Abstract
Type-2 isopentenyl diphosphate isomerase, which catalyzes the interconversion if isopentenyl diphosphate and dimethylallyl diphosphate, contains a tightly bound molecule of FMN. Incubation of the active enzyme•FMNH2 complex with an analog of isopentenyl diphosphate, where the methyl group has been replaced with a cyclopropane ring, results in isomerization of the analog to the corresponding allylic isomer without inactivation of the enzyme. In contrast, the related epoxide analog is a potent irreversible inhibitor that covalently modifies the flavin cofactor in a proton-initiated reaction. These results suggest that the mechanism for isomerization by the type-2 isopentenyl diphosphate isomerase may be similar to the protonation-deprotonation sequence of the type-1 enzyme and places limits on the lifetimes of radical intermediates in an alternative hydrogen atom addition/abstraction mechanism.
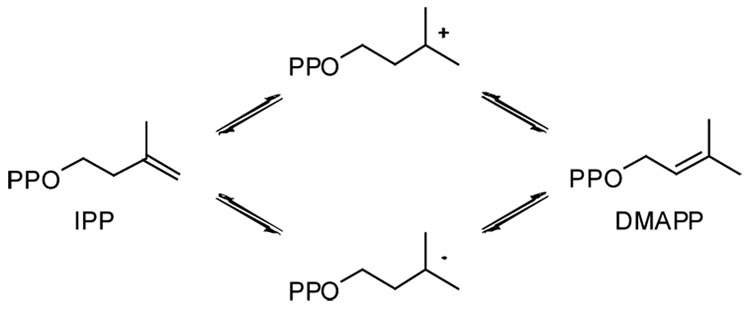
Isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) are the building blocks for biosynthesis of isoprenoid compounds.1 In Eukarya, Archaea, and some Bacteria, IPP is synthesized from acetate by the mevalonate pathway. Conversion of IPP to DMAPP is catalyzed by IPP isomerase (IDI), a mandatory step that generates the electrophilic allylic moiety required for subsequent prenyl transfer reactions.2 In other Bacteria and plant chloroplasts, IPP and DMAPP are synthesized from pyruvate and D-glyceraldehyde phosphate in the methylerythritol phosphate pathway.3 IDI activity, although not required, is typically found in these organisms.
Two forms of IDI are known. Type-1 IPP isomerase (IDI-1), discovered in the 1950s, is a metalloprotein that catalyzes interconversion of IPP and DMAPP by protonation of the carbon-carbon double bond, followed by elimination of a proton from the carbocationic intermediate.4 A second IPP isomerase (IDI-2) was reported in 2001.5 The absence of similarities in the amino acid sequences and X-ray structures of IDI-1 and IDI-2 indicate that the two forms of the enzyme evolved independently.6 Like IDI-1, IDI-2 requires divalent magnesium, presumably for binding the diphosphate moieties of the substrates.7 However, IDI-2 is a flavoprotein, and NADPH or other suitable reducing agents are required to reduce the FMN cofactor to generate the catalytically competent form of the enzyme.8 Although FMN is typically associated with oxidation-reduction chemistry, there are examples of flavins participating in isomerizations, with transient changes in oxidation states.9
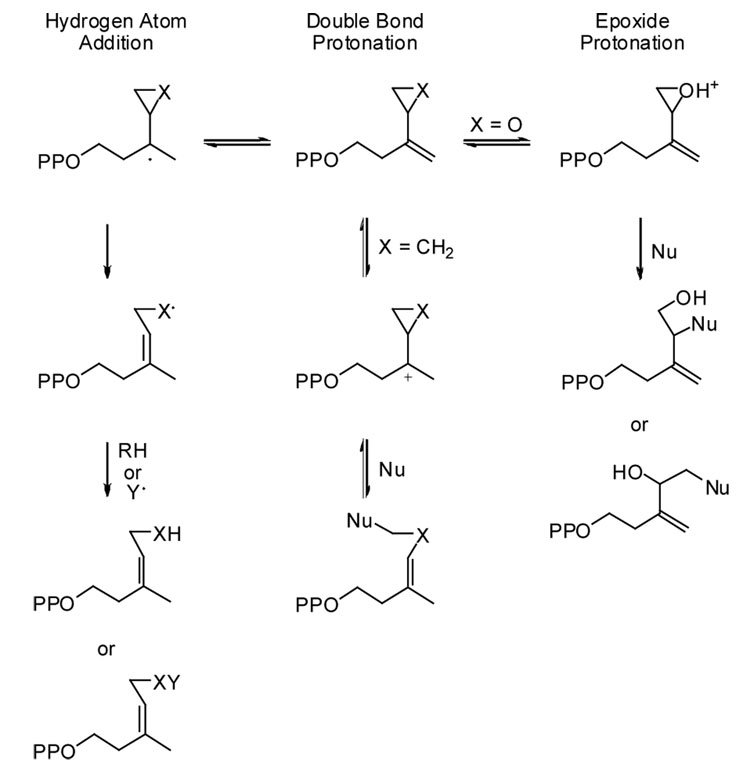
Two mechanisms have been proposed for the isomerization catalyzed by IDI-2 - a proton addition-elimination mechanism similar to IDI-17,9 and a hydrogen atom addition-elimination in which reduced FMN donates a hydrogen atom to generate a transient semiquinone/substrate radical pair9,10 (Scheme 1). We synthesized cyclopropyl (cIPP, X = CH2) and epoxy (oIPP, X = O) analogs as mechanism-based inhibitors of IDI-2 in an effort to distinguish between these two possible mechanisms. Potential reactions of the two compounds in proton initiated and hydrogen atom initiated scenarios for the mechanism of isomerization of IPP by IDI-2 are shown in Scheme 2. Proton-initiated isomerization could lead to inhibition with cIPP by nucleophilic attack