Abstract
Eukaryotic translation initiation factor 6 (eIF6) binds to the 60S ribosomal subunit and prevents its association with the 40S ribosomal subunit. In this paper, we devised a procedure for purifying eIF6 from rabbit reticulocyte lysates and immunochemically characterized the protein by using antibodies isolated from egg yolks of laying hens immunized with rabbit eIF6. By using these monospecific antibodies, a 1.096-kb human cDNA that encodes an eIF6 of 245 amino acids (calculated Mr 26,558) has been cloned and expressed in Escherichia coli. The purified recombinant human protein exhibits biochemical properties that are similar to eIF6 isolated from mammalian cell extracts. Database searches identified amino acid sequences from Saccharomyces cerevisiae, Drosophila, and the nematode Caenorhabditis elegans with significant identity to the deduced amino acid sequence of human eIF6, suggesting the presence of homologues of human eIF6 in these organisms.
It is now well established that the initiation of protein synthesis in eukaryotic cells begins with the recognition of the initiation AUG codon of mRNA by the small ribosomal subunit (40S) containing bound initiatior methionyl-tRNA (Met-tRNAf) to form the 40S initiation complex (40S⋅mRNA⋅Met-tRNAf). Subsequently, a 60S ribosomal subunit joins the 40S initiation complex to form the 80S initiation complex (80S⋅mRNA⋅Met-tRNAf) that is competent to undergo peptide bond synthesis during the elongation phase of protein synthesis (for a review, see refs. 1–5). After termination of translation, ribosomes are released from the polysomal complex as 80S particles that are in equilibrium with 40S and 60S ribosomal subunits (6, 7). Because a new round of initiation requires separated subunits, a mechanism must exist for maintaining a pool of both ribosomal subunits or for generating them from 80S ribosomes. Two protein factors, eukaryotic translation initiation factor 3 (eIF3) and eIF6, have been implicated in this process (1–5). It has been reported that eIF3, a multisubunit protein complex of Mr > 500,000, binds to the 40S ribosomal subunit and prevents its association with the 60S ribosomal subunit to form 80S ribosomes (8–11). In contrast to eIF3, eIF6, a monomeric protein of about 25 kDa, has been shown to bind to the 60S ribosomal subunit and prevents its association with the 40S ribosomal subunit (12–15). This ribosomal subunit anti-association property was used as an assay to identify and purify eIF6 from both wheat germ (12, 13) and mammalian cell extracts (14, 15). However, unlike eIF3 whose requirement for the binding of Met-tRNAf and mRNA to the 40S ribosomal subunit to form the 40S initiation complex is firmly established, the requirement of eIF6 in the initiation of protein synthesis both in vivo and in vitro has not yet been defined. This is in part because of the relatively low abundance of eIF6 in eukaryotic cells (14, 15) and the inability to obtain sufficient quantities of the pure protein in stable form to carry out structural and detailed functional studies.
To facilitate the characterization of eIF6, we have devised a rapid procedure for the isolation of eIF6 from rabbit reticulocyte lysates and prepared specific antibodies in chicken egg yolks directed against purified rabbit reticulocyte eIF6. These antibodies were used to characterize eIF6 in crude cell lysates as well as to isolate and characterize a human cDNA encoding active eIF6. The ORF of the eIF6-cDNA has been expressed in a functionally active form in Escherichia coli. The purified bacterially expressed recombinant protein has been shown to have properties similar to eIF6 isolated from mammalian cell extracts. Database searches suggest that homologues of human eIF6 are present in other organisms including the yeast Saccharomyces cerevisiae.
MATERIALS AND METHODS
Purification of eIF6 from Rabbit Reticulocyte Lysates.
