Abstract
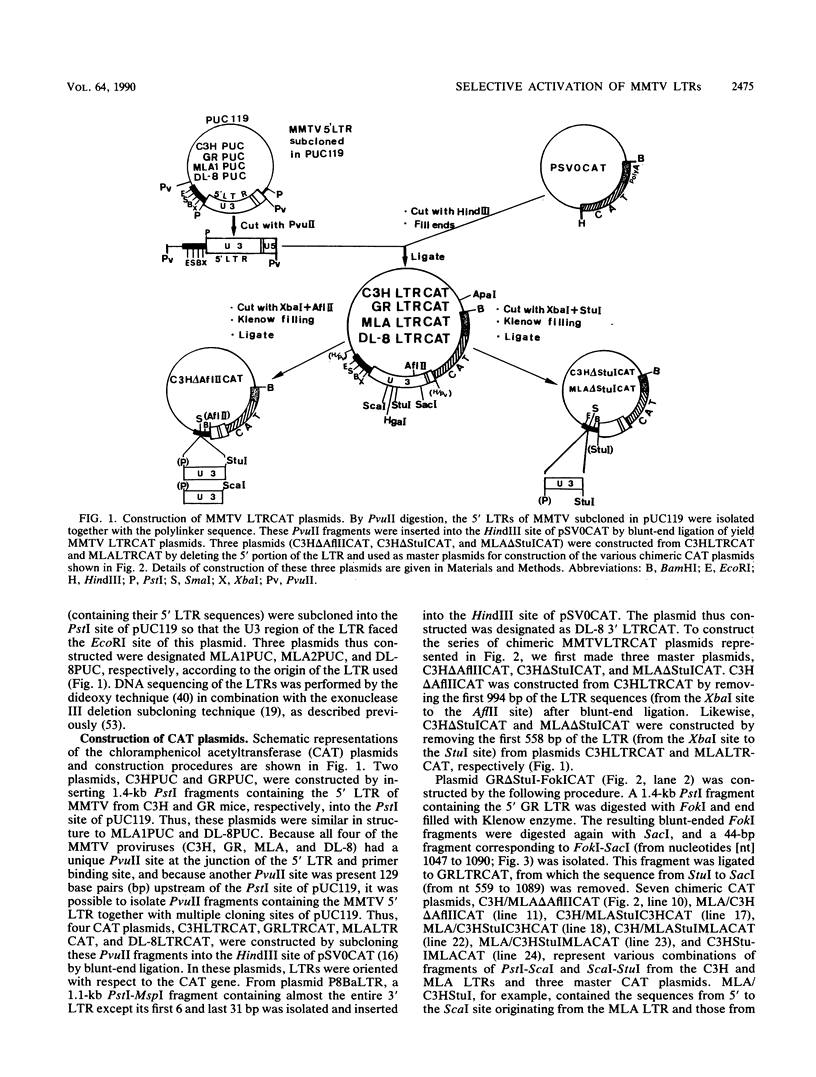
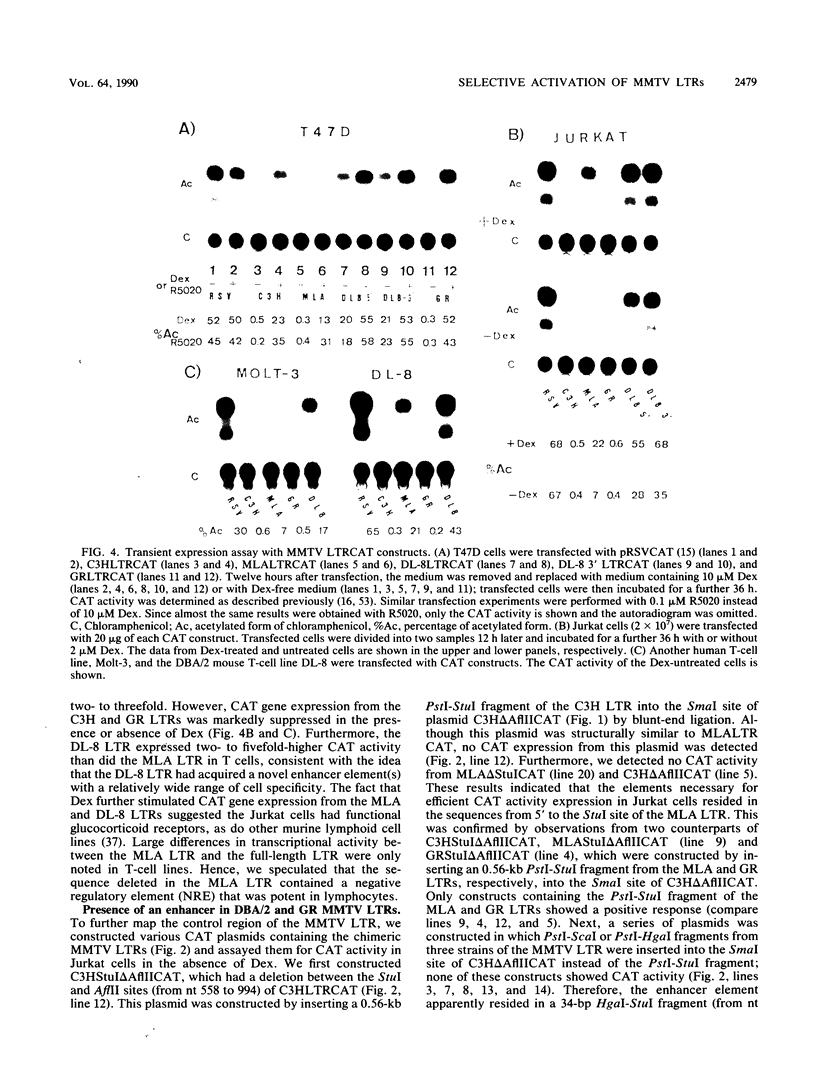
We determined the nucleotide sequences of the long terminal repeats (LTRs) from mouse mammary tumor virus (MMTV) proviruses acquired in two DBA/2 mouse lymphoma cell lines, MLA and DL-8. Proviruses from MLA contained a 352-base-pair deletion from nucleotides 669 to 1020 in the U3 region of the LTR, whereas the LTR alteration of the DL-8 provirus involved both a similar 360-base-pair deletion and generation of a tandem repeat region consisting of sequences of flanking deletions. To assess the function of the rearranged LTRs, we constructed plasmids in which normal and rearranged LTRs drove the reporter chloramphenicol acetyltransferase gene and transfected them into T-cell lines (Jurkat, Molt-3, and DL-8) and the mammary tumor cell line T47D. Both rearranged LTRs were transcriptionally active, but normal LTRs were not active in either the presence or absence of glucocorticoids in all T-cell lines. In T47D cells, however, the MLA provirus LTR showed the same glucocorticoid- or progestin-dependent transcriptional activity as did normal LTRs. The DL-8 provirus LTR acquired a novel enhancer(s) by rearrangement and thus had a high basal transcriptional activity in T47D cells. The results of chloramphenicol acetyltransferase assays using plasmids with various chimeric MMTV LTRs revealed that the rearranged LTRs had lost their negative regulatory element and contained an enhancer element that was highly homologous to the enhancer A element of polyomavirus (from nucleotides 525 to 558). GR but not C3H mouse MMTV contained this enhancer. These results elucidate some of the molecular mechanisms involved in the selection of mutant MMTVs with rearranged LTRs in lymphoma cells.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ball J. K., Arthur L. O., Dekaban G. A. The involvement of a type-B retrovirus in the induction of thymic lymphomas. Virology. 1985 Jan 15;140(1):159–172. doi: 10.1016/0042-6822(85)90455-6. [DOI] [PubMed] [Google Scholar]
- Ball J. K., Dekaban G. A. Characterization of early molecular biological events associated with thymic lymphoma induction following infection with a thymotropic type-B retrovirus. Virology. 1987 Dec;161(2):357–365. doi: 10.1016/0042-6822(87)90128-0. [DOI] [PubMed] [Google Scholar]
- Ball J. K., Diggelmann H., Dekaban G. A., Grossi G. F., Semmler R., Waight P. A., Fletcher R. F. Alterations in the U3 region of the long terminal repeat of an infectious thymotropic type B retrovirus. J Virol. 1988 Aug;62(8):2985–2993. doi: 10.1128/jvi.62.8.2985-2993.