Abstract
Cholesterol feeding reduces the mRNAs encoding multiple enzymes in the cholesterol biosynthetic pathway and the low density lipoprotein receptor in livers of hamsters. Here we show that cholesterol feeding also reduces the levels of the nuclear NH2-terminal domains of sterol regulatory element binding proteins (SREBPs), which activate transcription of sterol-regulated genes. We show that livers of hamsters, like those of mice and humans, predominantly produce SREBP-2 and the 1c isoform of SREBP-1. Both are produced as membrane-bound precursors that must be proteolyzed to release the transcriptionally active NH2-terminal domains. Diets containing 0.1% to 1.0% cholesterol decreased the amount of nuclear SREBP-1c without affecting the amount of the membrane precursor or its mRNA, suggesting that cholesterol inhibits the proteolytic processing of SREBP-1 in liver as it does in cultured cells. Cholesterol also appeared to reduce the proteolytic processing of SREBP-2. In addition, at high levels of dietary cholesterol the mRNA encoding SREBP-2 declined and the amount of the precursor also fell, suggesting that cholesterol accumulation also may inhibit transcription of the SREBP-2 gene. The high-cholesterol diets reduced the amount of low density lipoprotein receptor mRNA by 30% and produced a more profound 70–90% reduction in mRNAs encoding 3-hydroxy-3-methylglutaryl CoA synthase and reductase. Treatment with lovastatin and Colestipol, which increases hepatic demands for cholesterol, increased the amount of SREBP-2 mRNA as well as the precursor and nuclear forms of the protein. This treatment caused a reciprocal decline in SREBP-1c mRNA and protein. Considered together, these data suggest that SREBPs play important roles in controlling transcription of sterol-regulated genes in liver, as they do in cultured cells.
Keywords: cholesterol biosynthesis, low density lipoprotein receptors, 3-hydroxy-3-methylglutaryl CoA synthase and reductase, transcriptional regulation
Since the pioneering work of Gould, carried out more than 40 years ago (1, 2), scientists have known that high-cholesterol diets suppress cholesterol synthesis in the livers of experimental animals. More recently, the converse also was shown to be true, i.e., manipulations that deplete the liver of cholesterol lead to an increase in cholesterol synthesis (see ref. 3 for review). Much of this control is attributable to coordinate changes in the levels of mRNAs encoding multiple enzymes in the cholesterol biosynthetic pathway, including 3-hydroxy-3-methylglutaryl CoA (HMG CoA) synthase, HMG CoA reductase, farnesyl diphosphate synthase, squalene synthase, and others (see ref. 4 for review). The mRNA for the low density lipoprotein (LDL) receptor also is reduced by cholesterol feeding and increased by cholesterol depletion, although the amplitude of these changes is not as profound as that of the cholesterol biosynthetic enzymes, and the changes do not necessarily occur in parallel (4, 5). The changes in hepatic LDL receptors contribute to the elevation in blood cholesterol levels induced by high-cholesterol diets and to the reduction that follows hepatic cholesterol depletion (6).
A potential mechanism for this regulation was disclosed recently through studies of nonhepatic cells in tissue culture. In these cells the transcription of genes encoding cholesterol biosynthetic enzymes and the LDL receptor is controlled by a family of transcription factors designated SREBPs (sterol regulatory element binding proteins) (see ref. 7 for review). The SREBPs are proteins of ≈1,150 amino acids that are bound to membranes of the endoplasmic reticulum. In sterol-depleted cells, proteases release the NH2-terminal domains of the SREBPs, which are transcription factors of the basic-helix-loop-helix-leucine zipper (bHLH-Zip) family. These soluble domains, designated the mature forms of the SREBPs, enter the nucleus where they activate transcription by binding to 10-bp sterol regulatory elements in the enhancer regions of target genes. When cultured cells are overloaded with sterols, the proteolytic process is inhibited, the SREBPs remain bound to endoplasmic reticulum membranes, and transcription of the target genes declines (7).
The three known members of the SREBP family are produced by two genes (7). The SREBP-1 gene gives rise to two transcripts designated SREBP-1a and SREBP-1c, which differ only in the first exon that encodes an acidic transcription activation domain. This domain is much longer in SREBP-1a than in SREBP-1c, and therefore SREBP-1a is a much stronger activator of transcription (8). The third member of the family, designated SREBP-2, has a long activation domain, and its action resembles that of SREBP-1a. Tissue culture cells produce predominantly SREBP-1a and SREBP-2 (9), and the proteolytic processing of the two proteins is regulated in parallel (7).