The postribosomal supernatant of rabbit reticulocyte lysates (1,300-ml volume), prepared as described previously (16), was fractionated with solid ammonium sulfate. The proteins precipitating between 40 and 60% saturation (2,700 mg of protein) were dissolved in about 100 ml of buffer A [20 mM Tris⋅HCl, pH 7.5/1 mM DTT/1 mM EDTA/10% (vol/vol) glycerol] containing 150 mM KCl, dialyzed for about 14 hr against the same buffer, and then loaded onto a 175-ml DEAE-Sephacel column (35 × 2.5-cm bed volume) equilibrated in buffer A/150 mM KCl. After being washed with 1.1 liter of buffer A/150 mM KCl, bound proteins were eluted with a 1.1-liter linear gradient of KCl (0.15–0.5 M) in buffer A. Fractions containing eIF6 activity (eluting between 0.24 and 0.28 M KCl) were pooled, and the proteins were precipitated by adding solid ammonium sulfate to 70% saturation. The precipitated proteins were dissolved in about 4 ml of buffer A/100 mM KCl, dialyzed for 2 hr against the same buffer, and then concentrated to 2 ml by Centricon-10 filtration. The concentrated protein sample (12.6 mg of protein) was then applied to a 135-ml bed volume column of Sephadex G-75 superfine (1.4 × 89 cm) equilibrated in buffer A/100 mM KCl. The column was developed with the same buffer, and fractions of 2 ml were collected. Fractions 37–48 containing eIF6 activity (6.1 mg of protein) were pooled, diluted with an equal volume of buffer B (20 mM potassium phosphate, pH 7.0/1 mM DTT/0.1 M EDTA/0.5 mM phenylmethylsulfonyl fluoride) containing 1 M (NH4)2SO4, and then applied to a 25-ml bed volume column of phenyl-Sepharose (7 × 2 cm) equilibrated in buffer B/1 M(NH4)2SO4. After being washed with 75 ml of the same buffer, the bound proteins were eluted with a decreasing salt gradient (500-ml total volume) from buffer B/1 M (NH4)2SO4 to buffer B/50% ethylene glycol. Fractions of 10 ml were collected, and elution of eIF6 from the column was monitored by SDS/PAGE. Fractions 21–30 containing eIF6 were pooled, dialyzed against buffer A/100 mM KCl for about 7 hr, and then purified by gradient elution (buffer A/100 mM KCl to buffer A/500 mM KCl) from a fast protein liquid chromatography-Mono Q column (1-ml bed volume). eIF6 activity eluted at about 330 mM KCl. Active fractions (2-ml total volume) were pooled, dialyzed for about 6 hr against buffer A/100 mM KCl/55% (vol/vol) glycerol, and stored in small aliquots at −70°C. The total yield was about 220 μg of homogeneous eIF6 protein.
Immunological Methods and cDNA Cloning.
IgY antibodies, specific for eIF6, were isolated from egg yolks of a laying hen immunized with purified rabbit reticulocyte eIF6 by an adaptation of the procedure of Carroll and Stollar (17) as described previously for the preparation of anti-eIF5 antibodies (16). These antibodies were affinity-purified against purified rabbit reticulocyte eIF6 blotted onto aminophenylthioether paper (Schleicher & Schuell) following the procedure described by Ghosh et al. (18). Immunoscreening of a HeLa cell λZAPII cDNA library (Stratagene) was performed by using affinity-purified monospecific anti-eIF6 antibodies as probes as described by Snyder et al. (19). Screening with 32P-labeled DNA probes was carried out as described by Sambrook et al. (20).
Expression of eIF6 in E. coli and Purification of the Bacterially Expressed Recombinant Protein.
The ORF of eIF6 cDNA in pBluescript SK(+) was amplified by PCR with Pyrococcus DNA polymerase (Stratagene) and two primer sequences as follows: N terminus, 5′-dCGGGATCCCATATGGCGGTCCGAGCTTC-3′ having a BamHI/NdeI overhang; C terminus, a T7 primer. The PCR product was sequenced to ensure error-free DNA synthesis, digested with NdeI and EcoRI, and cloned into the same sites of pET-5a plasmid (Novagen) to yield the recombinant pET-5a-eIF6 expression plasmid. The recombinant plasmid was transformed into E. coli BL21 (DE3) cells, and the expression of the plasmid-encoded eIF6 protein was then induced by the addition of 1 mM isopropyl β-d-thiogalactopyranoside to an exponentially growing bacterial culture (3 liters). The cells were harvested by centrifugation at 3 hr postinduction. The frozen cells (15 g) were suspended in 50 ml of 20 mM Tris⋅HCl, pH 8.0/5 mM EDTA/100 mM KCl/10 mM 2-mercaptoethanol/0.5 mM phenylmethylsulfonyl fluoride, treated with 400 μg of lysozyme for 30 min at 4°C, and then disrupted by sonication. After centrifugation at 15,000 × g for 15 min, the supernatant was adjusted to 0.5 M KCl by the addition of 2 M KCl and centrifuged at 150,000 × g for 2 hr. The ribosomal supernatant (300 mg of protein in 60 ml) was loaded onto a 100-ml bed volume of a column of DEAE-cellulose (30 × 2.1 cm) equilibrated in buffer A/150 mM KCl. After washing the column with 500 ml of the same buffer, bound proteins were eluted with buffer A/300 mM KCl. Recombinant eIF6 was further purified from the DEAE-cellulose eluate (40 mg of protein) by a succession of purification steps as described above for isolation of eIF6 from rabbit reticulocytes except that the size of columns and the gradient volumes used in each step were reduced proportionately. The yield of homogeneous recombinant eIF6 was about 30 μg.
For the isolation of 35S-labeled recombinant eIF6, eIF6 was labeled biosynthetically by adding 5 mCi of [35S]methionine and [35S]cysteine to a 500-ml induced culture of BL21 (DE3) cells harboring the eIF6 expression plasmid pET-5a-eIF6 by using a procedure similar to that used previously (21) for the isolation of 35S-eIF1A. Homogeneous 35S-eIF6 was isolated from labeled cells by using the purification protocol described above for isolation of unlabeled recombinant eIF6.
