Abstract
At its carboxylate-bridged diiron active site, the hydroxylase component of toluene/o-xylene monooxygenase activates dioxygen for subsequent arene hydroxylation. In an I100W variant of this enzyme, we characterized the formation and decay of two species formed by addition of dioxygen to the diiron(II) form by rapid-freeze quench (RFQ) EPR, Mössbauer, and ENDOR spectroscopy. The dependence of the formation and decay rates of this mixed-valent transient on pH and the presence of phenol, propylene, or acetylene, was investigated by double-mixing stopped-flow optical spectroscopy. Modification of the α-subunit of the hydroxylase after reaction of the reduced protein with dioxygen-saturated buffer was investigated by tryptic digestion coupled mass spectrometry. From these investigations, we conclude that (i) a diiron(III,IV)–W• transient, kinetically-linked to a preceding diiron(III) intermediate, arises from the one-electron oxidation of W100; (ii) the tryptophan radical is deprotonated; (iii) rapid exchange of either a terminal water or hydroxide ion with water occurs at the ferric ion in the diiron(III,IV) cluster; and (iv) the diiron(III,IV) core and W• decay to the diiron(III) product by a common mechanism. No transient radical was observed by stopped-flow optical spectroscopy for reactions of the reduced hydroxylase variants I100Y, L208F, and F205W with dioxygen. The absence of such species, and the deprotonated state of the tryptophanyl radical in the diiron(III,IV)–W• transient, allow for a conservative estimate of the reduction potential of the diiron(III) intermediate as lying between 1.1 and 1.3 V. We also describe the X-ray crystal structure of the I100W variant of ToMOH.
Keywords: toluene monooxygenase, carboxylate-bridged diiron proteins, oxygen activation, hydrocarbon oxidation
Metal-activated dioxygen species are capable of oxidizing a broad range of substrates (1–4). In synthetic systems, these units are generated in a solvent that is inert to oxidation. The peptide matrix surrounding the active site protects reactive intermediates in protein systems. Enzymes are able to coordinate reduction of an active metal center and dioxygen activation with substrate binding, thereby assuring that reactive metal-oxygen units are generated only when the substrate is available (5, 6). The active sites in carboxylate-bridged diiron (CBDI) proteins are housed within a four-helix bundle and shielded by the protein framework (7–9). As a consequence, reactive intermediates such as Q, an oxo-bridged diiron(IV) center, in MMOH and X, a mixed-valent diiron(III,IV) cluster, in RNR-R2 can accumulate and be characterized by a number of spectroscopic methods (10, 11).
The primary coordination spheres of the diiron centers in the CBDI enzyme superfamily are remarkably similar, yet the high-valent intermediates Q and X are observed only in MMOH and RNR-R2, respectively (12). Although the capacity to form these iron(IV)-containing units is reserved for these two enzymes, proteins in the superfamily family activate dioxygen by analogous mechanisms. The reduced diiron(II) core reacts rapidly with O2 to generate a peroxo-bridged diiron(III) species as the first observable intermediate. No other oxygenated intermediates are observed in Ft and Δ9D, and the peroxodiiron(III) unit is detectable in Δ9D only when the substrate-carrier protein conjugate is bound to the desaturase. This behavior has been ascribed to stearoyl-ACP blocking access to the active site via a large substrate channel (13). To date, no oxygenated intermediates have been reported for reactions of diiron(II) or diiron(III) forms of rubrerythrin with either dioxygen or hydrogen peroxide, respectively. From its crystal structure, the active site of rubrerythrin appears to be more accessible to solvent and buffer components than those of other enzymes in the family (14). The accumulation of reactive oxygenated iron(III) and iron(IV) intermediates would therefore appear to be correlated with accessibility of the diiron center to potential quenching moieties in the medium.
At the beginning of the present study, no transient species with UV-visible absorption bands had been detected kinetically for the reaction of reduced ToMOH with dioxygen. In the crystal structure of the native enzyme, a large channel for substrate access and/or product egress, extending from the protein surface to the active site pocket, was identified (15). Site-directed mutagenesis studies of residues within the channel of ToMOH were therefore undertaken to evaluate the possibility that reactive intermediates might accumulate if solvent or buffer components were occluded from the active site in this system. Residues I100, F198, F205, and H96 form a hydrophobic portal to the active site at the end of the solvent-accessible channel in ToMOH (Figure 1). Of these residues, I100 is especially noteworthy. The analogous residue in MMOH, L110, has been proposed to function as a gate for substrate entry to, and/or product egress from, the active site during catalysis (7). Residue L98 in hemerythrin might perform a similar function, controlling solvent access to the diiron core (16). We therefore wondered whether mutation of I100 would sufficiently isolate the active site pocket from the channel to shield the dimetallic center from buffer components or solvent, allowing accumulation of reactive intermediates to observable levels. Residue F205, which is on the opposite side of the channel from I100, and L208, which is farther from the active site pocket, were also selected as targets for mutagenesis (Figure 2). We substituted I100 with tyrosine and tryptophan, L208 with phenylalanine, and F205 with tryptophan. Modeling structures of these variant hydroxylases using the native crystal structure suggested that they would afford maximal closure of the channel without disrupting the protein fold of the surrounding matrix.
Figure 1.

View of the active site pocket of ToMOH from the substrate access channel. The diiron atoms and the hydroxylase are depicted as orange spheres and as a grey ribbon diagram, respectively. The opening to the pocket is bordered by residues I100, T201, F205 and F196. I100 is analogous to the proposed leucine gate, L110, in MMOH.
Figure 2.