In liver, the pattern of SREBP expression and regulation differs from that observed in cultured cells. In livers of mice and humans, the SREBP-1c mRNA is at least 9-fold more abundant than the SREBP-1a mRNA (9). The abundance of the SREBP-2 transcript appears to be intermediate between these extremes (9). In hamster liver, depletion of cholesterol by treatment with a bile acid binding resin (Colestipol) and an HMG CoA reductase inhibitor (lovastatin) caused a paradoxical decline in the amount of total SREBP-1 protein and in the efficiency of its processing to the mature form (7, 10). At the same time, the total amount of SREBP-2 increased, and its processing to the mature form increased. If hamster liver produces SREBP-1c as its predominant isoform, then the net effect of the lovastatin/Colestipol regimen would be the replacement of a weak activator, SREBP-1c, with a strong activator, SREBP-2.
The current studies were designed to determine whether hamster liver produces SREBP-1c as the predominant isoform and to assess the effects of cholesterol feeding on the pattern of expression and processing of the SREBPs in livers of these animals. We chose to perform these studies in hamsters because of the extensive previous literature documenting an inhibition of cholesterol synthesis and LDL receptor activity elicited by cholesterol feeding in this species (11, 12). Our results indicate that hamster liver does produce SREBP-1c and that cholesterol feeding reduces the amount of nuclear SREBP-1c and SREBP-2. These changes appear to explain the reduction in mRNAs for the cholesterol biosynthetic enzymes in response to cholesterol feeding.
METHODS
Materials and Procedures.
Standard molecular biology techniques were used (13). We obtained all restriction enzymes and modifying enzymes from New England Biolabs, [α-32P]CTP (3,000 Ci/mmol) from Amersham, and other chemicals from Sigma. Plasmid DNA was prepared with Plasmid Maxi kits (Qiagen, Chatsworth, CA). Total RNA was prepared by the guanidinium thiocyanate/phenol/chloroform method (14). The content of cholesterol in plasma and liver was measured as described (15, 16).
Animals.
Male and female Golden Syrian hamsters (100–120 g), obtained from Sasco (Omaha, NE), were exposed to a 12-hr light/12-hr dark cycle and fed one of the following diets: standard Teklad 4% Mouse/Rat Diet 7001 (Harlan Teklad, Madison, WI), the same diet supplemented with powdered cholesterol (0.1–1%, wt/wt), or the same diet containing powdered lovastatin (0.05%) (Merck Sharp and Dohme) plus 4% Colestipol (Upjohn). Hamsters had free access to diet and water during the experimental period and were sacrificed without fasting at the midpoint of the dark cycle.
Immunoblot Analysis.
Membranes (105 g pellet) and nuclear extracts from hamster livers were prepared immediately after exsanguination as described (10). Aliquots of membranes and nuclear extracts were subjected to 8% SDS/PAGE and transferred to Hybond C extra membranes (Amersham). The following primary antibodies were used: mouse mAb (IgG-2A4) against amino acids 301–407 of human SREBP-1a (17) at 5 μg/ml, and a 1:4,000 dilution of a rabbit IgG fraction of antiserum against amino acids 32–250 of hamster SREBP-2 (18). Immunoblot analysis was carried out with the ECL Western Blotting Detection System Kit (Amersham) by using conditions for the horseradish-peroxidase reaction and wash as described (10, 19). Protein concentrations were determined with the BCA Kit (Pierce).
cDNA Cloning of Hamster Sequences Corresponding to 5′ Ends of SREBP-1a and SREBP-1c.
The 5′ ends of hamster SREBP-1a and SREBP-1c cDNAs were cloned from Syrian hamster liver poly(A)+ RNA by the 5′ rapid amplification of cDNA ends (RACE) method (20) by using a 5′ RACE system kit (GIBCO/BRL), AmpliTaq polymerase (Perkin–Elmer/Cetus), and nested primers derived from the Chinese hamster ovary SREBP-1a cDNA sequence (17). The primer for first-strand synthesis was 5′-TGGACCTGGGTGTGTAAAGAGATGGGCG-3′, which corresponds to a sequence in exon 2. The nested primer was 5′-GAGAAGCCTGAAGGAAGGCTAGAATAC-3′. The 5′ RACE library was probed with 32P-labeled oligonucleotides derived from exon 1a (5′-CGCCATGGACGAGCTGGCCTTCGGT-3′) or exon 1c (5′-AATGTGCAATCCATGGCTCCGTGGTCCGCG-3′) of mouse SREBP-1 (9). The sequences of the 5′ ends of the Syrian hamster liver SREBP-1a and SREBP-1c cDNAs were used to generate SREBP-1a and SREBP-1c specific DNA templates for generation of cRNA probes as described below.