Other Methods.
Ribosomal subunits were isolated from Artemia salina eggs as described previously (14). eIF6 was assayed by its ability to bind to 60S ribosomal subunits and prevent their association with 40S ribosomal subunits (14) as described in the legend to Fig. 5. Protein was determined by the method of Bradford (22) with reagents obtained from Bio-Rad.
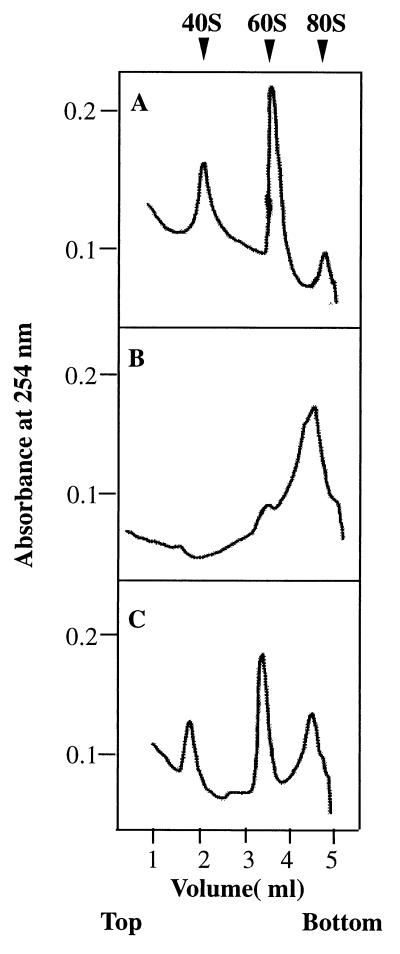
Figure 5.
Ribosomal subunit anti-association activity of bacterially expressed recombinant eIF6. eIF6 activity was measured by its ability to bind to a 60S ribosomal subunit and prevent its association with the 40S ribosomal subunit to form an 80S ribosome as described (14, 15). Reaction mixtures (100 μl each) contained 20 mM Tris⋅HCl, pH 7.5/100 mM KCl/1 mM MgCl2/1 mM DTT/0.4 A260 unit (8 pmol) of 80S ribosomes. In addition, in the reaction in C, 300 ng (12 pmol) of purified recombinant eIF6 was added whereas the other two reactions in A and B did not contain eIF6. After incubation at 30°C for 5 min, the Mg2+ concentration of reaction mixtures in B and C was raised to 5 mM whereas the Mg2+ concentration of the reaction in A was kept at 1 mM, and the incubation continued for another 5 min at 37°C. Under these conditions, ribosomal subunits generated by dissociation of 80S ribosomes at 1 mM Mg2+ during the first incubation should reassociate during the second incubation at 5 mM Mg2+ to 80S ribosomes provided no eIF6 was present in the incubation mixture. In contrast, when eIF6 was present during the first incubation, 60S⋅eIF6 particles formed would fail to associate with 40S particles to form 80S ribosomes during the second incubation. Reaction mixtures were then chilled in an ice–water bath, layered onto 5-ml linear 5–30% (wt/vol) sucrose gradients containing 20 mM Tris⋅HCl, pH 7.5, 100 mM KCl, 1 mM DTT, 5 mM MgCl2 (for reactions in B and C) and 1 mM MgCl2 (for reaction in A only), and centrifuged in a SW 50.1 rotor for 90 min at 48,000 rpm. The gradients were analyzed in an ISCO model 640 gradient fractionator, and the absorbance profile at 254 nm was analyzed by using the UA-5 absorbance monitor.
RESULTS
Purification and Immunochemical Characterization of eIF6 from Rabbit Reticulocyte Lysates.
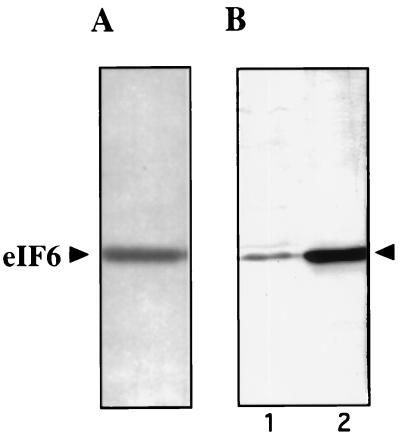
To facilitate studies on eIF6, we first devised a modified procedure to purify eIF6 from rabbit reticulocyte lysates to obtain a higher yield of the protein (see Materials and Methods). The final preparation showed a single polypeptide of about 25 kDa by SDS/PAGE (Fig. 1A). The yield of homogeneous eIF6 obtained by this procedure was severalfold higher than that obtained previously (15). This difference permitted us to raise antibodies in laying hens against purified eIF6 as well as to obtain amino acid sequences of CNBr peptides of eIF6. Fig. 1 shows that the immune IgYs, affinity-purified against purified eIF6, reacted strongly with a single polypeptide in HeLa cell extracts (Fig. 1B, lane 1) that migrated with the same mobility as purified eIF6 immunoblotted with these antibodies (Fig. 1B, lane 2). In contrast, preimmune IgY did not react with eIF6 (data not shown). These results indicated anti-eIF6 antibodies are monospecific and could be used as a cloning reagent. It should, however, be noted that although these antibodies are highly specific for eIF6 in immunoblot analysis, they do not immunoprecipitate native eIF6, indicating that denatured conformation of eIF6 has the antigenic determinants (data not shown). These observations suggest that eIF6 may be highly conserved through evolution.