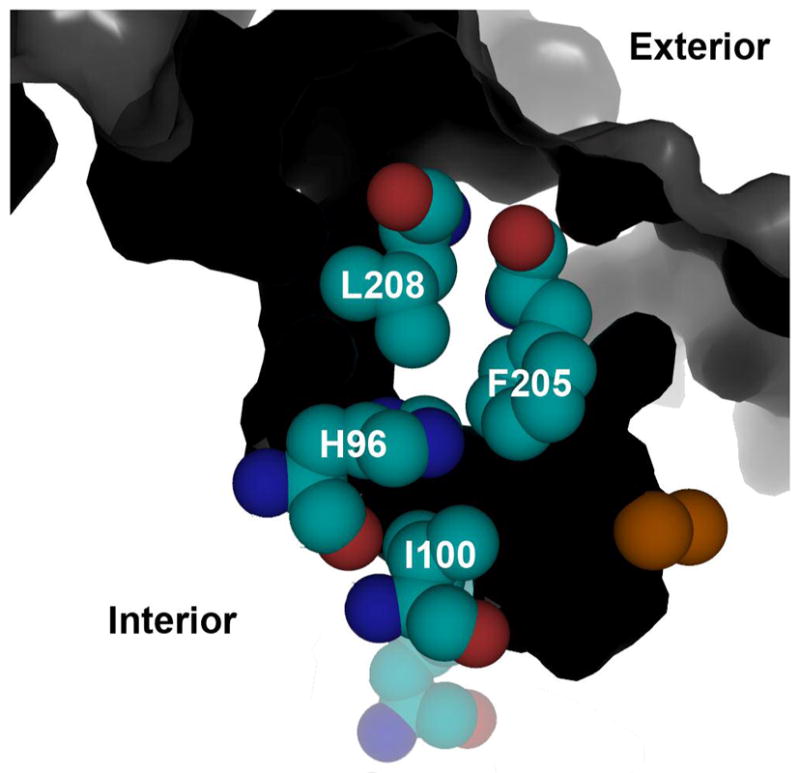
A substrate access channel (black) in ToMOH extends from the protein surface to the diiron active site. Residues H96, I100, and F205 form a hydrophobic region that joins the active site pocket to the channel. Residue L208 resides farther from the diiron active site, forming part of the channel wall. The iron atoms are depicted as orange spheres.
Preliminary investigations of reaction of ToMOH I100W with dioxygen were reported previously (17). From this initial study, dioxygen activation at the diiron(II) core was discovered to produce an optically tranparent diiron(III) intermediate that could be identified by Mössbauer spectroscopy. This intermediate oxidizes the nearby W100 to form a mixed-valent diiron(III,IV) unit coupled to a protein-based tryptophanyl radical. This species subsequently decays to the resting, oxidized diiron(III) state. The process by which the diiron(III) intermediate oxidizes the nearby W residue, and the subsequent decay of the mixed-valent center and the protein-based radical, have been investigated in considerably detail and form the basis for this report. The reactivity of other channel-blocking mutations, I100Y, L208F, and F205W was also investigated under pre-steady-state and steady-state conditions. We include these results included because they provide additional insight into the nature of the important diiron(III) transient.
Experimental Methods
General Considerations
Plasmids containing the genes for the ToMO components were supplied by the laboratory of Professor Alberto Di Donato, Naples, Italy. Recombinant expression and preparation of the ToMO component proteins were carried out as described elsewhere (15). ToMOH variant proteins were prepared by the same protocol as used for the native enzyme. The iron content, measured by the ferrozine assay, ranged from 4.2 to 4.6 iron atoms per ToMOH dimer for all samples. Isotopically enriched ToMOH I100W protein for Mössbauer and ENDOR spectroscopy was obtained by expression in LeMaster’s media containing 57FeCl3 or tryptophan-d5, selectively labeled on the indole ring (Cambridge Isotope Labs, Andover, MA) (18). The solution of 57FeCl3 was prepared by dissolving 57Fe powder (96.7% isotopic purity, Advanced Materials Technologies Ltd., Nes-Ziona, Israel) in concentrated hydrochloric acid. Deuterium oxide was purchased from Cambridge Isotope Labs and all other reagents were acquired from Aldrich Chemical Company. HPLC experiments were carried out with a Vydac protein & peptide C18 column connected to a Waters 600s controller and a Waters 2487 dual wavelength absorbance detector. Optical absorption spectra were recorded with an HP8452 diode-array spectrophotometer.
Crystallization and Data Collection
Crystallization conditions for ToMOH I100W were as published (19). Purified ToMOH I100W protein was exchanged into buffer containing 10 mM MES, pH 7.5, and 10% (v/v) glycerol to a final concentration of 50 μM. A 2 μL aliquot of this solution was mixed with 1 μL of a native ToMOH micro seed stock solution containing 2.3 M (NH4)2SO4, 100 mM HEPES, pH 7.5, and 2% (v/v) PEG 400, as well as 1 μL of precipitant solution. The precipitant solution contained 2.0–3.0 M (NH4)2SO4, 100 mM HEPES, pH 7.5, and 2% (v/v) PEG 400. Cryogenic solution for data collection contained the precipitant solution with added 20% (v/v) glycerol. X-ray diffraction data were collected at the SSRL on beam line 9-2 using the BLU-ICE data collection suite (20). Crystal annealing was conducted as described for 1.85 Å native ToMOH, using a 5.0-s pause in the cryo stream flow (19). Diffraction data were integrated and scaled in HKL2000 (21).