RNase Protection Assay.
cDNA fragments for Syrian hamster SREBP-1a, SREBP-1c, SREBP-2, LDL receptor, HMG CoA synthase, HMG CoA reductase, and β-actin were amplified by PCR from first-strand cDNA prepared from Syrian hamster liver poly(A)+ RNA by using the following primers: SREBP-1a, 5′ primer, 5′-GCGCCATGGAGGAGCTGCCCTTCG-3′, and 3′ primer, 5′-GTCACTGTCTTGGTTGTTGATG-3′ (this paper); SREBP-1c, 5′ primer, 5′-TGCGGACGCAGTCTGGGCAAC-3′, and 3′ primer, 5′-GTCACTGTCTTGGTTGTTGATG-3′ (this paper); SREBP-2, 5′ primer, 5′-GACCACAATGCCTGTGATGATG-3′, and 3′ primer, 5′-GTCCACATCACTGTCCACCAG-3′ (18); LDL receptor, 5′ primer, 5′-GAGTGCTTGGACAACAATGGTGGCTGTTCC-3′, and 3′ primer, 5′-ACAGCCTTGCAGACCCTGGTGTGAGGGTCCAT-3′ (21, 22); HMG CoA synthase, 5′ primer, 5′-CTTTGC(A/C)TGACTGTGGTTCAGAATCT-3′, and 3′ primer, 5′-ACAGCATTGAA- GACAGCAGCTGTGGC-3′ (23, 24); HMG CoA reductase, 5′ primer, 5′-TGTGCCATGGCTGGGAGCATAGGAGGC-3′, and 3′ primer, 5′-GCTCCTTGAACACCTAGCATCTGC-3′ (25, 26); β-actin; 5′ primer, 5′-CACCAGGGCGTGATGGTGGG-3′, and 3′ primer, 5′-GATGCCTCTCTTGCTCTGGGC-3′ (27, 28). In cases where Syrian hamster cDNA sequences were not available, the primer sequences were derived from known Chinese hamster, mouse, rat, and human cDNA sequences as indicated in the above references. HindIII and EcoRI sites were added to all 5′ and 3′ primers, respectively. All first-strand cDNAs were prepared with a SuperscriptII kit (GIBCO/BRL). Amplified cDNA fragments were subcloned into the pGEM-3Zf(+) vector (Promega). After linearization of plasmid DNA with HindIII, antisense RNA was synthesized with [α-32P]CTP (20 mCi/ml) by using bacteriophage T7 RNA polymerase (Ambion, Austin, TX). Specific activities of the cRNAs were measured in each experiment and were in the range of 1.7–2.6 × 109 cpm/μg except for β-actin, which was 5.3–8.1 × 108 cpm/μg as a result of dilution of the [α-32P]CTP.
Aliquots of total RNA (10–15 μg) were assayed by RNase protection by using a HybSpeed RPA kit (Ambion) as described (9). Each assay tube contained a cRNA probe for the mRNA to be tested plus cRNA complementary to the β-actin mRNA. In preparing the probes, we adjusted the specific activity of the [α-32P]CTP to give a β-actin signal comparable to the test mRNAs. After digestion with RNase, protected fragments were separated on 8 M urea/4.8% polyacrylamide gels, which were dried and subjected to autoradiography by using reflection film and intensifying screens (DuPont). The dried gels also were analyzed quantitatively with a Bio-Imaging analyzer by using BAS 1000 MacBAS software (Fuji Medical System, Standish, ME). The level of β-actin mRNA in each RNA sample was used to normalize signals obtained for the test mRNAs.