Figure 1.
SDS/PAGE of purified eIF6 and characterization of anti-eIF6 antibodies. (A) Approximately 400 ng of Mono Q–eIF6 was subjected to SDS/15% polyacrylamide gel electrophoresis followed by silver staining. A set of molecular weight marker proteins was run in a parallel lane of the same gel (not shown). (B) Approximately 200 ng of purified eIF6 (lane 2) and 10 μg of crude HeLa cell lysates prepared by lysing HeLa cells directly into 3% SDS buffer (lane 1) were electrophoresed in a SDS/15% polyacrylamide gel followed by electroblotting onto a pol(vinylidene difluoride) membrane. The membrane was probed with affinity-purified chicken anti-eIF6 antibodies.
Molecular Cloning and Characterization of Human eIF6 cDNA.
To clone the cDNA encoding eIF6, affinity-purified anti-eIF6 antibodies were used as probes to immunoscreen a HeLa cell λZAPII cDNA library. A single immunoreactive phage clone containing a cDNA insert of about 1 kb was obtained. Analysis of the nucleotide sequence (nucleotides −6 to +1011 of Fig. 2) of this cDNA revealed that the longest ORF contains the peptide sequence VGNRHGLLVPNNTTD obtained by microsequencing a CNBr peptide of purified calf liver eIF6, indicating that we have isolated an authentic eIF6 cDNA. However, because the first ATG codon in this cDNA was not preceded by any in-frame translational stop codon and the 5′-untranslated region was relatively short (6 nucleotides in length), we further characterized the isolated cDNA as follows.
Figure 2.
Characterization of eIF6 cDNA encoding eIF6. (A) The ATG assigned as the translation start codon (numbered +1) was found in-frame with the partial amino acid sequence of a CNBr peptide of purified eIF6 (underlined). There is no other ATG in the 5′-untranslated region. The translation stop codon TGA at nucleotide position +736 is marked with an asterisk. The position of the transcription initiation site of eIF6 mRNA determined from primer extension analysis of Fig. 3B is shown by a downward arrow (↓) at position −78 of the eIF6 cDNA. [The length of the poly(A) tail at the 3′ end of the cDNA varies between 20 and 30 in different clones and is indicated by (A)n.] (B) Restriction map of eIF6 clones obtained from human cDNA libraries. a, the 1.045-kb partial HeLa cDNA clone (clone a); b, the 1.096-kb complete human skeletal muscle cDNA clone (clone b). The black bar indicates the HeLa cell cDNA fragment used as a probe for screening λgt 10 skeletal muscle library as well as for Northern analysis in Fig. 3.
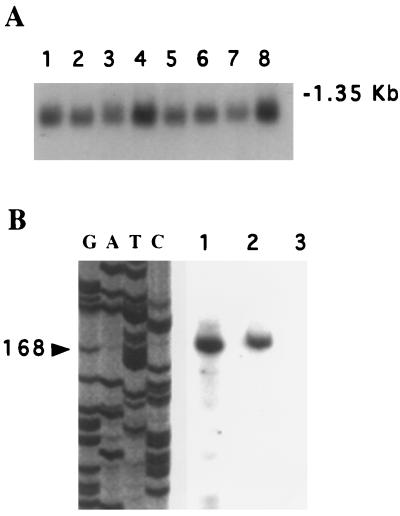
First, Northern blot analysis was carried out by using a 250-bp KpnI/EcoRI fragment derived from clone “a” of HeLa cell eIF6 cDNA (Fig. 2B) as a hybridization probe. In all tissues examined, one major size class of mRNA of about 1.1 kb was observed, which was nearly of the same size as the cDNA (Fig. 3A). Second, we carried out a primer extension analysis by using a 20-nucleotide-long oligonucleotide primer complementary to nucleotides +71 to +90 of the eIF6 cDNA (see Fig. 2) and HeLa cell poly(A)+ RNA as a template. A single 168-nt-long cDNA was found to be the only major reaction product (Fig. 3B), indicating that all eIF6 mRNAs were initiated at nucleotide position −78 upstream of the first ATG codon in the isolated cDNA. These results indicated that the cDNA isolated in the initial screen lacks 72 nucleotides in the 5′-untranslated region of complete eIF6 cDNA.