Structure Determination and Refinement
Phasing of the native ToMOH data was accomplished by using EPMR and 1.85Å native ToMOH coordinates (PDB code 2INC) in which all non-protein atoms and the side chains of the iron coordinating ligands and residues I100, T201, N202, Q228, S232, and R233 removed as a starting model (15, 22). Subsequent models were built in Coot and refined using REFMAC5 in CCP4 and CNS (23–25). Simulated annealing composite omit maps were generated in CNS (26). MSDchem ideal coordinates as well as CNS topology and parameter files for glycerol, PEG 400, and MOPS heterocompounds were obtained from the HIC-UP database (residue codes GOL, P6G, and MPO, respectively) (27).
Site Directed Mutagenesis of the ToMOH α-Subunit
The mutations were introduced by site directed mutagenesis on the parent pET-22b(+)/touBEA plasmid with an MJ Research MiniCycler using DNA polymerase pfU Turbo, dNTPs, and reaction buffer (Stratagene, La Jolla, CA) according to the manufacturer’s protocol. The sequences for oligonucleotides (Invitrogen, Carlsbad, CA) used as primers are given in Table S1. Primers for the mutation I100W were as reported previously (17). PCR products were transformed into E. coli XL-1 Blue supercompetent cells (Strategene, La Jolla, CA) by heat-shock as described by the manufacturer and grown overnight on LB-Agar plates containing ampicillin (300 μg/mL). Five colonies from each plate were picked and grown in 5 mL cultures (LB media, 300 μg ampicillin/mL) for 20 h. Cells were pelleted at 3500 rpm for 15 min and the plasmids were isolated with a Qiagen Mini-Prep kit. Isolated plasmids were submitted for sequencing in the forward and reverse directions to the Biopolymers core facility in the Center for Cancer Research (M.I.T.).
Steady-State Activity Assays and Product Determinations for ToMOH Variants
A colorimetric assay was used to detect catechol that formed during steady-state hydroxylation of phenol by these variant hydoxylases (28). Catechol-2,3-dioxygenase cleaves catechol to form 2-hydroxymuconic semialdehyde, which can be monitored by measuring the absorption at 410 nm (ε = 1260 M−1cm−1). Assays were conducted at 25 °C in 0.1 M Tris/HCl pH 7.5 in a final volume of 1 mL. Reaction mixtures contained 0.15 μM ToMOH, 10 μM ToMOD, 4 μM ToMOC, 30 nM ToMOF, and saturating amounts of catechol-2,3-dioxygenase. Steady-state hydroxylation of phenol (1 mM) was initiated with NADH to a final concentration of 1 mM.
To determine if the regiospecificity of hydroxylation was altered by these mutations, products of steady-state turnover for phenol were identified by HPLC. The ToMO protein component concentrations were 0.3 μM ToMOH, 2 μM ToMOD, 4 μM ToMOC, and 30 nM ToMOF in 0.1 M Tris/HCl pH 7.5 (150 μL). Assay solutions contained 1 mM phenol, and were initiated with NADH to a final concentration of 2 mM. Reaction mixtures were incubated at 25 °C for 15 min, quenched with 50 μL TFA, centrifuged for 10 min at 14000 × g, frozen in liquid nitrogen, and stored at 20 °C. Samples were thawed and 100 μL of the supernatant was injected on to the Vydac column. HPLC conditions for separation of hydroxylated products from phenol were 0% buffer B for 7 min, 0% to 40% B for 1 min (linear gradient), 40% to 100% for 7 min (linear gradient), and 100% B for 3 min (A: 1% acetonitrile, 98.8% ddH2O, 0.2% TFA; B: 49.9% acetonitrile, 49.9% ddH2O, 0.2% TFA). Absorption at 280 nm was monitored with time for all samples. Retention times for catechol, resorcinol, and hydroquinone were determined under these conditions.
Stopped-Flow Optical Spectroscopy
A HiTech DX2 stopped flow instrument was made anaerobic by treatment with an anaerobic solution of sodium dithionite (> 4 mM). This solution was allowed to stand in the drive syringes and flow lines for at least 15 min to ensure complete scavenging of dioxygen. The instrument was then flushed with anaerobic 25 mM MOPS buffer at the appropriate pH immediately prior to use. Solutions containing the hydroxylase and regulatory protein were made anaerobic by cycles of vacuum gas exchange with nitrogen and transferred to a Vacuum Atmospheres MO-20 anaerobic chamber, where they were reduced with excess sodium dithionite in the presence of methyl viologen. The reduced protein was dialyzed (8000 MWCO) twice against 25 mM MOPS at specific pH values. Samples were then loaded either into tonometers or Hamilton gas-tight sample-lock syringes and loaded into the anaerobic stopped-flow instrument and mixed against O2-saturated 25 mM MOPS buffer at a specific pH. The software packages KinetAsyst 3.14 (HiTech Scientific, UK) or Kaleidagraph 3.5 (Synergy Software, Reading, PA) was used to fit the time dependence of the optical data.
Reaction of ToMOHred I100Y, F205W, and L208F with O2
Concentrations of the variant hydroxylases after mixing varied from 25 μM to 150 μM. Reaction mixtures contained three equivalents of ToMOD to one equivalent of ToMOH. The reduced hydroxylase pre-complexed with the regulatory protein was dialyzed against buffer, pH 7.0, and allowed to react with dioxygen-saturated buffer of the same pH. The temperature of the stopped-flow instrument was maintained at 4.0 ± 0.1 °C. Multi-wavelength data were collected between 350 and 750 nm with a xenon arc lamp and a diode array detector.
