Abstract
F- and V-type ATPases are central enzymes in energy metabolism that couple synthesis or hydrolysis of ATP to the translocation of H+ or Na+ across biological membranes. They consist of a soluble headpiece that contains the catalytic sites and an integral membrane-bound part that conducts the ion flow. Energy coupling is thought to occur through the physical rotation of a stalk that connects the two parts of the enzyme complex. This mechanism implies that a stator-like structure prevents the rotation of the headpiece relative to the membrane-bound part. Such a structure has not been observed to date. Here, we report the projected structure of the V-type Na+-ATPase of Clostridium fervidus as determined by electron microscopy. Besides the central stalk, a second stalk of 130 Å in length is observed that connects the headpiece and membrane-bound part in the periphery of the complex. This additional stalk is likely to be the stator.
In oxidative phosphorylation, F-type ATPases synthesize ATP at the expense of the proton gradient across the membrane, and in fermentative bacteria F- and V-type ATPases generate a proton or sodium ion motive force upon ATP hydrolysis. F-type and V-type ATPases, operating in either the ATP synthesis or hydrolysis mode, have similar architectures, and it is generally accepted that the catalytic mechanism is the same (1). The enzymes consist of a soluble headpiece, F1 or V1, that contains the ATP binding sites and an integral membrane part, F0 or V0, that translocates H+ or Na+. The two parts are connected by a central stalk. Detailed structural information is available on the F-type ATPases (2). The 2.8-Å atomic resolution structure of bovine heart mitochondrial F1 shows alternating α and β subunits arranged as the segments of an orange (α3β3). The central cavity is occupied with two long α-helices of the γ subunit, which is the major stalk subunit (2). In the rotary catalysis mechanism, the three nucleotide (ATP/ADP) binding sites on the β subunits cycle alternately through the open, loose, and tight states according to the binding change mechanism (3). Concomitant changes in the conformation of the β subunits drive the physical rotation of the central γ subunit (ATP hydrolysis mode). In support of the model, ATP-driven rotation of γ relative to α3β3 was recently demonstrated experimentally (4, 5). In turn, the rotating γ subunit is thought to drive H+ translocation in F0 by an unknown mechanism. However, efficient transduction of the torque exerted by the rotating stalk on F0 in pumping activity requires that a stator-like structure is present that prevents dissipation of energy in a rotation of F1 relative to F0 (6, 7). Here, we report for the first time the visual demonstration of the stator by electron microscopy.
METHODS
Sample Preparation.
The V-ATPase of Clostridium fervidus was purified by anion exchange chromatography (MonoQ), followed by size exclusion chromatography (Superose 6) as described (8). The protein complex was purified in a buffer containing 50 mM N-2-morpholine propanesulfonic acid (MOPS), pH 7.0, 150 mM KCl, 5 mM MgCl2, 5% glycerol, 0.1% Triton X-100, 2 mM DTT, and 0.1 mg/ml phospholipids (phosphatidylethanolamine and phosphatidylcholine at a ratio of 3:1). A sample was dialyzed at room temperature for 2 hr against buffer without detergent and glycerol and prepared for electron microscopy (Philips CM10) using the droplet method with uranyl acetate as negative stain.
Image Analysis.
A total of 829 membrane-incorporated and 44 nonincorporated V1V0 projections were analyzed using imagic software (9). Band pass-filtered images were subjected to multireference alignment, multivariate statistical analysis, and classification (10, 11). In the data set of the membrane-incorporated V1V0, a first classification was focused on the area occupied by the V1 headpiece plus stalk region. It revealed a class of 240 V1 projections that was characterized by a symmetrical projected density with high densities in the center and at the sides of the headpiece (trilobed view). This suggests that these molecules were oriented in one and the same orientation toward the carbon grid. This data set was subjected again to the classification procedure, but this time a mask was used that was focused on the stalk area. The best separation was obtained by decomposition into six classes. After this procedure, the normalized images belonging to these classes were summed without the band-pass filter imposed.