RESULTS
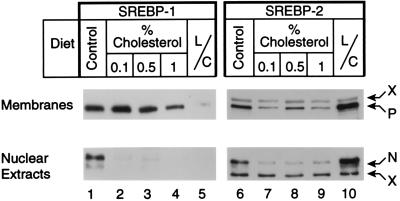
Fig. 1 shows immunoblots of the precursor and mature forms of SREBP-1 and SREBP-2 in livers of male hamsters fed varying amounts of cholesterol for 10 days. The amounts of the mature forms of SREBP-1 and SREBP-2 in nuclear extracts declined markedly at the lowest cholesterol concentration (0.1%), and there was no further change at cholesterol concentrations as high as 1%. The amounts of the full-length precursor forms of SREBP-1 in cell membranes were unchanged at 0.1% cholesterol and declined by about 50% at 1% cholesterol. The SREBP-2 precursor was visualized as the lower band of a doublet. This declined somewhat at 0.1% cholesterol and remained at about the same level at 1% cholesterol. For comparative purposes, we fed a companion group of hamsters a diet containing a mixture of lovastatin and Colestipol, which is designed to increase the hepatic demand for cholesterol. As shown previously (10), this treatment caused the expected increase in the nuclear form of SREBP-2, but it produced a paradoxical decline in the nuclear form of SREBP-1. The changes in the precursor forms paralleled the changes in the nuclear forms of SREBP-1 and SREBP-2 in the animals fed lovastatin and Colestipol.
Figure 1.
Levels of the precursor and nuclear forms of SREBP-1 and SREBP-2 in membranes and nuclear extracts from livers of male hamsters fed the indicated diet for 10 days. Each lane contained a pooled sample from livers of four hamsters that were fed normal chow (lanes 1 and 6), 0.1% cholesterol (lanes 2 and 7), 0.5% cholesterol (lanes 3 and 8), 1% cholesterol (lanes 4 and 9), or lovastatin/Colestipol (lanes 5 and 10) (same animals as in Fig. 5). Aliquots (30 μg protein) of membranes (Upper) and nuclear extracts (Lower) were subjected to SDS/PAGE. Immunoblot analysis was performed with either 5 μg/ml of mouse mAb (IgG-2A4) against amino acids 301–407 of human SREBP-1 (lanes 1–5) or 1:4,000 dilution of rabbit IgG against amino acids 32–250 of hamster SREBP-2 (lanes 6–10). Bound antibodies were visualized with the ECL system. Blots were exposed to film for 60 sec (lanes 1–5) or 120 sec (lanes 6–10). P, precursor form of SREBP; N, nuclear form of cleaved SREBP. X denotes crossreactive proteins whose levels did not change with these dietary manipulations. L/C, lovastatin/Colestipol.
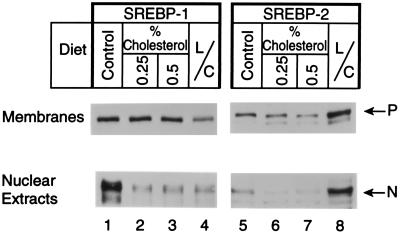
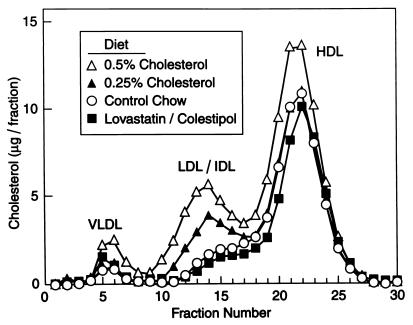
Fig. 2 shows the results of a similar cholesterol-feeding experiment performed with female hamsters. The results were similar to those observed with the males. In this experiment we measured the levels of cholesterol in plasma and liver (Table 1). As expected, the high-cholesterol diet increased the cholesterol content of liver and plasma. Lovastatin/Colestipol treatment decreased the cholesterol content of plasma by a small amount, but there was no detectable decrease in that of the liver. Separation of plasma lipoproteins by fast performance liquid chromatography revealed that the 0.25% cholesterol diet selectively increased cholesterol in the LDL/intermediate density lipoprotein size range, whereas 0.5% cholesterol increased the cholesterol content of very low density lipoprotein and high density lipoprotein as well (Fig. 3). We observed no significant difference in the fast performance liquid chromatography profiles of control animals and animals fed the lovastatin/Colestipol mixture.
Figure 2.
Levels of the precursor and nuclear forms of SREBP-1 and SREBP-2 in membranes and nuclear extracts from livers of female hamsters fed the indicated diet for 12 days (same animals as in Table 1 and Figs. 3 and 6). Each lane contained pooled samples from livers of five hamsters that were fed normal chow (lanes 1 and 5), 0.25% cholesterol (lanes 2 and 6), 0.5% cholesterol (lanes 3 and 7), and lovastatin/Colestipol (lanes 4 and 8). Aliquots (30 μg protein) of membranes (Upper) and nuclear extracts (Lower) were subjected to SDS/PAGE. Immunoblot analysis was performed with either 5 μg/ml of mouse mAb (IgG-2A4) against amino acids 301–407 of human SREBP-1a (lanes 1–4) or 1:4,000 dilution of rabbit IgG against amino acids 32–250 of hamster SREBP-2 (lanes 5–8). Bound antibodies were visualized with the ECL system. Blots were exposed to film for 60 sec (lanes 1–4) or 120 sec (lanes 5–8). P, precursor form of SREBP; N, nuclear form of cleaved SREBP. L/C, lovastatin/Colestipol.