Solvent Kinetic Isotope Effect (SKIE) for Formation and Decay of the ToMOH I100W Transient
After reduction, protein mixtures were dialyzed anaerobically against either 25 mM MOPS buffer pD 6.61 in D2O or pH 7.0 in H2O and allowed to react with dioxygen. The reaction was carried out over the range 4.0 and 36.0 °C. The concentration of ToMOH I100W:3ToMOD varied from 25 to 60 μM in the optical cell. The optical traces collected in single-wavelength mode at 500 nm were fit to an A→B→C model to determine the formation and decay rate constants.
Effect of pH on the Reaction of ToMOHred I100W with O2
In these experiments, the pH of the reaction mixture was adjusted either by using double-mixing mode to set the pH after dialysis and prior to oxygenation in the second push, or in single-mixing mode with dialysis carried out using buffers at specific pH values. For double-mixing experiments, the concentrations of the hydroxylase and regulatory protein after mixing were 94 μM and 282 μM, respectively. The reduced protein was dialyzed against buffer with a pH value of 6.6. In the first push, the solution of the hydroxylase and regulatory protein was mixed with anaerobic buffer, pH 6.6 or 8.0, and allowed to age for 1 min. In the second push, the aged protein solution was allowed to react with oxygenated buffer, pH 6.6 or 7.2, and data were collected in multi- and single-wavelength modes. The instrument temperature was maintained at 4.0 °C. For single-mixing experiments, the concentrations of the hydroxylase and regulatory protein in the optical cell were 18 μM and 45 μM, respectively. After reduction, the first dialysis was carried out against 25 mM MOPS pH 7.0. The pH of the buffer was 6.5, 7.0, or 7.5 for the second dialysis. The reduced anaerobic samples were mixed at 4.0 °C with oxygenated 25 mM MOPS buffer at the appropriate pH values. Data were collected at 500 nm for 100 to 200 s.
Effect of Substrates on the Decay Rate of the ToMOH I100W Transient
This double-mixing experiment was carried out at 4.0 °C monitoring the absorption 500 nm. The concentration of ToMOH I100W:3ToMOD after mixing was 25 μM. Reduced ToMOH I100W:3ToMOD was dialyzed against buffer at pH 7.0, and loaded into the anaerobic stopped-flow instrument. In the first push, the reduced hydroxylase was reacted with dioxygen-saturated buffer (pH 7.0) for 3.9 s. The aged solution was subsequently mixed with 25 mM MOPS pH 7.0 containing phenol, acetylene, or propylene. The concentration of phenol after mixing ranged from 25 μM to 5.2 mM. For acetylene and propylene, an aliquot (10 mL) of buffer was sparged with the gas for 1.5 h to obtain saturated solutions of the substrate (7.7 mM or 42.4 mM, respectively) (29). Optical data were collected for 10 s to 200 s and fit to either one or two exponential functions.
Rapid-Freeze Quench Sample Preparation
Protein solutions of ToMOH I100W:3ToMOD were reduced and dialyzed against 25 mM MOPS buffer pH 7.0 as described for stopped-flow experiments. Reduced mixtures were loaded into gas-tight syringes for an Update Instruments 1000 ram drive system connected to a model 705A computer controller. The RFQ instrumentation has been described in detail elsewhere (30). Reduced protein was mixed with dioxygen-saturated buffer, and allowed to age for reaction times between 0.03 s and 900 s, after which the protein solution was quenched in isopentane at −140 °C. The frozen protein solutions were packed into X-band EPR tubes and Mössbauer sample cups. Samples for ENDOR spectroscopy were quenched 4 s after mixing with dioxygen-saturated buffer, and packed into Q-band EPR tubes. All RFQ samples were stored in liquid nitrogen until spectra were acquired.
Mössbauer Spectroscopy
ToMOH I100W isotopically enriched with 57Fe was used to generate RFQ samples for Mössbauer spectroscopy. The concentration of ToMOH I100W:3ToMOD after mixing was 290 μM. Mössbauer spectra were recorded at 4.2 K in an applied magnetic field of either 50 mT or between 1 T and 8 T applied parallel to the γ-beam on instrumentation described elsewhere (30). The zero velocity refers to the centroid of a room temperature spectrum of an Fe foil.
X-Band EPR Spectroscopy
Samples were generated with ToMOH I100W that contained either 56Fe or 57Fe. The concentration of ToMOH I100W:3ToMOD in the quenched reaction mixtures was 198 μM. EPR spectra at g = 2.00 were recorded at 30 K with the following parameters: power = 0.02 mW; frequency = 9.65 GHz; modulation frequency = 100 kHz; modulation amplitude = 5 G; gain = 6.3 × 104. EPR spectra at g = 16 were recorded at 8 K with the following parameters in parallel mode: power = 20 mW; frequency = 9.39 GHz; modulation frequency = 100 kHz; modulation amplitude = 10 G; gain = 6.3 ×104.
ENDOR Spectroscopy
Samples were prepared either with 25 mM MOPS in D2O (pD 6.6) or H2O (pH 7.0), and the hydroxylase contained either natural abundance 56Fe or was isotopically enriched with 57Fe. For samples prepared with D2O-containing buffers, the reduced protein was dialyzed against deuterated buffers. The concentration of ToMOH I100W:3ToMOD in ENDOR samples ranged from 250 μM to 350 μM. 1H- and 2H-Mims ENDOR spectra were recorded on instrumentation described elsewhere (31).