RESULTS AND DISCUSSION
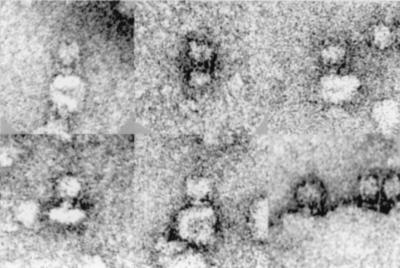
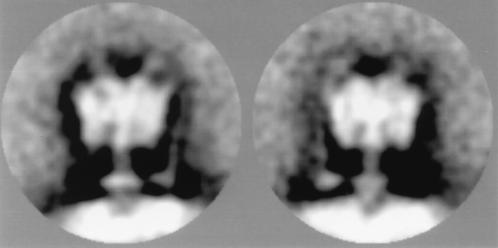
C. fervidus is a thermophilic organism that relies completely on Na+ as the energy coupling ion (12). The Na+ ion gradient across the cytoplasmic membrane is generated by an ATPase that is one out of three V-type ATPases described in the bacterial kingdom to date (8). Recently, the ATPase complex was purified to homogeneity and functionally reconstituted in liposomes (8). Electron micrographs of the solubilized enzyme purified in the presence of phospholipids showed that most of the ATPase molecules were incorporated in small liposome-like structures of variable size. The side view projections show V1 attached to the membrane via a stalk region (Fig. 1). Single particle averaging techniques revealed that a large fraction of the particles was oriented in the same way on the grid. Statistical analysis of this group of particles (see Materials and Methods) revealed six classes in all of which the central stalk was clearly visible (Fig. 2). The stalk has a length of about 60 Å, which is significantly longer than the stalk of F-type ATPases (13–15). The stalk is substantially thicker (about 50 Å) at the base just above the membrane, another characteristic that seems to be specific for V-type ATPases (16, 17). Remarkably, in two of the six classes a second stalk that connects the V1 head and V0 membrane part in the periphery is clearly visible (Fig. 2). The main difference between the two classes is that this structure appears at either the left or right side of the central stalk, which corresponds to views of the molecules from opposite sites. The peripheral stalk has a length of about 130 Å and runs up almost to the top of the V1 headpiece. The thickness is about 10–15 Å, and at a similar position, as observed in the central stalk, it widens to approximately 30 Å.
Figure 1.
A gallery of electron microscopy images of the V-ATPase of C. fervidus.
Figure 2.
Side view projections of the V-ATPase of C. fervidus incorporated in liposomes. Shown are two out of six classes of a subgroup of 240 side view projections of the V-type ATPase of C. fervidus incorporated in liposomes. They show V1 in a trilobed view and, besides a central stalk, a peripheral second stalk in two positions. For the left and right images, 39 and 42 images were summed, respectively. The diameter of the area shown is 320 Å.
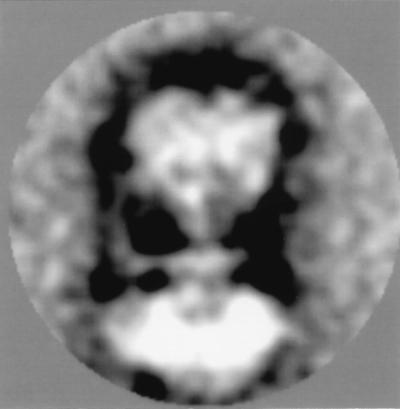
Additional structural information was obtained from the analysis of V1V0 molecules, which were strongly reduced in attached lipid and detergent molecules and not incorporated in liposome-like structures. These V1V0 molecules were present at a lower frequency in the electron micrographs. Single particle averaging of 44 side views followed by classification revealed one class consisting of 12 particles in which the central and peripheral stalks were clearly visible (Fig. 3). The exact dimensions of V0 cannot be determined precisely from this view because the negative stain outlines protein plus attached lipid and detergent molecules. Nevertheless, the image indicates that V0 is not a symmetrical structure with respect to the central stalk; V0 is wider at the side of the peripheral stalk.
Figure 3.
Side view projection of nonincorporated V1V0-ATPase. One class representing 12 particles out of a total of 44 particles is shown in which a peripheral stalk and the asymmetry of V0 with respect to the central stalk are clearly visible.