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Cato A. C., Miksicek R., Schütz G., Arnemann J., Beato M. The hormone regulatory element of mouse mammary tumour virus mediates progesterone induction. EMBO J. 1986 Sep;5(9):2237–2240. doi: 10.1002/j.1460-2075.1986.tb04490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celander D., Haseltine W. A. Tissue-specific transcription preference as a determinant of cell tropism and leukaemogenic potential of murine retroviruses. Nature. 1984 Nov 8;312(5990):159–162. doi: 10.1038/312159a0. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- DesGroseillers L., Jolicoeur P. The tandem direct repeats within the long terminal repeat of murine leukemia viruses are the primary determinant of their leukemogenic potential. J Virol. 1984 Dec;52(3):945–952. doi: 10.1128/jvi.52.3.945-952.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson C., Smith R., Brookes S., Peters G. Tumorigenesis by mouse mammary tumor virus: proviral activation of a cellular gene in the common integration region int-2. Cell. 1984 Jun;37(2):529–536. doi: 10.1016/0092-8674(84)90383-0. [DOI] [PubMed] [Google Scholar]
- Dickson C., Smith R., Peters G. In vitro synthesis of polypeptides encoded by the long terminal repeat region of mouse mammary tumour virus DNA. Nature. 1981 Jun 11;291(5815):511–513. doi: 10.1038/291511a0. [DOI] [PubMed] [Google Scholar]
- Dudley J. P., Arfsten A., Hsu C. L., Kozak C., Risser R. Molecular cloning and characterization of mouse mammary tumor proviruses from a T-cell lymphoma. J Virol. 1986 Jan;57(1):385–388. doi: 10.1128/jvi.57.1.385-388.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley J., Risser R. Amplification and novel locations of endogenous mouse mammary tumor virus genomes in mouse T-cell lymphomas. J Virol. 1984 Jan;49(1):92–101. doi: 10.1128/jvi.49.1.92-101.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T., Shibuya H., Ohashi T., Yamanishi K., Taniguchi T. Regulation of human interleukin-2 gene: functional DNA sequences in the 5' flanking region for the gene expression in activated T lymphocytes. Cell. 1986 Aug 1;46(3):401–405. doi: 10.1016/0092-8674(86)90660-4. [DOI] [PubMed] [Google Scholar]
- Garcia M., Wellinger R., Vessaz A., Diggelmann H. A new site of integration for mouse mammary tumor virus proviral DNA common to BALB/cf(C3H) mammary and kidney adenocarcinomas. EMBO J. 1986 Jan;5(1):127–134. doi: 10.1002/j.1460-2075.1986.tb04186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C., Padmanabhan R., Howard B. H. High efficiency DNA-mediated transformation of primate cells. Science. 1983 Aug 5;221(4610):551–553. doi: 10.1126/science.6306768. [DOI] [PubMed] [Google Scholar]