Enzymatic Digestion and Mass Spectrometry of ToMOH I100W
Reduced ToMOH I100W was prepared as described above for the stopped-flow experiments. A 10-μL control aliquot of the ToMOH I100W:3ToMOD mixture was removed prior to making the protein anaerobic. The remainder of the protein was reduced with excess sodium dithionite and dialyzed under anaerobic conditions against 25 mM MOPS pH 7.0. The reduced protein was mixed with buffer oxygenated with either natural abundance 16O2 or enriched 18O2 (95%, Cambridge Isotope Labs, Andover, MA) in the stopped-flow instrument. The polypeptide chains of reacted and unreacted oxidized protein solutions were separated on a 4–20% Tris/HCl SDS-PAGE gel run at 200 V for 45 min. The band corresponding to the γ-subunit was excised and digested with trypsin according to the manufacturer’s protocol (New England Biolabs, Ipswich, MA). Positive ion MALDI-TOF mass spectrometry of digests was carried out with a Voyager DE-STR MALDI-TOF mass spectrometer (Applied Biosystems, Foster City, CA), installed in the Biopolymers Core Facility of the M.I.T. Center for Cancer Research. The instrument was operated in reflector mode with an accelerating potential of 20 kV and mass resolution of at least 1:10000. The MALDI matrix was α-cyano-4-hydroxybenzoic acid. Protein digest samples were mixed with a 10 mg/mL matrix solution in a 1:1 ratio and were deposited on the MALDI plate. Mass spectra were obtained using a nitrogen UV laser (337 nm). Each mass spectrum is the average of 50 laser shots. The mass spectrometer was calibrated as per the manufacturer’s standard operating protocols, with a mixture of peptides of known mass that were spotted on the MALDI plate adjacent to the sample. Mass spectra were processed using the Applied Biosystems Data Explorer software.
LC-MS analyses of the protein digests were carried out using a Tempo nano HPLC system (Applied Biosystems, Foster City, CA) coupled on-line to a QSTAR Elite quadrupole-time-of-flight tandem mass spectrometer (MDS Sciex/Applied Biosystems, Foster City, CA), installed in the Proteomics Core Facility of the M.I.T. Center for Cancer Research. The mass spectrometer was calibrated as per the manufacturer’s standard operating protocols, with the fragment ions from a peptide of known sequence. Separation of proteolytic peptides was carried out on a C18 capillary HPLC column (Michrom Bioresources, Auburn, CA) and a water-acetonitrile (with 0.1% formic acid) solvent gradient at a flow rate of 300 nL/min. Mass spectral data were acquired and processed with the Applied Biosystems Analyst QS software. Data acquisition was performed using the software “Information Dependent Acquisition” mode with each MS scan, where peptide ion m/z values were measured, followed by four MS/MS scans, where fragment ion spectra of the four most abundant precursor ions were acquired. A temporary exclusion list was generated by the software after each MS/MS data acquisition to minimize the generation of duplicate MS/MS spectra from the same precursor ion.
Results
X-Ray Crystal Structure of ToMOH I100W
ToMOH I100W crystallization time and morphology matched that published for native and Mn(II)-ToMOH (19). The best-diffracting I100W crystal yielded a 2.1-Å resolution data set, which was used for the structure determination. Successful phasing followed by multiple rounds of model fitting and refinements afforded the current structure with refinement statistics shown in Table S2. The diffraction data exhibit a moderately low overall completeness of 84.7%, but there are high I/σ ratios and redundancy counts in all resolution shells. The global folds of the α-, β-, and γ-subunits of ToMOH I100W are within error the same as those for the native protein, except for predicted differences in the channel interior, with a Cα-to-Cβ r.m.s.d of 0.183 Å. The indole ring of the W100 side chain was modeled in two different rotameric form as required by the observed electron density. One position orients the plane of the indole ring almost parallel to the Fe-Fe vector (Figure 3, Position A), whereas the second positions Cζ3 through Cε3 of the indole side chain toward the diiron active site (Figure 3, Position B). The two rotamers were assigned equal occupancies of 50%, which led to average refined Debye-Waller factors for the two W100 side chains of 43.1 Å2 for Position A and 40.7 Å2 for Position B; the average B-value for all other atoms in the α-subunit is 50.4 Å3. Both positions of the W100 side chain block direct access to the active site, as can be seen from van der Waals surface renderings of the channel interior (Figure 4). Distances between the indole ring and the active site iron atoms range from 6.1 to 12.1 Å, with average values of 10.6 and 8.2 Å for W100 positions A and B, respectively (Table S3). The shortest distance is that between Cζ3 (Position B) and Fe1 at 6.1 Å, and the longest is that between Cζ2 (Position A) and Fe2 at 12.1 Å.
Figure 3.
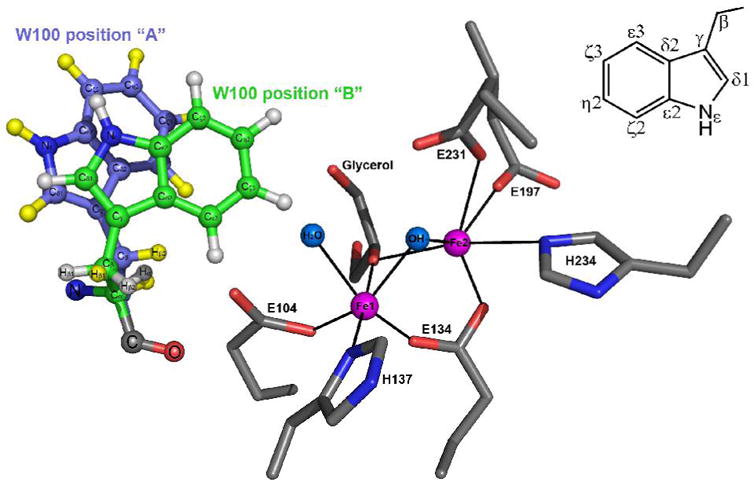
Conformations adopted by W100 in the crystal structure. The aromatic side-chain occupies two positions with populations of 50% for positions A and position B. The indole ring in A is oriented parallel to the diiron center whereas Cζ3 to Cε3 are pointed towards the diiron center in second conformation. The inset shows the labeling scheme referred to for the atoms in the indole ring.