An interesting new feature observed in the averaged side views of both the V1V0 particle incorporated in the vesicles (Fig. 2) and the free particle (Fig. 3) is an accumulation of stain just below the central stalk. This feature can be explained as originating from the ring-shaped multimeric complex of the proteolipid subunit of the membrane-bound part (subunit c in F0). Other studies have revealed a central dimple in the isolated V-type multimer that had a radius of about 75 Å (18, 19). The similar dimple-like structure observed in our side view projections suggests that, in agreement with other topological studies on F1F0 (19, 20), the proteolipid multimer is situated symmetrically under the central stalk and that the other integral membrane subunits (a and b in F0) are outside of the ring. This suggestion is in agreement with the asymmetry observed in the image of V0 (Fig. 3) and is most strongly supported by the peripheral stalk observed in this study. The most likely candidate for the peripheral stalk is the b subunit, with a mass between 13 and 17 kDa in V- and F-ATPases (21). It has a hydrophilic amino acid extension, predicted to be α-helical, that is long enough to fit the 130-Å-long peripheral stalk observed in our images. In prokaryotic F-ATPases, two copies of subunit b are present that may interact (1, 22). In V-ATPases, the exact subunit stoichiometry remains to be established. The wide spacing between the central and peripheral stalks (65 Å), together with the diameter of the V0 proteolipid multimer (75 Å; ref. 18) or F0 subunit c multimer (62 Å; ref. 23), suggests that subunit a is between b and c, in contrast to other EM data (20).
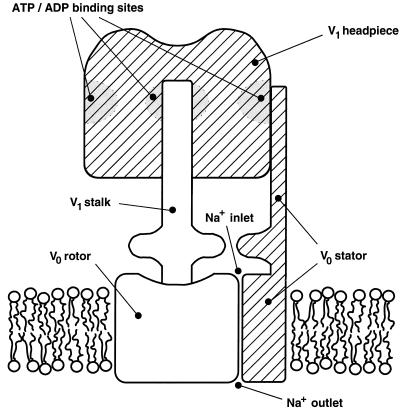
The structures of F0/V0 are not precisely understood, but the projection maps of the V-type Na+-ATPase of C. fervidus favor the models that place the a and b subunits outside the multimeric ring of c subunits (15, 19, 20) and strongly suggest that the b subunit forms a peripheral stalk that prevents rotations of the headpiece relative to the membrane part. The position of the peripheral stalk closely matches some hypothetical models (6, 24, 25). Fig. 4 presents a model of the molecular V-type ATPase motor as is suggested by the present studies. In the hydrolysis mode, hydrolysis of ATP drives the physical rotation of the central stalk via chemomechanic coupling. The central stalk transmits the rotation to the rotor in the V0 part that rotates against the stator structure that is fixed to the V1 engine. The rotation results in the pumping of Na+ ions at the interface via an osmomechanic coupling mechanism. In the ATP synthesis mode, the events happen in the reversed direction.
Figure 4.
Model of the molecular V-ATPase motor. ATP hydrolysis in the V1 headpiece drives rotation of the central stalk. The stalk transmits the rotation to the V0 rotor. Rotation of the rotor against the stator structure results in the pumping of Na+ ions across the membrane at the interface. Hatched parts are static, and open parts are rotating.
