitated by protonation of one of the hydroxyl groups of the tetrahedral intermediate, yielding water as the actual leaving group. Because ADPB now occupies the ATP/ADP site of domain B, it cannot act as a nucleotide switch for domain C. Thus, domain C cannot attain the switched-on conformation necessary for Eq. 3⋆. Instead, a standard N-ligase reaction yielding ADPC, Pi, and carbamate occurs on domain C (Eqs. 1-3), and triggers a “switched-off” conformation that allows release of the products bound to both domains (CP, Pi, carbamate, and 2 ADP). Upon dissociation from the enzyme, carbamate would form NH3 and HCO3−, the substrates for the overall reaction. For the CPSs where NH3 is delivered to domain B from the glutaminase domain (rather than free NH3 binding directly), it is likely that NH3 will not also be delivered to domain C and that water will instead serve to break down the CxP intermediate, yielding HCO3− and phosphate.
As outlined in Table 2, our proposed coupled scheme reconciles the many previous experimental findings on the CPS mechanism, whereas a number of the previous findings are not consistent with the sequential scheme. The coupled scheme predicts functionally equivalent domains (point 1) because the distinction between the functions carried out by domains B and C is determined only by which of these domains first binds ATP. Our present assignment of dedicated functions for domains B and C within intact CPS was based on the domain localizations of partial CPS activities (points 2–5), but is further supported by the recent finding that the glutaminase domain of E. coli CPS is near domain B and removed from domain C (29). The existence of the two partial reactions and their domain localizations (points 2–5) are consistent with both proposed schemes. In both schemes, the HCO3−-dependent ATPase activity is thought to reflect CxP formation at domain B. The ability of CPS to catalyze the formation of ATP from CP and ADP is interpreted as reflecting the reverse of Eq. 4 (47) in the sequential mechanism. In the coupled scheme it is interpreted as the reversal of Eq. 1, CxP formation, with CP acting as a CxP analog (16) and with the ADP binding at domain C, because this is the site for initial ATP binding. However, only the coupled scheme can provide a rationale for the finding (point 6) that many of the site-directed mutations at the ATPC site have a significant effect on the Km for ATPB (measured in the assay for HCO3−-dependent ATPase activity) whereas the corresponding ATPB site mutations fail to appreciably affect the Km for ATPC (measured in the assay for ATP synthesis from ADP plus CP). These kinetic effects would be consistent with the required binding of ATPC before ATPB in Fig. 2 but not with the independent and sequential utilization of the two ATP molecules previously proposed in Fig. 1. The previous observation (55, 56) that allosteric control of CPS is centered on binding of ATPC would also be consistent with the coupled scheme where ATPC binds first in the intact CPS.
Table 2.
Consistency of experimental evidence with the proposed mechanisms for CPSase
| Experimental evidence | Consistent with proposed mechanisms
|
||
|---|---|---|---|
| Sequential model (Fig. 1) | Coupled model (Fig. 2) | ||
| 1. | Functional equivalence of domains B and C (30) | No | Yes |
| 2. | Catalysis of bicarbonate-dependent cleavage of ATP to ADP + Pi (47) | Yes | Yes |
| 3. | Domain B localization of bicarbonate-dependent ATPase activity (22, 23) | Yes | Yes |
| 4. | Catalysis of ATP formation from CP + ADP (47) | Yes | Yes |