Figure 4.

Diiron center blocking effect of W100 in ToMOH I100W (Bottom) in comparison to MMOH (Top, left) and native ToMOH (Top, right). Amino acids in the region of the active site pocket and iron atoms are shown as spheres in grey (carbon), red (oxygen), blue (nitrogen), and green (iron). The two partially occupied positions of the W100 side chain in the variant ToMOH structure are highlighted in yellow.
Flanking the W100 side chain are two molecules of glycerol, presumably derived from the purification or cryo buffers used in sample preparation. These glycerol molecules, together with the W100 indole ring, occupy the space containing hexaethylene glycol encountered in the recent crystal structures of ToMOH (19). One of these two glycerol molecules resides in the active site cavity and, in a manner similar to that observed for ethanol in MMOH, asymmetrically bridges the active site iron atoms, replacing the hydroxide ion typically found syn to the ligating histidine residues (Figure 3) (32). With exception of this {μ-−OCH(CH2OH)2} bridging glycerol anion, the diiron center in ToMOH I100W is otherwise identical to that in native ToMOHox (15, 19). The B-factors for atoms in the coordinated and fully occupied glycerol molecule average 42.7 Å2.
Steady-State Product Distribution and Activity of ToMOH Variants with Phenol as Substrate
The steady-state specific activities of the ToMOH variants for phenol were lower than that observed for wild-type hydroxylase as determined by the coupled assay with catechol-2,3-dioxygenase. The I100W variant had a specific activity of 80 mU/mg compared to 1250 mU/mg for the wild-type protein. ToMOH I100Y, L208F, and F205W showed no activity under the conditions of this assay. Catechol, resorcinol, hydroquinone, and phenol are well separated by the method described in the Experimental Section, with retention times of 13, 11, 6.5, and 15 min, respectively. The peak corresponding to the enzymatic product had a retention time coincident with that of catechol (Figure S1). No peaks corresponding to resorcinol or hydroquinone were observed. Product analyses for the variant hydroxylases revealed catechol to be the only product formed. A trace amount of catechol was observed in assay mixtures for ToMOH I100Y, F205W, and L208F.
Stopped-Flow Optical Study of the Reaction of ToMOH Variants with O2-Saturated Buffer
No transient absorption bands between 350 and 750 nm were observed after mixing solutions of chemically reduced hydroxylase variants I100Y, F205W, or L208F in complex with ToMOD against dioxygen-saturated buffer.
Reaction of Reduced ToMOH I100W with O2 Monitored by Mössbauer Spectroscopy
An initial spectroscopic characterization of the dioxygen activation reaction by the reduced I100W ToMOH variant revealed two transient species, an EPR-silent, optically transparent diiron(III) intermediate that exhibits Mössbauer spectra characteristic of ferric iron (δ= 0.54 mm/s and ΔEQ = 0.67 mm/s) and a spin-coupled diiron(III,IV)–W• chromophore that displays an absorption band at 500 nm, an EPR signal in the g = 2.0 region, and Mössbauer spectral characters indicative of a mixed-valent diiron(III,IV) cluster (17). The magnetic field dependence of the Mössbauer spectrum of the diiron(III,IV)–W• intermediate confirmed a dipolar spin-spin interaction between the diiron(III,IV) cluster and W•. To gain further insight into dioxygen activation by reduced ToMOH I100W variant and to obtain time evolution profiles of Fe species generated in the reaction, Mössbauer and EPR spectra of RFQ samples freeze-quenched during the reaction were collected and analyzed.
Figure 5 shows Mössbauer spectra of reduced protein before (A) and after (B-F) mixing with O2. Because the parameters of these iron intermediates had been determined previously (17), decomposition of the spectra into components corresponding to different Fe species was possible (Figure 5, colored lines). Percentages of Fe-absorption corresponding to species generated at different time points, including others not depicted here, were thereby obtained. On the basis of the Fe/protein ratio determined for the freeze-quenched samples, 3.88 Fe atoms/ToMOH dimer, these relative percentages were converted to accumulation amounts of diiron cluster/protomer for the diiron species at various time points. The results (diamonds) are presented in Figure 6, which shows clearly the formation and decay of the various diiron species. Before mixing with O2, the reduced protein sample contains mainly diiron(II) clusters, 0.96 diiron clusters/ToMOH protomer, the spectrum of which can be modeled as two unresolved quadrupole doublets (Figure 5A, green lines) with parameters given in the figure caption. After mixing with O2-saturated buffer, approximately 40% of the diiron(II) sites react rapidly to form the diiron(III) intermediate (Figure 5, red lines) with a rate constant of ~ 18 s−1. Accumulation of the diiron(III) intermediate reaches a maximum of 0.32 clusters/protomer at 0.14 s and then decays at a rate of ~ 1.1 s−1 (Figure 6, red line and diamonds). The decay of the diiron(III) intermediate parallels the formation of the diiron(III,IV)–W• species (Figure 6, blue line and diamonds), while the unreacted diiron(II) sites stay relatively stable (Figure 6, green line and diamonds) during this decay phase of the diiron(III) intermediate. This result establishes unambiguously that the diiron(III) transient is a true precursor to diiron(III,IV)–W•. The data also reveal that the diiron(III,IV)–W• species reaches a maximum accumulation of 0.33 clusters/protomer at 3.5 s and decays with a rate constant of ~ 0.08 s−1 to generate the diiron(III) product (Figure 6, orange line). The formation and decay rates of the optically silent diiron(III,IV) cluster are linked to that of the optically active W•, because the rates of the former, determined from Mössbauer spectroscopy, agree with those of the latter, determined from optical studies (17, also, see Table 1). Slow oxidation (~ 0.01 s−1) of residual unreacted diiron(II) species accompanies the second phase formation of the diiron(III) product and the generation of other minor, unidentified ferric species. A previously unreported, minor oxidation product (Figure 5, cyan lines) can detected as forming around 1 to 2 s after mixing with O2, after 5 s its accumulation reaches a constant value of ~ 8% of the total iron in the samples. Its Mössbauer parameters (δ= 0.56 mm/s and ΔEQ = 1.77 mm/s) and diamagnetism, revealed by high-field Mössbauer measurements, indicate a diiron(III) cluster. Its kinetic profile suggests that it is a final product. A definitive identification cannot be made, however, due to the small amount of material that accumulates.