at the cyclopropane ring of the cyclopropylcarbinyl cationic intermediate11 in analogy to the inactivation of IDI-1 by vinyl-IPP derivatives,12 while protonation of the oxirane ring in oIPP could inactivate the enzyme by nucleophilic attack at the activated epoxide by a nucleophile in the active site similar to irreversible inhibition by epoxide analogs previously reported for IDI-1.13 Hydrogen atom initiated isomerization would generate cyclopropylcarbinyl14 or epoxycarbinyl radicals,15 which could isomerize to their allylic counterparts and inactivate the enzyme through hydrogen abstraction or recombination reactions.
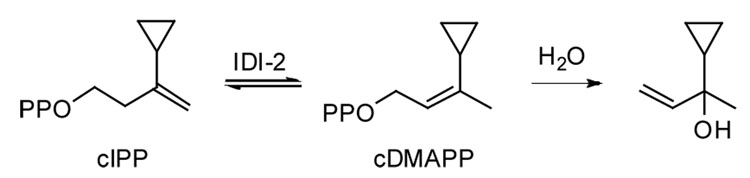
A 2.5 mM solution of cIPP in 50 mM Na2HPO4 buffer at 37 °C, pH 7.2, containing 2 mM NADPH was incubated with IDI-2 from Thermus thermophilus and the course of the reaction was followed by 1H NMR spectroscopy. Over a period of 5 h, the cyclopropyl analog isomerized to its allylic isomer, which was unstable to the incubation conditions and solvolyzed to give a rearranged tertiary vinyl alcohol as shown in Scheme 3. cIPP did not inhibit IDI-2 in a time-dependent irreversible manner when preincubated with the enzyme. In contrast, preincubation of IDI-2 with oIPP at 37 °C, pH 7.0, resulted in rapid, time-dependent 1st-order inactivation of the enzyme. A double reciprocal plot of the rate of inactivation versus [oIPP] gave kinact = 0.37 ± 0.07 min−1 and KI = 1.4 ± 0.3 µM. The enzyme was not inactivated during a control experiment without NADPH in the buffer.
Scheme 3.
IDI-2 catalyzed isomerization of cIPP and spontaneous solvolysis of cDMAPP
In a large-scale inactivation experiment, flavin-bound IDI-2 was incubated with oIPP. The sample was analyzed by RP-HPLC on a C18 column with UV detection and by negative ion electrospray HPLC-MS. Samples introduced by directed infusion were also analyzed by negative ion mode electrospray-MS. A peak was seen at m/z 731 in both the LC-MS and direct infusion experiments, consistent with formation of a covalent adduct between reduced FMN and oIPP. The peak at m/z 731 was also accompanied by peaks at m/z 456 and 274, apparently from a fragmentation of the oIPP·flavin adduct. Control experiments with IPP or without substrate instead of oIPP gave an ESI peak at m/z 455 for oxidized FMN. When enzyme that had been incubated with IPP was washed under aerobic conditions, the sample gave a UV-vis spectrum characteristic of oxidized FMN.8 In contrast, enzyme inhibited with oIPP gave a spectrum consistent with a modified flavin (See Supporting Information). In addition, a transient absorbance was seen at 600 nm during guanidinium-HCl denaturation and ultrafiltration. Similar behavior was previously attributed to a semiquinone intermediate during the oxidation of N5 flavin adducts. 16
To further delineate the mechanism for inhibition by oIPP, parallel inactivation experiments were carried out in D2O and H2O buffers. In parallel experiments, samples of the enzyme were prepared in Tris buffer, pH 7.8 (H2O) or pD 7.8 (99% D2O), both containing NADPH. The mixtures were pre-incubated without inhibitor for 5 min to reduce bound FMN before addition of oIPP or IPP and then were incubated for 2 h. Following the incubations, the samples were washed under aerobic conditions in H2O buffer and analyzed in protic solvents.