Table 1.
Cholesterol content of plasma and liver in hamsters on different diets
Figure 3.
Fast performance liquid chromatography profiles of plasma lipoproteins from female Syrian hamsters fed the indicated diet for 12 days. The animals used in the study are the same as in Table 1 and Figs. 2 and 6. Pooled plasma from each group of five animals was subjected to gel filtration on fast performance liquid chromatography, and the cholesterol content of each fraction was measured as described in Methods. VLDL, very low density lipoprotein; HDL, high density lipoprotein; IDL, intermediate density lipoprotein.
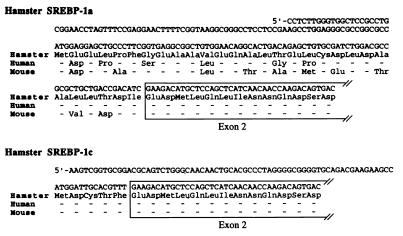
In humans (29) and mice (9), SREBP-1 can contain either of two first exons, giving rise to versions called SREBP-1a and SREBP-1c. To determine whether these two transcripts exist in hamster liver, we cloned the 5′ ends of the SREBP-1 cDNA by using the 5′ rapid amplification of cDNA ends method (see Methods). These studies yielded two populations of cDNAs. In terms of amino acid sequence, one of these clones was 79% and 59% identical to human and mouse SREBP-1a, respectively, and the other clone was 100% identical to both human and mouse SREBP-1c (Fig. 4). The amino acid differences among human, mouse, and hamster SREBP-1a are highly conservative, and they preserve the overall negative charge of this region of the protein.
Figure 4.
Nucleotide sequences and deduced amino acid sequences of the acidic amino acid domain of Syrian hamster SREBP-1a and SREBP-1c, and comparison with the amino acid sequences of human and mouse. Exon 2 is boxed.
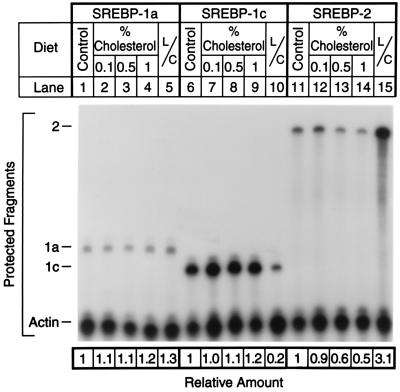
We used a quantitative RNase protection assay to determine whether diets containing cholesterol or lovastatin/Colestipol altered the amounts of mRNA for SREBP-1a, SREBP-1c, or SREBP-2 (Fig. 5). The livers correspond to those of the male hamsters shown in Fig. 1. As reported previously for mouse and human liver (9), the livers of hamsters had much more SREBP-1c mRNA than SREBP-1a mRNA. Cholesterol feeding had no significant effect on the amounts of mRNA encoding either isoform of SREBP-1, but lovastatin/Colestipol specifically decreased the amount of mRNA encoding SREBP-1c. The mRNA encoding SREBP-2 was suppressed by cholesterol feeding, and it was increased markedly by lovastatin/Colestipol.
Figure 5.
Amounts of mRNA for SREBP-1a, SREBP-1c, and SREBP-2 as measured by RNase protection in livers of male hamsters fed the indicated diet for 10 days. Total RNA was isolated from the livers of the same hamsters shown in Fig. 1. Aliquots (15 μg) of total RNA from pooled samples of four livers from the indicated source were hybridized in solution for 10 min at 68°C to 32P-labeled cRNA probes for SREBP-1a, SREBP-1c, or SREBP-2, all in the presence of a cRNA probe for β-actin as described in Methods. After RNase digestion, the protected fragments were separated by gel electrophoresis and exposed to film with an intensifying screen for 24 hr at −80°C. The radioactivity in the gels was quantified by PhosphorImaging, normalized to the β-actin signal, and expressed relative to the mRNA level in hamsters fed the chow diet, which was arbitrarily set at 1 for each transcript. L/C, lovastatin/Colestipol.