Figure 5.
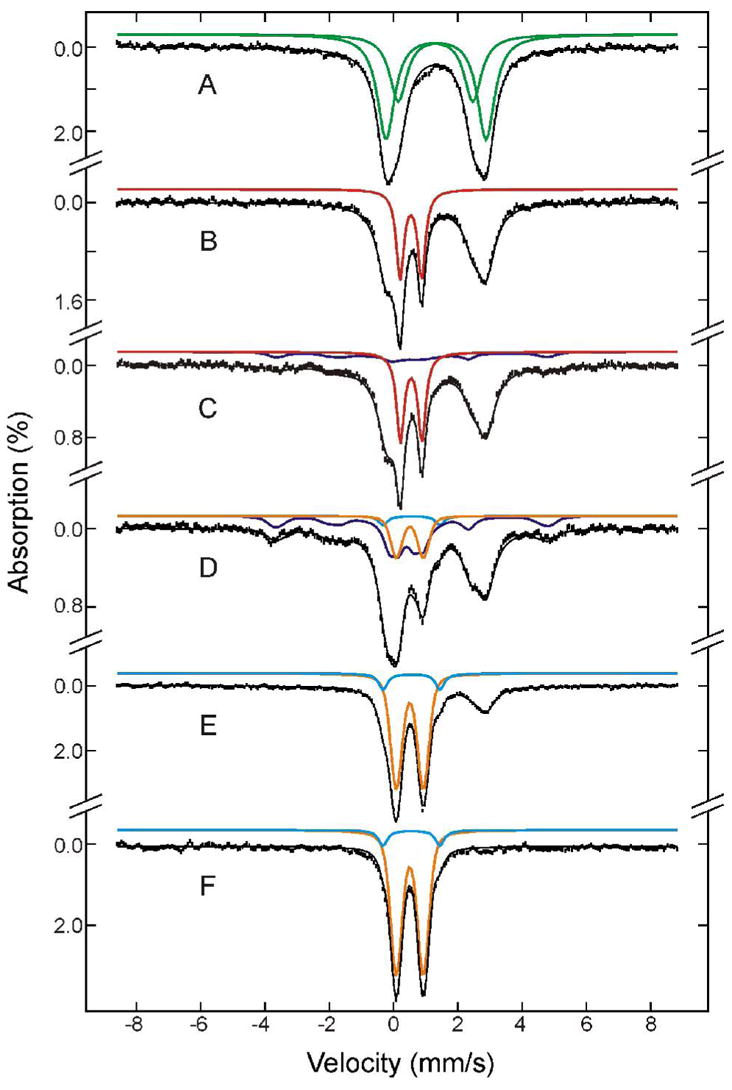
Mössbauer spectra of freeze -quenched samples from the reaction of ToMOHred I100W:3ToMOD with O2. The samples were frozen before mixing (A) and 0.07 s (B), 0.44 s (C), 3.5 s (D), 37 s (E) and 900 s (F) after mixing. The spectra (vertical bars) are collected at 4.2 K in a 50 mT field applied parallel to the γ-beam. The diiron(II) spectrum (A) was simulated as a superposition of two unresolved quadrupole doublets (green lines in A) with an intensity ratio of 1.6:1 for doublet 1:doublet 2. The parameters are δ = 1.32 mm/s, ΔEQ = 3.11 mm/s, and line width = 0.65 mm/s for doublet 1, and δ = 1.31 mm/s, ΔEQ = 2.32 mm/s, and line width = 0.65 mm/s for doublet 2. In B-F, the red, blue, orange and cyan lines are simulated spectra of the diiron(III) transient, diiron(III,IV)-W• intermediate, major diion(III) and minor diiron(III) products, respectively. The spectra of the diiron(III) transient (
 ) and diiron(III,IV)-W• intermediate (
) and diiron(III,IV)-W• intermediate (
 ) are simulated with parameters reported previously (17). The spectrum of the major diiron(III) product (
) are simulated with parameters reported previously (17). The spectrum of the major diiron(III) product (
 ) is modeled with a single quadrupole doublet with δ = 0.51 mm/s, ΔEQ = 0.84 mm/s, and line width = 0.35 mm/s. The spectrum of the minor diiron(III) product (
) is modeled with a single quadrupole doublet with δ = 0.51 mm/s, ΔEQ = 0.84 mm/s, and line width = 0.35 mm/s. The spectrum of the minor diiron(III) product (
 ) is simulated with δ = 0.56 mm/s, ΔEQ = 1.77 mm/s, and line width = 0.33 mm/s. For clarity, the diiron(II) spectral component is not shown specifically in B-F. The simulated spectra are plotted at the following absorption intensity: red, 29% and 27% in B and C, respectively; blue, 16% and 34% in C and D, respectively; orange, 14%, 60% and 65% in D, E, and F, respectively; cyan, 3%, 8% and 8% in D, E, and F, respectively. The black lines overlaid with the experimental spectra are composite spectra including the diiron(II) and all other species mentioned above. Approximately 25% of the total Fe in F appears in the form of broad, featureless absorptions, indicative of ill-defined paramagnetic ferric species. We therefore did not include this iron in the composite spectra.