If o-IPP were activated by addition of a proton or a hydrogen atom to the double bond of oIPP, the hydrogen would be retained in the oIPP-FMN adduct, and the oIPP-FMN adduct generated by inactivation of IDI-2 in D2O buffer would contain at least one atom of deuterium. Conversely, inactivation initiated by protonation of the epoxide moiety in oIPP in D2O, followed by exchange in a protic solvent, would not result in deuterium incorporation into the final adduct. Negative ion electrospray mass spectra for the oIPP-FMN adducts from incubations in H2O and D2O gave a peak at m/z 731, demonstrating the absence of deuterium incorporation (see Supplemental Information). This result is consistent with protonation of the epoxide oxygen in oIPP, followed by ring opening. There is no evidence for rearrangement of cyclopropylcarbinyl or epoxycarbinyl radicals from cIPP or oIPP, which are well-documented radical “clocks” with rearrangement rates of ~1.3 × 108 s−114 and 3.2 × 1010 s−1,15 respectively. Our results with oIPP are similar to those recently reported for IPP epoxide, a mechanism-based inhibitor of type-1 IPP isomerase,13 where high concentrations of the inhibitor irreversibly inhibit IDI-2 with concomitant formation of a covalent adduct with the flavin cofactor.17
In conclusion, we have established that oIPP is a potent active-site-directed, irreversible inhibitor of IDI-2. The enzyme catalyzes formation of a stable covalent adduct between oIPP and FMNH2 in a reaction catalyzed by protonation of the epoxide moiety in oIPP followed by nucleophilic addition of the cofactor. At this point the structure of the adduct has not been established, although the UV-visible spectrum and the mass spectral fragmentation patterns are consistent with addition of the inhibitor to the N5 nitrogen of the isoalloxazine ring in FMN. These results suggest that isomerization by IDI-2 involves protonation of the substrates in a manner similar to the reaction catalyzed by IDI-1. It is evident from our inactivation studies that the reduced flavin resides in close proximity to bound substrate, where it can potentially provide a function critical to catalysis. Isomerization of cIPP to cDMAPP indicates that a tertiary cyclopropylcarbinyl radical, generated by hydrogen transfer to cIPP from FMNH2 or by single electron transfer (SET) from FMNH− to a tertiary cyclopropylcarbinyl cationic intermediate, is not an intermediate or hydrogen atom transfer from FMNH· to the intermediate cyclopropylcarbinyl radical is substantially faster than the cyclopropylcarbinyl to homoallyl radical rearrangement. At this point, the exact role of the flavin cofactor has not been established.
Supplementary Material
SUPPORTING INFORMATION AVAILABLE: NMR Spectra for conversion of cIPP to cDMAPP. Experimental details for large scale inactivation experiments. LC-MS chromatogram with ESI for the flavin adduct. ESI spectra for oIPP/flavin adducts formed in H2O and D2O. Spectra of enzyme complexes following incubation and following guanidium-HCl denaturation. This material is available free of charge via the Internet at http://pubs.acs.org
ACKNOWLEDGMENTS
J.B.J was supported by NIH pre-doctoral training grant GM 08537. S.C.R. was supported by NIH postdoctoral fellowship GM 071114. This work was also funded by NIH grant GM 25521. We wish to thank Drs. E. Rachlin and J. Muller for assistance with mass spectrometry experiments.
Footnotes
Publisher's Disclaimer: This PDF receipt will only be used as the basis for generating PubMed Central (PMC) documents. PMC documents will be made available for review after conversion (approx. 2–3 weeks time). Any corrections that need to be made will be done at that time. No materials will be released to PMC without the approval of an author. Only the PMC documents will appear on PubMed Central -- this PDF Receipt will not appear on PubMed Central.