We also used the RNase protection assay to quantify the amounts of mRNA produced by genes that are targets of SREBPs (Fig. 6). The mRNA sample was obtained from the female animals whose protein results are shown in Fig. 2. Cholesterol feeding decreased the LDL receptor mRNA by only about 30% even at the highest dose, whereas it decreased the amount of HMG CoA synthase mRNA by 90%, even at the lowest dose. The mRNA for HMG CoA reductase also was reduced substantially (60–70% reduction). The lovastatin/Colestipol diet increased all three mRNAs by 2.8- to 6-fold.
Figure 6.
Amounts of mRNA for the LDL receptor, HMG CoA synthase, and HMG CoA reductase in livers of female Syrian hamsters fed the indicated diet for 12 days. Total RNA was isolated from livers of the same hamsters shown in Table 1 and Figs. 2 and 3. Aliquots of total RNA (10 μg) from pooled samples of five livers were hybridized in solution for 10 min at 68°C to 32P-labeled cRNA probes for the LDL receptor, HMG CoA synthase, and HMG CoA reductase, all in the presence of a cRNA probe for β-actin as described in Methods. After RNase digestion, the protected fragments were separated by gel electrophoresis and exposed to film at −80°C with an intensifying screen for 16 hr for LDL receptor and for 8 hr for HMG CoA synthase and reductase. The radioactivity in the gels was quantified by PhosphorImaging, normalized to the β-actin signal, and expressed relative to the mRNA level of control chow-fed hamsters. L/C, lovastatin/Colestipol; LDLR, LDL receptor.
DISCUSSION
The major conclusion of the current studies is that high- cholesterol diets reduce the amounts of SREBP-1c and SREBP-2 in hamster liver nuclei. This reduction likely explains the concomitant reduction in the mRNAs encoding enzymes of cholesterol biosynthesis and the LDL receptor. The reduction in mRNAs, together with posttranscriptional events that reduce HMG CoA reductase activity (4, 30), lead to a profound fall in cholesterol synthesis when hamsters consume cholesterol (31).
The high-cholesterol diets reduced the nuclear forms of the two SREBPs by somewhat different mechanisms. The mechanism for down-regulation of SREBP-1c appeared to be relatively simple. Dietary cholesterol produced a clear reduction in the mature nuclear form of SREBP-1c without a significant change in the amount of precursor (Figs. 1 and 2) or the amount of SREBP-1c mRNA (Fig. 5). The most likely mechanism for this change is a reduction in the rate of proteolytic processing of the precursor to the mature form, as occurs in cultured cells that are treated with sterols (7).
The regulation of SREBP-2 appeared to be more complex. Low levels of dietary cholesterol elicited a profound reduction in the amount of nuclear SREBP-2, but there was also a drop in the amount of precursor and a slight fall in the amount of SREBP-2 mRNA. At high levels of cholesterol the amount of SREBP-2 mRNA declined by about 50% (Fig. 5). These findings suggest that cholesterol may control the amount of nuclear SREBP-2 by two mechanisms: (i) regulation of proteolytic processing; and (ii) regulation of the level of SREBP-2 mRNA and hence the rate of precursor production. Evidence for this dual control also is supplied by the cholesterol depletion experiments. Lovastatin/Colestipol increased the mRNA for SREBP-2 (Fig. 5), and this was associated with an increase in both the precursor form and the mature nuclear form of the protein (Figs. 1 and 2).
The current studies also establish that hamster liver, like previously studied livers of mice and humans (9), produces SREBP-1c as the predominant form of SREBP-1. The conservation of the SREBP-1a/1c dichotomy in three species attests to the physiologic importance of the two forms of the proteins. The sequence comparisons show that the 1c-specific amino acids are completely conserved in the three species, and the overall acidic nature of the 1a region also is preserved (Fig. 4). One can infer, therefore, that in hamsters, as in humans and mice (8), the SREBP-1c transcript is a weaker transcriptional activator than is the SREBP-1a transcript. Previous studies have shown that SREBP-1a is produced predominantly in cultured cells and in testis, spleen, jejunum, and ileum, whereas SREBP-1c predominates in most organs, including the liver, adrenal, and both white and brown adipose tissue (9).
Studies with transgenic mice have shown that overexpression of nuclear SREBP-1a stimulates fatty acid synthesis as well as cholesterol synthesis in liver (19). This is likely attributable to the ability of SREBP-1a to bind to elements in the promoters of the fatty acid synthase and acetyl CoA carboxylase genes (7, 32–34). Animals overexpressing the nuclear form of SREBP-1c had a moderate increase in fatty acid synthesis and no demonstrable increase in cholesterol synthesis, suggesting that SREBP-1c is a relatively specific activator of the fatty acid biosynthetic pathway (8).