Figure 3.
Analysis of eIF6 mRNA. (A) Northern blot analysis. Northern blots (CLONTECH) containing 2 μg of electrophoretically separated poly(A)+ RNA isolated from human organs were hybridized to a 32P-labeled 250-bp KpnI/EcoRI fragment of eIF6 cDNA (Fig. 2B, black bar). Conditions for hybridization and washing of blots were according to the procedure of Sambrook et al. (20), as described in a previous paper from this laboratory (23). The blot was analyzed by autoradiography. Lanes 1–8 represent poly(A)+ RNA isolated from human heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas, respectively. The blot also contained a set of RNA size markers, of which the position of 1.35-kb RNA is indicated. (B) Primer extension analysis of eIF6 mRNA. Poly(A)+ RNA (1 μg, lane 1) or 5 μg of total RNA (lane 2), isolated from HeLa cells, was used for primer extension reactions at 42°C by using Moloney murine leukemia virus reverse transcriptase and 5′ 32P-labeled 20-mer deoxyoligonucleotide (5′-GCCTCCGATCGCTACCAGAC-3′) complementary to nucleotides +71 to +90 of the eIF6 cDNA (about 5 × 106 cpm of radioactivity) following a protocol similar to that described by George and Kadonaga (24). A control reaction containing poly(A)+ RNA and no reverse transcriptase was analyzed in lane 3. The reaction products were ethanol-precipitated, dissolved in a formamide-loading buffer containing 33 mM NaOH, boiled for 3 min, and then analyzed on a 6.6% polyacrylamide/8 M urea sequencing gel. Molecular weight sequencing markers were run in parallel lanes by using the cloned human cDNA as the template and the same 20-mer oligodeoxynucleotide primer (lanes G, A, T, and C). The arrow shows the 168-nucleotide primer extension product. The position of the transcription initiation site of eIF6 mRNA as determined by primer extension analysis is shown in Fig. 2 by an arrow (↓).
To obtain the entire human cDNA encoding eIF6, we rescreened a human skeletal muscle λgt 10 cDNA library by using a 32P-labeled 250-bp KpnI/EcoRI fragment derived from the 5′ end of HeLa cell–eIF6 cDNA (clone a, Fig. 2B) as a probe. Several independent positive clones were identified. The largest cDNA insert present in one clone was sequenced completely on both strands. The nucleotide sequence is presented in Fig. 2. Based on the results of primer extension analysis in Fig. 3B, it appears that this sequence represents the complete nucleotide sequence of eIF6 cDNA. Furthermore, the nucleotide sequence of the HeLa cell partial cDNA of eIF6 present in clone a is identical to that of human skeletal muscle eIF6 cDNA present in clone b in the overlapping region (Fig. 2B).
Analysis of the nucleotide sequence of Fig. 2 shows that ATG at position +1 is the first ATG codon at the 5′ end of the cDNA and is thus presumed to be the initiating ATG codon for eIF6 mRNA. The longest ORF of this cDNA encodes a protein of 245 amino acids with a predicted molecular mass of 26,558 daltons. This value is in close agreement with the experimentally determined size of purified mammalian eIF6 (apparent Mr 25,500) (14, 15).
In Vitro Translation of eIF6 cDNA.
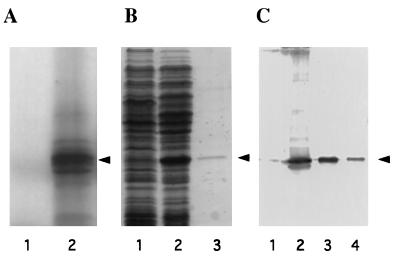
To demonstrate that the cloned cDNA encodes the full-length eIF6 polypeptide, the cDNA was cloned in vector pBluescript SK(+) under the transcriptional control of phage T3 RNA polymerase and translated in a rabbit reticulocyte-coupled transcription/translation system. Analysis of the 35S-labeled translation products by SDS/PAGE showed that the major polypeptide synthesized migrated with the same mobility as purified eIF6 from rabbit reticulocyte lysates (Fig. 4A, lane 2). Several lower molecular weight 35S-labeled polypeptides observed in the autoradiogram presumably arose by limited proteolysis of the eIF6 polypeptide product. Alternatively, the generation of smaller size products could be because of translational starts at internal AUG codons present in the in vitro transcribed RNA.
Figure 4.