Acknowledgments
We thank Profs. R. Kraayenhof and W. Junge for discussion and Dr. W. Keegstra for help with the image analysis.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Fillingame R H. Curr Opin Struct Biol. 1996;6:491–498. doi: 10.1016/s0959-440x(96)80114-x. [DOI] [PubMed] [Google Scholar]
- 2.Abrahams J P, Leslie A G W, Lutter R, Walker J E. Nature (London) 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 3.Boyer P D. Biochim Biophys Acta. 1993;1140:215–250. doi: 10.1016/0005-2728(93)90063-l. [DOI] [PubMed] [Google Scholar]
- 4.Sabbert D, Engelbrecht S, Junge W. Nature (London) 1996;381:623–625. doi: 10.1038/381623a0. [DOI] [PubMed] [Google Scholar]
- 5.Noji H, Yasuda R, Yoshida M, Kinosita K. Nature (London) 1997;368:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 6.Junge W, Sabbert D, Engelbrecht S. Ber Bunsen-Ges Phys Chem. 1996;100:2014–2019. [Google Scholar]
- 7.Kagawa Y, Hamamoto T. J Bioenerg Biomembr. 1996;28:421–431. doi: 10.1007/BF02113984. [DOI] [PubMed] [Google Scholar]
- 8.Höner zu Bentrup K, Ubbink-Kok T, Lolkema J S, Konings W N. J Bacteriol. 1997;179:1274–1279. doi: 10.1128/jb.179.4.1274-1279.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harauz G, Boekema E, van Heel M. Methods Enzymol. 1988;164:35–49. doi: 10.1016/s0076-6879(88)64033-x. [DOI] [PubMed] [Google Scholar]
- 10.Boekema E J, Böttcher B. Biochim Biophys Acta. 1992;1098:131–143. doi: 10.1016/0005-2728(92)90073-b. [DOI] [PubMed] [Google Scholar]
- 11.Van Heel M, Frank J. Ultramicroscopy. 1981;6:187–194. doi: 10.1016/0304-3991(81)90059-0. [DOI] [PubMed] [Google Scholar]
- 12.Speelmans G, Poolman B, Abee T, Konings W. Proc Natl Acad Sci USA. 1993;90:7975–7979. doi: 10.1073/pnas.90.17.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boekema E J, Schmidt G, Gräber P, Berden J A. Z Naturforsch. 1988;43C:219–225. doi: 10.1515/znc-1988-3-412. [DOI] [PubMed] [Google Scholar]
- 14.Lücken U, Gogol E P, Capaldi R A. Biochemistry. 1990;29:5339–5343. doi: 10.1021/bi00474a019. [DOI] [PubMed] [Google Scholar]
- 15.Capaldi R A. Nat Struct Biol. 1994;1:660–663. doi: 10.1038/nsb1094-660. [DOI] [PubMed] [Google Scholar]
- 16.Dschida W J, Bowman B J. J Biol Chem. 1992;267:18783–18789. [PubMed] [Google Scholar]
- 17.Getz H P, Klein M. Bot Acta. 1995;108:14–23. [Google Scholar]
- 18.Finbow M E, Eliopoulos E E, Jackson P J, Keen J N, Meagher L, Thompson P, Jones P, Findlay J B C. Protein Eng. 1992;5:7–15. doi: 10.1093/protein/5.1.7. [DOI] [PubMed] [Google Scholar]
- 19.Singh S, Turina P, Bustamante C J, Keller D J, Capaldi R. FEBS Lett. 1996;397:30–34. doi: 10.1016/s0014-5793(96)01127-1. [DOI] [PubMed] [Google Scholar]
- 20.Birkenhäger R, Hoppert M, Deckers-Hebestreit G, Mayer F, Altendorf K. Eur J Biochem. 1995;230:58–67. [PubMed] [Google Scholar]
- 21.Supekova L, Sbia M, Supek F, Ma Y, Nelson N. J Exp Biol. 1996;199:1147–1156. doi: 10.1242/jeb.199.5.1147. [DOI] [PubMed] [Google Scholar]
- 22.Cox G B, Jans D A, Fimmel A L, Gibson F, Hatch L. Biochim Biophys Acta. 1984;768:201–208. doi: 10.1016/0304-4173(84)90016-8. [DOI] [PubMed] [Google Scholar]
- 23.Boekema E, J, Fromme P, Gräber P. Ber Bunsen-Ges Phys Chem. 1988;92:1031–1036. [Google Scholar]
- 24.Duncan T M, Bulygin V V, Zhou Y, Hutcheon M L, Cross R L. Proc Natl Acad Sci USA. 1995;92:10964–10968. doi: 10.1073/pnas.92.24.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilkens S, Dunn S, Chandler J, Dahlquist F W, Capaldi R A. Nat Struct Biol. 1997;4:198–201. doi: 10.1038/nsb0397-198. [DOI] [PubMed] [Google Scholar]