| 5. | Domain C localization of ATP formation from CP + ADP (22, 23) | Yes | Yes |
| 6. | Significant increases in the Km for ATPB observed in ATPC site mutants but not in the Km for ATPC in response to corresponding mutations at the ATPB site (22, 32, 46) | No | Yes |
| 7. | Structural equivalence of domains B and C to each other and to the N-ligases (29) | No | Yes |
| 8. | Absence of significant sequence identity for domain B or C with carbamate kinase | No | Yes |
| 9. | Expected lability of carbamate during movement from domain B to domain C occurring in the sequential mechanism | No | Yes |
| 10. | Chemical and kinetic competency of carboxy-phosphate (14–21) | Yes | Yes |
| 11. | Absence of ATP/ADP or ATP/Pi exchange, indicating ADP remains bound as a component of the enzyme–carboxy–phosphate complex (18–20) | Yes | Yes |
| 12. | ATP positional isotope exchange evidence for an alternative pathway forming carboxy-phosphate in addition to the main CP-forming pathway (18, 21) | No | Yes |
| 13. | Binding of both molecules of ATP before ammonia is bound (16, 52–54) | No | Yes |
As discussed above, points 7–9 of Table 2 fit only the coupled scheme. Generally the experimental evidence supporting the intermediacy of CxP is consistent with both proposed mechanisms because they both involve its formation (points 10 and 11). However, the isotopic exchange evidence for a pathway forming CxP which is distinct from its formation via the main CP-forming pathway (point 12) is only consistent with the coupled scheme. The finding in a number of studies that both molecules of ATP bind before NH3 (point 13) is also consistent only with the coupled scheme. This order of binding is also predicted by one kinetic analysis (57), although two others (21, 58) find that the best fit to their data is given by NH3 binding after the first ATP. Rubio and Grisolia (16) have previously noted the inconsistency of the direct binding studies with the sequential mechanism and suggested a concerted reaction between CxP, NH3, and the second molecule of ATP to form CP with no intermediate formation of carbamate. This mechanism, however, would not be consistent with points 1, 6, 7, or 13 of Table 2. An additional alternative CPS mechanism has also been considered (30), where each domain can catalyze the overall reaction by the sequential mechanism but only when associated with another domain in a pseudodimer. However, this mechanism would be inconsistent with points 3, 5, 6, and 7 of Table 2. It thus seems that the existing experimental evidence supports our present proposal that CPS functions via coupled usage of the two molecules of ATP as depicted in Fig. 2. The availability of this novel proposed mechanism and of a solved three-dimensional structure for CPS should greatly facilitate the rational design and interpretation of future experiments, and yield significant insights into the CPS structure/function relationship.
Acknowledgments
We gratefully acknowledge Dr. Angela L. Lim for her helpful advice and Dr. Richard Deth for his assistance with homology modeling. These studies were supported by research grants from the National Institutes of Health (DK-33460) and from the Northeastern University Research and Scholarship Development Fund.
ABBREVIATIONS
- AA
d-alanine:d-alanine ligase
- BN
biotin carboxylase
- CP
carbamoyl-phosphate
- CPS
carbamoyl-phosphate synthetase
- CxP
carboxy-phosphate
- N-ligase group
enzymes catalyzing amide ligation and sharing structural folding pattern
- GS
glutathione synthetase
References
- 1.Anderson P M. In: Nitrogen Metabolism and Excretion. Walsh P J, Wright P, editors. Boca Raton, FL: CRC; 1995. pp. 33–49. [Google Scholar]