Expression of the eIF6 cDNA clone. (A) SDS/15% polyacrylamide gel electrophoresis of [35S]methionine-labeled translation products in a rabbit reticulocyte transcription–translation system (TnT system, Promega). Lanes 1 and 2, in vitro translation in the presence of parental vector pBluescript SK(+) (lane 1) or recombinant vector pBluescript SK(+) containing cloned eIF6 cDNA under the control of T3 RNA polymerase promoter (lane 2). The dried gel was analyzed by autoradiography. The position of purified eIF6 is shown by an arrowhead. (B) Expression of eIF6 cDNA in E. coli. Cell-free extracts, prepared from IPTG-induced cultures of E. coli BL21 (DE3) cells harboring either the parental plasmid pET-5a (lane 1) or recombinant plasmid, pETIF6 containing the eIF6 coding region under the control of T7 RNA polymerase promoter (lane 2), and purified recombinant eIF6 (lane 3) were electrophoresed in SDS/15% polyacrylamide gel followed by Coomassie blue staining. The position of migration of eIF6 is indicated by an arrowhead. (C) Immunoblot analysis of bacterially expressed eIF6. Cell-free extracts of isopropyl β-d-thiogalactopyranoside-induced cultures of BL21 (DE3) cells harboring either the parental vector pET-5a (lane 1) or the pETIF6 recombinant expression plasmid (lane 2) were subjected to Western blot analysis by using monospecific anti-eIF6 antibodies as probes. Purified recombinant eIF6 (lane 3) and purified rabbit reticulocyte eIF6 (lane 4) were also analyzed by Western blot. A set of molecular weight marker proteins were run in a separate lane of each gel (not shown). The position of eIF6 is indicated by an arrowhead.
Expression of eIF6 in E. coli.
To demonstrate that the cDNA-encoded 26,558-dalton polypeptide was indeed eIF6, the ORF of the eIF6 cDNA was expressed in E. coli as described under Materials and Methods, and the cell lysates were analyzed by SDS/PAGE. A polypeptide of about 26 kDa that migrated with the same mobility as purified mammalian eIF6 was observed (Fig. 4B, compare lanes 1 and 2). This polypeptide, which reacted strongly with chicken anti-eIF6 antibodies (Fig. 4C, lanes 2 and 3), was not observed in E. coli cells containing the parental nonrecombinant vector (Fig. 4C, lane 1). However, when the whole cell extract of E. coli cells overexpressing human eIF6 was centrifuged at 10,000 × g for 10 min and both the supernatant and pellet fractions were analyzed by Western blotting, a major fraction (about 90%) of the immunoreactive 26-kDa polypeptide was found in the pellet fraction as an insoluble protein, whereas a small fraction (approximately 10%) of immunoreactive 26-kDa eIF6 polypeptide was observed in the soluble fraction (data not shown).
Functional Characterization of Bacterially Expressed Recombinant eIF6.
To characterize recombinant eIF6, we purified the protein from the soluble fraction as described under Materials and Methods. eIF6 was monitored at different purification steps by SDS/PAGE followed by Coomassie blue staining as well as immunoblot analysis of eIF6 fractions by using polyclonal anti-eIF6 antibodies (data not shown). The final purified eIF6 fraction was >90% pure (Fig. 4B, lane 3).
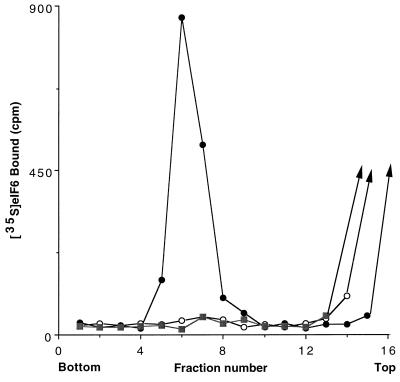
The biochemical properties of bacterially expressed purified eIF6 were investigated. As shown in Fig. 5B, when isolated 40S and 60S ribosomal subunits were first incubated at 1 mM Mg2+ and the Mg2+ concentration was then raised to 5 mM, a major fraction of the subunits associated to form 80S ribosomes as analyzed by sucrose gradient centrifugation. In contrast, when the ribosomal subunits were incubated with recombinant eIF6 at 1 mM Mg2+ and the Mg2+ concentration was then raised to 5 mM, a substantial fraction of the 40S and 60S ribosomal subunits failed to associate to form 80S ribosomes (Fig. 5C). A similar extent of dissociation was also observed when ribosomal subunits were incubated in the absence of eIF6 and sedimented at 1 mM Mg2+ (Fig. 5A). These results demonstrate that bacterially expressed recombinant eIF6 has ribosomal subunit anti-association activity. Evidence that the recombinant eIF6, like the protein isolated from both mammalian cells (14), and wheat germ extracts (13) also interacted specifically with 60S ribosomal subunits came from the following experiment. When 35S-labeled eIF6 was incubated with 60S ribosomal subunits and the mixture subjected to sucrose gradient centrifugation, a substantial fraction of radioactivity cosedimented with 60S particles (Fig. 6). In contrast, little binding was observed when labeled eIF6 was incubated with 40S subunits before sucrose gradient analysis. It should be noted that the assay described in Fig. 6 was carried out in the presence of an excess of ribosomal subunits. Based on the specific activity of 35S-labeled eIF6, it can be calculated that only about 30% of the input eIF6 (≈4 pmol) was bound to 60S ribosomal subunits.