- Gowland P. L., Buetti E. Mutations in the hormone regulatory element of mouse mammary tumor virus differentially affect the response to progestins, androgens, and glucocorticoids. Mol Cell Biol. 1989 Sep;9(9):3999–4008. doi: 10.1128/mcb.9.9.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearing P., Shenk T. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell. 1983 Jul;33(3):695–703. doi: 10.1016/0092-8674(83)90012-0. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hsu C. L., Fabritius C., Dudley J. Mouse mammary tumor virus proviruses in T-cell lymphomas lack a negative regulatory element in the long terminal repeat. J Virol. 1988 Dec;62(12):4644–4652. doi: 10.1128/jvi.62.12.4644-4652.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryszke M. H., Piette J., Yaniv M. Induction of a factor that binds to the polyoma virus A enhancer on differentiation of embryonal carcinoma cells. Nature. 1987 Jul 16;328(6127):254–256. doi: 10.1038/328254a0. [DOI] [PubMed] [Google Scholar]
- Kwon B. S., Weissman S. M. Mouse mammary tumor virus-related sequences in mouse lymphocytes are inducible by 12-O-tetradecanoyl phorbol-13-acetate. J Virol. 1984 Dec;52(3):1000–1004. doi: 10.1128/jvi.52.3.1000-1004.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. T., Prakash O., Klein D., Sarkar N. H. Structural alterations in the long terminal repeat of an acquired mouse mammary tumor virus provirus in a T-cell leukemia of DBA/2 mice. Virology. 1987 Jul;159(1):39–48. doi: 10.1016/0042-6822(87)90345-x. [DOI] [PubMed] [Google Scholar]
- Lee W., Mitchell P., Tjian R. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell. 1987 Jun 19;49(6):741–752. doi: 10.1016/0092-8674(87)90612-x. [DOI] [PubMed] [Google Scholar]
- Lenz J., Celander D., Crowther R. L., Patarca R., Perkins D. W., Haseltine W. A. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. 1984 Mar 29-Apr 4Nature. 308(5958):467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- Lopata M. A., Cleveland D. W., Sollner-Webb B. High level transient expression of a chloramphenicol acetyl transferase gene by DEAE-dextran mediated DNA transfection coupled with a dimethyl sulfoxide or glycerol shock treatment. Nucleic Acids Res. 1984 Jul 25;12(14):5707–5717. doi: 10.1093/nar/12.14.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequencing of an apparent proviral copy of env mRNA defines determinants of expression of the mouse mammary tumor virus env gene. J Virol. 1983 Sep;47(3):495–504. doi: 10.1128/jvi.47.3.495-504.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalides R., Wagenaar E., Hilkens J., Hilgers J., Groner B., Hynes N. E. Acquisition of proviral DNA of mouse mammary tumor virus in thymic leukemia cells from GR mice. J Virol. 1982 Sep;43(3):819–829. doi: 10.1128/jvi.43.3.819-829.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalides R., Wagenaar E. Site-specific rearrangements in the long terminal repeat of extra mouse mammary tumor proviruses in murine T-cell leukemias. Virology. 1986 Oct 15;154(1):76–84. doi: 10.1016/0042-6822(86)90431-9. [DOI] [PubMed] [Google Scholar]