In the experiment with male hamsters, treatment with lovastatin/Colestipol led to a selective down-regulation of the amount of SREBP-1c mRNA without affecting the mRNA for SREBP-1a (Fig. 5). This was associated with a profound fall in the amount of both the precursor and mature nuclear forms of SREBP-1 (Fig. 1). In the experiment with females the changes in SREBP-1 protein were in a similar direction, but not as profound as those in males (Fig. 2). These data indicate that the promoter that gives rise to the SREBP-1c transcript responds to a regulatory signal that is not detected by the promoter that gives rise to the 1a transcript. The nature of this signal and the mechanism of its suppression by lovastatin and Colestipol are presently unknown.
An important aspect of these studies was the finding that cholesterol feeding suppressed the LDL receptor mRNA by only 30% at a time when the HMG CoA synthase and reductase mRNAs were suppressed by 90% and 70%, respectively (Fig. 6). The incomplete suppression of LDL receptor mRNA presumably explains the failure of the plasma cholesterol to rise much above 300 mg/dl in the cholesterol-fed hamsters (Table 1). When the LDL receptor is reduced profoundly, as in mice homozygous for an LDL receptor gene knockout, high-cholesterol diets raise the plasma cholesterol level to over 2,000 mg/dl (16).
The current findings on LDL receptor mRNA are consistent with those of Spady et al. (11), who showed that cholesterol suppresses LDL receptor activity in animal livers only after it suppresses cholesterol synthesis markedly. This finding is in contrast to the finding in cultured cells, where LDL receptors and cholesterol biosynthetic enzymes decline in parallel (4, 35). It is possible that transcription of the LDL receptor gene in liver is maintained by the low levels of nuclear SREBPs, especially SREBP-2, that persist after cholesterol feeding. Alternatively, the transcription of the LDL receptor gene in liver may be driven by another factor, in addition to SREBPs, whose activity persists after cholesterol feeding. We note that the LDL receptor mRNA increased by 2.8-fold in response to the lovastatin/Colestipol treatment (Fig. 6), a finding that is consistent with the sensitivity of this gene to increased levels of nuclear SREBP-2.
Although the current paper reports only two independent studies, one in males and one in females, we have repeated the essential features of these experiments on six other occasions, and the major conclusions were similar to those shown here. Considered together with previous data, the experiments support the notion that SREBPs are important control elements for the enzymes of cholesterol biosynthesis and LDL receptors in livers of experimental animals.
Acknowledgments
We thank Richard Gibson for invaluable help with animals; Scott Clark and Robin Craddock for excellent technical assistance; and Jeff Cormier and Michelle Laremore for DNA sequencing and oligonucleotide synthesis. This work was supported by research funds from the National Institutes of Health (HL20948), the Moss Heart Foundation, and the Perot Family Foundation. I.S. is the recipient of a research fellowship from the Manpei Suzuki Diabetes Foundation of Tokyo, Japan. J.D.H. is the recipient of a Postdoctoral Fellowship for Physicians from the Howard Hughes Medical Institute.