- 2.Simmer J P, Kelly R E, Rinker A G, Jr, Scully J L, Evans D R. J Biol Chem. 1990;265:10395–10402. [PubMed] [Google Scholar]
- 3.van den Hoff M J B, Jonker A, Beintema J J, Lamers W H. J Mol Evol. 1995;41:813–832. doi: 10.1007/BF00173161. [DOI] [PubMed] [Google Scholar]
- 4.Walker J E, Saraste M, Runswick M J, Gay N J. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lusty C J, Widgren E E, Broglie K E, Nyuonya H. J Biol Chem. 1983;258:14466–14472. [PubMed] [Google Scholar]
- 6.Powers-Lee S G, Corina K. J Biol Chem. 1987;262:9052–9056. [PubMed] [Google Scholar]
- 7.Powers-Lee S G, Corina K. J Biol Chem. 1986;261:15349–15352. [PubMed] [Google Scholar]
- 8.Marshall M A, Fahien L A. Arch Biochem Biophys. 1988;262:455–470. doi: 10.1016/0003-9861(88)90397-9. [DOI] [PubMed] [Google Scholar]
- 9.Evans D R, Balon M A. Biochim Biophys Acta. 1988;953:185–196. doi: 10.1016/0167-4838(88)90023-4. [DOI] [PubMed] [Google Scholar]
- 10.Mareya S M, Raushel F M. Bioorg Med Chem. 1995;3:525–532. doi: 10.1016/0968-0896(95)00042-f. [DOI] [PubMed] [Google Scholar]
- 11.Kim H, Lee L, Evans D R. Biochemistry. 1991;30:10322–10329. doi: 10.1021/bi00106a033. [DOI] [PubMed] [Google Scholar]
- 12.Potter M D, Powers-Lee S G. J Biol Chem. 1992;267:2023–2031. [PubMed] [Google Scholar]
- 13.Potter M D, Powers-Lee S G. Arch Biochem Biophys. 1993;306:377–382. doi: 10.1006/abbi.1993.1526. [DOI] [PubMed] [Google Scholar]
- 14.Anderson P M, Meister A. Biochemistry. 1965;4:2803–2809. doi: 10.1021/bi00888a034. [DOI] [PubMed] [Google Scholar]
- 15.Powers S G, Meister A. Proc Natl Acad Sci USA. 1976;73:3020–3024. doi: 10.1073/pnas.73.9.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubio V, Grisolia S. Biochemistry. 1977;16:321–329. doi: 10.1021/bi00621a025. [DOI] [PubMed] [Google Scholar]
- 17.Powers S G, Meister A. J Biol Chem. 1978;253:1258–1265. [PubMed] [Google Scholar]
- 18.Wimmer M J, Rose I A, Powers S G, Meister A. J Biol Chem. 1979;254:1854–1859. [PubMed] [Google Scholar]
- 19.Raushel F M, Villafranca J J. Biochemistry. 1979;18:3424–3429. doi: 10.1021/bi00582a033. [DOI] [PubMed] [Google Scholar]
- 20.Rubio V, Britton H G, Grisolia S, Sproat B S, Lowe G. Biochemistry. 1981;20:1969–1973. doi: 10.1021/bi00510a036. [DOI] [PubMed] [Google Scholar]
- 21.Meek T D, Karsten W E, DeBrosse C W. Biochemistry. 1987;26:2584–2593. doi: 10.1021/bi00383a026. [DOI] [PubMed] [Google Scholar]
- 22.Post L E, Post D J, Raushel F M. J Biol Chem. 1990;265:7742–7747. [PubMed] [Google Scholar]
- 23.Alonso E, Cervera J, Garcia-Espana A, Bendala E, Rubio V. J Biol Chem. 1992;267:4524–4532. [PubMed] [Google Scholar]
- 24.Waldrop G L, Rayment I, Holden H M. Biochemistry. 1994;33:10249–10256. doi: 10.1021/bi00200a004. [DOI] [PubMed] [Google Scholar]
- 25.Knowles J R. Annu Rev Biochem. 1989;58:195–221. doi: 10.1146/annurev.bi.58.070189.001211. [DOI] [PubMed] [Google Scholar]
- 26.Artymiuk P J, Poirrette A R, Rice D W, Willett P. Nat Struct Biol. 1996;3:128–132. doi: 10.1038/nsb0296-128. [DOI] [PubMed] [Google Scholar]
- 27.Yamaguchi H, Kato H, Hata Y, Nishioka T, Kimura A, Oda A, Katsube Y. J Mol Biol. 1993;229:1083–1100. doi: 10.1006/jmbi.1993.1106. [DOI] [PubMed] [Google Scholar]
- 28.Fan C, Moew P C, Walsh C T, Knox J R. Science. 1994;266:439–443. doi: 10.1126/science.7939684. [DOI] [PubMed] [Google Scholar]
- 29.Thoden J B, Holden H M, Wesenberg G, Raushel F M, Rayment I. Biochemistry. 1997;36:6305–6316. doi: 10.1021/bi970503q. [DOI] [PubMed] [Google Scholar]
- 30.Guy H I, Evans D R. J Biol Chem. 1996;271:13762–13769. doi: 10.1074/jbc.271.23.13762. [DOI] [PubMed] [Google Scholar]