Figure 6.
Binding of 35S-labeled recombinant eIF6 to 60S ribosomal subunits. Reaction mixtures (100 μl each) contained 20 mM Tris⋅HCl, pH 7.5, 100 mM KCl, 1 mM MgCl2, 1 mM DTT, and 2.0 A260 unit of either 40S or 60S ribosomal subunits. Reactions were initiated by the addition of 320 ng (≈13 pmol) of purified 35S-labeled recombinant eIF6 containing 5,000 cpm of radioactivity. After incubation at 37°C for 5 min, the Mg2+ concentration of each reaction was raised to 5 mM, and the reactions were analyzed by sucrose gradient centrifugation as described in the legend to Fig. 5, except that each fraction was assayed for 35S radioactivity by counting in Aquasol in a liquid scintillation spectrometer. •, 60S ribosomal subunits/35S-eIF6; ○, 40S ribosomal subunits/35S-eIF6; ▪, 35S-eIF6 alone.
DISCUSSION
Several lines of evidence presented in this paper indicate that we have cloned the human cDNA encoding mammalian eIF6. The coding region of the cDNA has been expressed in E. coli to yield a functionally active eIF6 protein (calculated Mr 25,556) that migrated with the same mobility as purified eIF6 isolated from mammalian cell extracts. Furthermore, purified recombinant eIF6 mimics natural eIF6 in binding specifically to 60S ribosomal subunits and preventing their association with 40S ribosomal subunits to form 80S ribosomes. The availability of purified functionally active recombinant protein in relatively larger quantities than previously obtained from natural sources will now allow us to carry out experiments on the structural and detailed functional aspects of eIF6, particularly on the role of this protein in the overall initiation of translation in eukaryotic cells.
Analysis of the derived amino acid sequence of human eIF6 indicated that eIF6 is an acidic protein of calculated pI 5.8. The amino acid sequence contains multiple potential phosphorylation sites for cyclic AMP-dependent protein kinase and epidermal growth factor receptor-like tyrosine kinase. It remains to be seen whether eIF6, like many other translation initiation factors, is phosphorylated in vivo and whether phosphorylation of eIF6 is involved in the regulation of its activity.
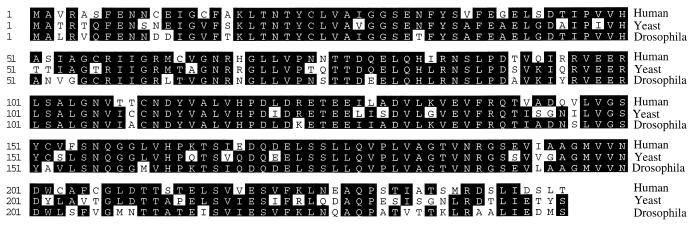
When the human eIF6 cDNA and its predicted amino acid sequence were used to search for homologous sequences in databases, several expressed sequence tags were identified in different species including Caenorhabditis elegans, mouse, rat, and human, suggesting that the eIF6 gene is conserved through evolution. When the predicted amino acid sequence of human eIF6 was used to search the yeast genomic sequence for homologous yeast eIF6 sequence, we identified an ORF in chromosome XVI of S. cerevisiae that encodes a protein of 245 amino acids, which is 72% identical to human eIF6 (Fig. 7). Likewise, another ORF that encodes a protein that is about 80% identical to human eIF6 was found in the Drosophila melanogaster gene bgen. It is highly likely that the yeast and the Drosophila sequences represent the homologues of human eIF6 in these organisms. A comparison of all three deduced amino acid sequences of human eIF6 and putative yeast and Drosophila proteins is presented in Fig. 7. The high degree of conservation in amino acid sequence among phylogenetically distant species indicates that eIF6 probably plays an essential role in a major metabolic process in the cell. It is to be noted that eIF6 has been purified and characterized from mammalian cells and wheat germ extracts based on an in vitro partial reaction, and the role of this protein in translation in vivo has not yet been determined. The presumed presence of the eIF6 gene in S. cerevisiae will allow us to clone and characterize the yeast gene and its product and use the molecular genetic approach to characterize the function of eIF6 in protein synthesis in vivo in yeast cells. Such an approach recently has been used successfully in this laboratory (25) to define the function of eIF5 in vivo and in vitro in the initiation of protein synthesis in yeast cells.
Figure 7.
Comparison of the predicted amino acid sequence of human eIF6 with those of S. cerevisiae and Drosophila obtained from protein database search. The nucleotide and derived amino acid sequences of human eIF6 were used to search the databases by using the blast search program to identify homologues of human eIF6. The putative homologues of mammalian eIF6 from the yeast S. cerevisiae and Drosophila are shown. The amino acid sequences are aligned for maximum sequence identity. The conserved residues are highlighted.