- Michalides R., Wagenaar E., Weijers P. Rearrangements in the long terminal repeat of extra mouse mammary tumor proviruses in T-cell leukemias of mouse strain GR result in a novel enhancer-like structure. Mol Cell Biol. 1985 Apr;5(4):823–830. doi: 10.1128/mcb.5.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley K. L., Toohey M. G., Peterson D. O. Transcriptional repression of a hormone-responsive promoter. Nucleic Acids Res. 1987 Sep 11;15(17):6973–6989. doi: 10.1093/nar/15.17.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R., van Ooyen A., Cox D., Fung Y. K., Varmus H. Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature. 1984 Jan 12;307(5947):131–136. doi: 10.1038/307131a0. [DOI] [PubMed] [Google Scholar]
- Nusse R., van der Ploeg L., van Duijn L., Michalides R., Hilgers J. Impaired maturation of mouse mammary tumor virus precursor polypeptides in lymphoid leukemia cells, producing intracytoplasmic A particles and no extracellular B-type virions. J Virol. 1979 Oct;32(1):251–258. doi: 10.1128/jvi.32.1.251-258.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piette J., Yaniv M. Two different factors bind to the alpha-domain of the polyoma virus enhancer, one of which also interacts with the SV40 and c-fos enhancers. EMBO J. 1987 May;6(5):1331–1337. doi: 10.1002/j.1460-2075.1987.tb02372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabindran S. K., Danielsen M., Stallcup M. R. Glucocorticoid-resistant lymphoma cell variants that contain functional glucocorticoid receptors. Mol Cell Biol. 1987 Dec;7(12):4211–4217. doi: 10.1128/mcb.7.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racevskis J., Beyer H. Amplification of mouse mammary tumor virus genomes in non-mammary tumor cells. J Virol. 1989 Jan;63(1):456–459. doi: 10.1128/jvi.63.1.456-459.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racevskis J., Sarkar N. H. ML antigen of DBA/2 mouse leukemias: expression of an endogenous murine mammary tumor virus. J Virol. 1982 Jun;42(3):804–813. doi: 10.1128/jvi.42.3.804-813.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart T. A., Hollingshead P. G., Pitts S. L. Multiple regulatory domains in the mouse mammary tumor virus long terminal repeat revealed by analysis of fusion genes in transgenic mice. Mol Cell Biol. 1988 Jan;8(1):473–479. doi: 10.1128/mcb.8.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Miyama-Inaba M., Masuda T., Uchino H., Murakami A., Tanaka H. Suppressive B-cell factor (SBF) produced by FcR-bearing B cells; suppression of B, but not non-B-cell proliferation. Immunology. 1983 Sep;50(1):149–157. [PMC free article] [PubMed] [Google Scholar]