- 31.Jones M E, Lipmann F. Proc Natl Acad Sci USA. 1960;46:1194–1205. doi: 10.1073/pnas.46.9.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stapleton M A, Javid-Majd F, Harmon M F, Hanks B A, Grahmann J L, Mullins L S, Raushel F M. Biochemistry. 1996;35:14352–14361. doi: 10.1021/bi961183y. [DOI] [PubMed] [Google Scholar]
- 33.Zheng W, Lim A L, Powers-Lee S G. Biochim Biophys Acta. 1997;1341:35–48. doi: 10.1016/s0167-4838(97)00058-7. [DOI] [PubMed] [Google Scholar]
- 34.Schindelin H, Kisker C, Schlessman J L, Howard J B, Rees D C. Nature (London) 1997;387:370–376. doi: 10.1038/387370a0. [DOI] [PubMed] [Google Scholar]
- 35.Ruvinov S B, Yang X J, Parris K D, Banik U, Ahmed S A, Miles E W, Sackett D L. J Biol Chem. 1995;270:6357–6369. doi: 10.1074/jbc.270.11.6357. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 37.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 38.Higuchi R. In: PCR Protocols. Innis M A, Gelfand D H, Sninsky J J, White T J, editors. New York: Academic; 1990. pp. 177–183. [Google Scholar]
- 39.Deng W P, Nickoloff J A. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 40.Lim A L, Powers-Lee S G. Arch Biochem Biophys. 1997;339:344–352. doi: 10.1006/abbi.1997.9887. [DOI] [PubMed] [Google Scholar]
- 41.Lim A L, Powers-Lee S G. J Biol Chem. 1996;271:11400–11409. doi: 10.1074/jbc.271.19.11400. [DOI] [PubMed] [Google Scholar]
- 42.Kondo H, Shiratsuhi K, Yoshimoto T, Masuda T, Kitazono A, Tsuru D, Anai M, Skiguchi M, Tanabe T. Proc Natl Acad Sci USA. 1991;88:9730–9733. doi: 10.1073/pnas.88.21.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brooks B R, Bruccoleri R E, Olafson B D, States D J, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 44.Pierard A, Schroter B. J Bacteriol. 1978;134:167–176. doi: 10.1128/jb.134.1.167-176.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guillou F, Liao M, Garcia-Espana A, Lusty C J. Biochemistry. 1992;31:1656–1664. doi: 10.1021/bi00121a012. [DOI] [PubMed] [Google Scholar]
- 46.Javid-Majd F, Stapleton M A, Harmon M F, Hanks B A, Mullins L S, Raushel F M. Biochemistry. 1996;35:14362–14369. doi: 10.1021/bi961184q. [DOI] [PubMed] [Google Scholar]
- 47.Anderson P M, Meister A. Biochemistry. 1966;5:3157–3163. doi: 10.1021/bi00874a012. [DOI] [PubMed] [Google Scholar]
- 48.Tate S S, Leu F Y, Meister A. J Biol Chem. 1972;247:5312–5321. [PubMed] [Google Scholar]
- 49.Buttlaire D H, Himes R H, Reed G H. J Biol Chem. 1976;251:4159–4161. [PubMed] [Google Scholar]
- 50.Baur H, Luethi E, Stalon V, Mercenier A, Haas D. Eur J Biochem. 1989;179:53–60. doi: 10.1111/j.1432-1033.1989.tb14520.x. [DOI] [PubMed] [Google Scholar]
- 51.Feng D F, Johnson M S, Doolittle R F. J Mol Evol. 1984;21:112–125. doi: 10.1007/BF02100085. [DOI] [PubMed] [Google Scholar]
- 52.Fahien L A, Cohen P P. J Biol Chem. 1964;239:1925–1934. [PubMed] [Google Scholar]
- 53.Powers S G, Meister A. J Biol Chem. 1978;253:800–803. [PubMed] [Google Scholar]
- 54.Rubio V, Britton H G, Grisolia S. Eur J Biochem. 1979;93:245–256. doi: 10.1111/j.1432-1033.1979.tb12817.x. [DOI] [PubMed] [Google Scholar]
- 55.Liu X, Guy H I, Evans D R. J Biol Chem. 1994;269:27747–27755. [PubMed] [Google Scholar]
- 56.Czerwinski R M, Mareya S M, Raushel F M. Biochemistry. 1995;34:13920–13927. doi: 10.1021/bi00042a025. [DOI] [PubMed] [Google Scholar]
- 57.Elliott K R F, Tipton K F. Biochem J. 1974;141:817–824. doi: 10.1042/bj1410817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raushel F M, Anderson P M, Villafranca J J. Biochemistry. 1978;17:5587–5591. doi: 10.1021/bi00619a001. [DOI] [PubMed] [Google Scholar]