Acknowledgments
This paper is dedicated to the loving memory of Dr. Jorge Chevesich. He was a superb scientist and an extraordinary human being, who made important contributions as a graduate student in this laboratory. This work was supported by Grants GM15399 from the National Institutes of Health, Cancer Core Support Grant P30CA13330 from the National Cancer Institute, and Diabetes Research and Training Center, National Institutes of Health 5P60-DK20541. The microsequencing of eIF6 peptides was carried out in the Laboratory of Macromolecular Analysis of this institution.
ABBREVIATION
- eIF
eukaryotic (translation) initiation factor
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF022229).
References
- 1.Maitra U, Stringer E A, Chaudhuri A. Annu Rev Biochem. 1982;51:869–900. doi: 10.1146/annurev.bi.51.070182.004253. [DOI] [PubMed] [Google Scholar]
- 2.Kozak M. Annu Rev Cell Biol. 1992;8:197–225. doi: 10.1146/annurev.cb.08.110192.001213. [DOI] [PubMed] [Google Scholar]
- 3.Merrick W C. Microbiol Rev. 1992;56:291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pain V M. Eur J Biochem. 1996;236:747–771. doi: 10.1111/j.1432-1033.1996.00747.x. [DOI] [PubMed] [Google Scholar]
- 5.Merrick W C, Hershey J W B. In: Translational Control. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 31–69. [Google Scholar]
- 6.Davis B D. Nature (London) 1971;231:153–157. doi: 10.1038/231153a0. [DOI] [PubMed] [Google Scholar]
- 7.Noll M, Hapke B, Schreier M H, Noll H. J Mol Biol. 1973;75:281–294. doi: 10.1016/0022-2836(73)90021-1. [DOI] [PubMed] [Google Scholar]
- 8.Trachsel H, Staehelin T. Biochim Biophys Acta. 1979;565:305–314. doi: 10.1016/0005-2787(79)90207-7. [DOI] [PubMed] [Google Scholar]
- 9.Thompson H A, Sadnik I, Scheinbuks J, Moldave K. Biochemistry. 1977;16:2221–2230. doi: 10.1021/bi00629a028. [DOI] [PubMed] [Google Scholar]
- 10.Goss D, Rounds D, Harrigan T, Woodley C L, Wahba A J. Biochemistry. 1988;27:1489–1494. doi: 10.1021/bi00405a014. [DOI] [PubMed] [Google Scholar]
- 11.Thomas A, Goumans H, Voorma H O, Benne R. Eur J Biochem. 1980;107:39–45. doi: 10.1111/j.1432-1033.1980.tb04621.x. [DOI] [PubMed] [Google Scholar]
- 12.Russell D W, Spremulli L L. J Biol Chem. 1979;254:8796–8800. [PubMed] [Google Scholar]
- 13.Russell D W, Spremulli L L. Arch Biochem Biophys. 1980;201:518–526. doi: 10.1016/0003-9861(80)90540-8. [DOI] [PubMed] [Google Scholar]
- 14.Valenzuela D M, Chaudhuri A, Maitra U. J Biol Chem. 1982;257:7712–7719. [PubMed] [Google Scholar]
- 15.Raychaudhuri P, Stringer E A, Valenzuela D M, Maitra U. J Biol Chem. 1984;259:11930–11935. [PubMed] [Google Scholar]
- 16.Chevesich J, Chaudhuri J, Maitra U. J Biol Chem. 1993;268:20659–20667. [PubMed] [Google Scholar]
- 17.Carroll S B, Stollar B D. J Biol Chem. 1983;258:24–26. [PubMed] [Google Scholar]
- 18.Ghosh S, Chevesich J, Maitra U. J Biol Chem. 1989;264:5134–5140. [PubMed] [Google Scholar]
- 19.Snyder M, Elledge S, Sweetser D, Young R A, Davis R W. Methods Enzymol. 1987;154:107–128. doi: 10.1016/0076-6879(87)54073-3. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 21.Chaudhuri J, Si K, Maitra U. J Biol Chem. 1997;272:7883–7891. doi: 10.1074/jbc.272.12.7883. [DOI] [PubMed] [Google Scholar]
- 22.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 23.Si K, Das K, Maitra U. J Biol Chem. 1996;271:16934–16938. doi: 10.1074/jbc.271.28.16934. [DOI] [PubMed] [Google Scholar]
- 24.George C P, Kadonaga J T. In: A Laboratory Guide to RNA Isolation, Analysis and Synthesis. Krieg P A, editor. New York: Wiley–Liss; 1996. pp. 133–136. [Google Scholar]
- 25.Maiti T, Maitra U. J Biol Chem. 1997;272:18333–18340. doi: 10.1074/jbc.272.29.18333. [DOI] [PubMed] [Google Scholar]