- Theunissen H. J., Paardekooper M., Maduro L. J., Michalides R. J., Nusse R. Phorbol ester-inducible T-cell-specific expression of variant mouse mammary tumor virus long terminal repeats. J Virol. 1989 Aug;63(8):3466–3471. doi: 10.1128/jvi.63.8.3466-3471.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze J., Tooze S., Haisma H., Hilgers J. AtT20 pituitary tumour cells contain mouse mammary tumour virus and intracisternal A-type particles in addition to murine leukemia virus. Eur J Cell Biol. 1985 Nov;39(1):224–231. [PubMed] [Google Scholar]
- Tsubura A., Inaba M., Imai S., Murakami A., Oyaizu N., Yasumizu R., Ohnishi Y., Tanaka H., Morii S., Ikehara S. Intervention of T-cells in transportation of mouse mammary tumor virus (milk factor) to mammary gland cells in vivo. Cancer Res. 1988 Nov 15;48(22):6555–6559. [PubMed] [Google Scholar]
- Vaidya A. B., Long C. A., Sheffield J. B., Tamura A., Tanaka H. Murine mammary tumor virus deficient in the major glycoprotein: biochemical and biological studies on virions produced by a lymphoma cell line. Virology. 1980 Jul 30;104(2):279–293. doi: 10.1016/0042-6822(80)90333-5. [DOI] [PubMed] [Google Scholar]
- Veldman G. M., Lupton S., Kamen R. Polyomavirus enhancer contains multiple redundant sequence elements that activate both DNA replication and gene expression. Mol Cell Biol. 1985 Apr;5(4):649–658. doi: 10.1128/mcb.5.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk C., Imler J. L., Perez-Mutul J., Wasylyk B. The c-Ha-ras oncogene and a tumor promoter activate the polyoma virus enhancer. Cell. 1987 Feb 13;48(3):525–534. doi: 10.1016/0092-8674(87)90203-0. [DOI] [PubMed] [Google Scholar]
- Wellinger R. J., Garcia M., Vessaz A., Diggelmann H. Exogenous mouse mammary tumor virus proviral DNA isolated from a kidney adenocarcinoma cell line contains alterations in the U3 region of the long terminal repeat. J Virol. 1986 Oct;60(1):1–11. doi: 10.1128/jvi.60.1.1-11.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Satake M., Ito Y. Two overlapping sequence motifs within the polyomavirus enhancer are independently the targets of stimulation by both the tumor promoter 12-O-tetradecanoylphorbol-13-acetate and the Ha-ras oncogene. J Virol. 1989 Mar;63(3):1040–1048. doi: 10.1128/jvi.63.3.1040-1048.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa S., Murakami A., Hoshino M., Tanaka H. Structural and functional analysis of long terminal repeats of Suncus murinus mammary tumor virus. J Virol. 1988 Apr;62(4):1235–1242. doi: 10.1128/jvi.62.4.1235-1242.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak-Nejmark T., Steuden J., Radzikowski C. Mammary leukaemia (ML) antigen isolated from L 1210 leukaemia cells. Int J Cancer. 1978 Apr 15;21(4):490–495. doi: 10.1002/ijc.2910210415. [DOI] [PubMed] [Google Scholar]