References
- 1.Thulasiram HV, Erickson HK, Poulter CD. Science. 2007;316:73–76. doi: 10.1126/science.1137786. [DOI] [PubMed] [Google Scholar]
- 2.Kuzuyama T, Seto H. Nat. Prod. Rep. 2003;20:171–183. doi: 10.1039/b109860h. [DOI] [PubMed] [Google Scholar]
- 3.Rohmer M. In: Comprehensive Natural Products Chemistry. Cane DE, editor. vol. 2. Pergamon Press; 1999. pp. 45–67. [Google Scholar]
- 4.(a) Carrigan CN, Poulter CD. J. Am. Chem. Soc. 2003;125:9008–9009. doi: 10.1021/ja0350381. [DOI] [PubMed] [Google Scholar]; (b) Lee S, Poulter CD. J. Am. Chem. Soc. 2006;128:11545–11550. doi: 10.1021/ja063073c. [DOI] [PubMed] [Google Scholar]; (c) RamosValdivia AC, vanderHeijden R, Verpoorte R. Nat. Prod. Rep. 1997;14:591–603. doi: 10.1039/np9971400591. [DOI] [PubMed] [Google Scholar]
- 5.Kaneda K, Kuzuyama T, Takagi M, Hayakawa Y, Seto H. Proc. Natl. Acad. Sci. USA. 2001;98:932–937. doi: 10.1073/pnas.020472198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) de Ruyck J, Rothman SC, Poulter CD, Wouters J. Biochem. Biophys. Res. Commun. 2005;338:1515–1518. doi: 10.1016/j.bbrc.2005.10.114. [DOI] [PubMed] [Google Scholar]; (b) Steinbacher S, Kaiser J, Gerhardt S, Eisenreich W, Huber R, Bacher A, Rohdich F. J Mol Biol. 2003;329:973–982. doi: 10.1016/s0022-2836(03)00527-8. [DOI] [PubMed] [Google Scholar]
- 7.Laupitz R, Hecht S, Amslinger S, Zepeck F, Kaiser J, Richter G, Schramek N, Steinbacher S, Huber R, Arigoni D, Bacher A, Eisenreich W, Rohdich F. Eur. J. Biochem. 2004;271:2658–2669. doi: 10.1111/j.1432-1033.2004.04194.x. [DOI] [PubMed] [Google Scholar]
- 8.Rothman SC, Helm TR, Poulter CD. Biochemistry. 2007;46:5437–5445. doi: 10.1021/bi0616347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bornemann S. Nat. Prod. Rep. 2002;19:761–772. doi: 10.1039/b108916c. [DOI] [PubMed] [Google Scholar]
- 10.Hemmi H, Ikeda Y, Yamashita S, Nakayama T, Nishino T. Biochem. Biophys. Res. Commun. 2004;322:905–910. doi: 10.1016/j.bbrc.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 11.Poulter CD, Winstein S. J. Am. Chem. Soc. 1970;92:4282–4288. [Google Scholar]
- 12.Wu Z, Wouters J, Poulter CD. J Am. Chem. Soc. 2005;127:17433–17438. doi: 10.1021/ja056187h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiang JL, Christensen DJ, Poulter CD. Biochemistry. 1992;31:9955–9960. doi: 10.1021/bi00156a014. [DOI] [PubMed] [Google Scholar]
- 14.Griller D, Ingold KU. Acc. Chem. Res. 1980;13:317–323. [Google Scholar]
- 15.Krishnamurthy V, Rawal VH. J. Org. Chem. 1997;62:1572–1573. [Google Scholar]
- 16.Ghisla S, Massey V, Choong YS. J. Biol. Chem. 1979;254:10662–10669. [PubMed] [Google Scholar]
- 17.Hoshino T, Tamegai H, Kakinuma K, Eguchi T. Bioorg. Med. Chem. 2006;14:6555–6559. doi: 10.1016/j.bmc.2006.06.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPORTING INFORMATION AVAILABLE: NMR Spectra for conversion of cIPP to cDMAPP. Experimental details for large scale inactivation experiments. LC-MS chromatogram with ESI for the flavin adduct. ESI spectra for oIPP/flavin adducts formed in H2O and D2O. Spectra of enzyme complexes following incubation and following guanidium-HCl denaturation. This material is available free of charge via the Internet at http://pubs.acs.org