ABBREVIATIONS
- HMG CoA
3-hydroxy-3-methylglutaryl CoA
- LDL
low density lipoprotein
- SREBP
sterol regulatory element binding protein
References
- 1.Gould R G. Am J Med. 1951;11:209–227. doi: 10.1016/0002-9343(51)90107-6. [DOI] [PubMed] [Google Scholar]
- 2.Gould R G, Taylor C B, Hagerman J S, Warner I, Campbell D J. J Biol Chem. 1953;201:519–523. [PubMed] [Google Scholar]
- 3.Turley S D, Daggy B P, Dietschy J M. J Cardiovasc Pharmacol. 1996;27:71–79. doi: 10.1097/00005344-199601000-00012. [DOI] [PubMed] [Google Scholar]
- 4.Goldstein J L, Brown M S. Nature (London) 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 5.Spady D K, Turley S D, Dietschy J M. J Lipid Res. 1985;26:465–472. [PubMed] [Google Scholar]
- 6.Brown M S, Goldstein J L. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 7.Brown M S, Goldstein J L. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 8.Shimano H, Horton J D, Shimomura I, Hammer R E, Brown M S, Goldstein J L. J Clin Invest. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimomura I, Shimano H, Horton J D, Goldstein J L, Brown M S. J Clin Invest. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheng Z, Otani H, Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1995;92:935–938. doi: 10.1073/pnas.92.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spady D K, Stange E F, Bilhartz L E, Dietschy J M. Proc Natl Acad Sci USA. 1986;83:1916–1920. doi: 10.1073/pnas.83.6.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horton J D, Cuthbert J A, Spady D K. J Clin Invest. 1993;92:743–749. doi: 10.1172/JCI116645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 14.Chomczynski P, Sacchi N. Analy Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 15.Yokode M, Hammer R E, Ishibashi S, Brown M S, Goldstein J L. Science. 1990;250:1273–1275. doi: 10.1126/science.2244210. [DOI] [PubMed] [Google Scholar]
- 16.Ishibashi S, Goldstein J L, Brown M S, Herz J, Burns D K. J Clin Invest. 1994;93:1885–1893. doi: 10.1172/JCI117179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato R, Yang J, Wang X, Evans M J, Ho Y K, Goldstein J L, Brown M S. J Biol Chem. 1994;269:17267–17273. [PubMed] [Google Scholar]
- 18.Yang J, Sato R, Goldstein J L, Brown M S. Genes Dev. 1994;8:1910–1919. doi: 10.1101/gad.8.16.1910. [DOI] [PubMed] [Google Scholar]
- 19.Shimano H, Horton J D, Hammer R E, Shimomura I, Brown M S, Goldstein J L. J Clin Invest. 1996;98:1575–1584. doi: 10.1172/JCI118951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frohman M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee L Y, Mohler W A, Schafer B L, Freudenberger J S, Byrne-Connolly N, Eager K B, Mosley S T, Leighton J K, Thrift R N, Davis R A, Tanaka R D. Nucleic Acids Res. 1989;17:1259–1260. doi: 10.1093/nar/17.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bishop R W. J Lipid Res. 1992;33:549–557. [PubMed] [Google Scholar]
- 23.Rokosz L L, Boulton D A, Butkiewicz E A, Sanyal G, Cueto M A, Lachance P A, Hermes J D. Arch Biochem Biophys. 1994;312:1–13. doi: 10.1006/abbi.1994.1273. [DOI] [PubMed] [Google Scholar]
- 24.Gil G, Goldstein J L, Slaughter C A, Brown M S. J Biol Chem. 1986;261:3710–3716. [PubMed] [Google Scholar]
- 25.Chin D J, Gil G, Russell D W, Liscum L, Luskey K L, Basu S K, Okayama H, Berg P, Goldstein J L, Brown M S. Nature (London) 1984;308:613–617. doi: 10.1038/308613a0. [DOI] [PubMed] [Google Scholar]
- 26.Luskey K L, Stevens B. J Biol Chem. 1985;260:10271–10277. [PubMed] [Google Scholar]
- 27.Ponte P, Ng S-Y, Engel J, Gunning P, Kedes L. Nucleic Acids Res. 1984;12:1687–1696. doi: 10.1093/nar/12.3.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tokunaga K, Taniguchi H, Yoda K, Shimizu M, Sakiyama S. Nucleic Acids Res. 1986;14:2829. doi: 10.1093/nar/14.6.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hua X, Wu J, Goldstein J L, Brown M S, Hobbs H H. Genomics. 1995;25:667–673. doi: 10.1016/0888-7543(95)80009-b. [DOI] [PubMed] [Google Scholar]
- 30.Brown M S, Goldstein J L. J Lipid Res. 1980;2l:505–517. [PubMed] [Google Scholar]
- 31.Spady D K, Dietschy J M. J Lipid Res. 1983;24:303–315. [PubMed] [Google Scholar]
- 32.Kim J B, Spiegelman B M. Genes Dev. 1996;10:1096–1107. doi: 10.1101/gad.10.9.1096. [DOI] [PubMed] [Google Scholar]
- 33.Magana M M, Osborne T F. J Biol Chem. 1996;271:32689–32694. doi: 10.1074/jbc.271.51.32689. [DOI] [PubMed] [Google Scholar]
- 34.Lopez J M, Bennett M K, Sanchez H B, Rosenfeld J M, Osborne T F. Proc Natl Acad Sci USA. 1996;93:1049–1053. doi: 10.1073/pnas.93.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldstein J L, Sobhani M K, Faust J R, Brown M S. Cell. 1976;9:195–203. doi: 10.1016/0092-8674(76)90110-0. [DOI] [PubMed] [Google Scholar]