) is simulated with δ = 0.56 mm/s, ΔEQ = 1.77 mm/s, and line width = 0.33 mm/s. For clarity, the diiron(II) spectral component is not shown specifically in B-F. The simulated spectra are plotted at the following absorption intensity: red, 29% and 27% in B and C, respectively; blue, 16% and 34% in C and D, respectively; orange, 14%, 60% and 65% in D, E, and F, respectively; cyan, 3%, 8% and 8% in D, E, and F, respectively. The black lines overlaid with the experimental spectra are composite spectra including the diiron(II) and all other species mentioned above. Approximately 25% of the total Fe in F appears in the form of broad, featureless absorptions, indicative of ill-defined paramagnetic ferric species. We therefore did not include this iron in the composite spectra.
Figure 6.

Speciation plot for the reaction of ToMOH red I100W:3ToMOD with dioxygen. The diiron(II) starting material (
 ) rapidly converts to a diiron(III) intermediate (
) rapidly converts to a diiron(III) intermediate (
 ) at ~ 18 s−1. The diiron(III) transient evolves to the mixed-valent species (
) at ~ 18 s−1. The diiron(III) transient evolves to the mixed-valent species (
 ) at ~ 1 s−1. The formation and decay rates of the g = 2.0 signal (
) at ~ 1 s−1. The formation and decay rates of the g = 2.0 signal (
 ) from RFQ EPR experiments are within error of the rates for mixed-valent species determined by Mössbauer. The diiron(III) resting state (
) from RFQ EPR experiments are within error of the rates for mixed-valent species determined by Mössbauer. The diiron(III) resting state (
 ) is reformed as the mixed-valent species decays at ~ 0.08 s−1. Ill-defined ferric species formed during reaction of the reduced hydroxylase with dioxygen (see caption to Figure 5) are omitted from this kinetic analysis. The diiron(II) starting material (
) is reformed as the mixed-valent species decays at ~ 0.08 s−1. Ill-defined ferric species formed during reaction of the reduced hydroxylase with dioxygen (see caption to Figure 5) are omitted from this kinetic analysis. The diiron(II) starting material (
 ), the diiron(III) intermediate (
), the diiron(III) intermediate (
 ), mixed-valent diiron(III,IV) transient (
), mixed-valent diiron(III,IV) transient (
 ), g = 2.0 EPR signal (
), g = 2.0 EPR signal (
 ), and the diiron(III) product (
), and the diiron(III) product (
 ) were fit to two-exponential functions.
) were fit to two-exponential functions.
Table 1.
Formation and Decay Rate Constants of the Mixed-Valent Diiron(III,IV)-W• Species Measured by Optical, Mössbauer, and EPR Spectroscopy
| Rate Constant | Optical | Mössbauer | EPR |
|---|---|---|---|
| kf (s−1) | 0.804 (1) | 1.1 | 0.77 |
| kd (s−1) | 0.054 (2) | 0.08 | 0.15 |
Reaction of Reduced ToMOH I100W with O2 Monitored by EPR Spectroscopy
As reported previously (17), two EPR signals were observed during the reaction of the reduced I100W variant hydroxylase with dioxygen, a signal at g = 16, corresponding to diiron(II) centers, and a transient g = 2.0 signal, associated with the diiron(III,IV)–W• intermediate. The g = 16 signal decays in a biphasic manner. The first phase is rapid and could not be simulated accurately with the data collected. This initial decay is complete by ~ 0.17 s, which is consistent with the decay kinetics of the rapidly interacting diiron(II) sites observed in the Mössbauer measurements (vide supra). The second phase was slower and incomplete by 150 s. Figure 7 displays EPR spectra of selected freeze-quenched samples in the g = 2.0 region, showing the rise and fall of the transient EPR signal. The intensity of this transient signal appeared and decayed with rate constants of 0.77 s−1 and 0.15 s−1, maximizing at approximately 4 s (Figure 6, ⋄ and −). The rate constants for formation and decay agree with those reported from the stopped-flow optical and Mössbauer experiments, confirming that this EPR active species corresponds to the spin coupled mixed-valent diiron(III,IV)–W• intermediate (Table 1). The time-dependent EPR data indicate that the fast-reacting diiron(II) protein and the mixed-valent species are not kinetically linked because the rate of decay of the former is much faster than the rate of formation of the latter. This result indicates the presence of an intervening EPR-silent species, namely, the diiron(III) intermediate observed in the Mössbauer spectra of RFQ samples quenched between 0.03 s and 4 s, described above.
Figure 7.

Selected EPR spectra for the transient formed during the reaction of ToMOHred I100W:3ToMOD with O2. The intensity of the signal at g = 2.0 maximizes 4 s (
 ) after mixing reduced protein with dioxygen-saturated buffer. Spectra for samples that were allowed to react for 520 ms (—), 10 s (
) after mixing reduced protein with dioxygen-saturated buffer. Spectra for samples that were allowed to react for 520 ms (—), 10 s (
 ), and 100 s (
), and 100 s (
 ) are also shown.
) are also shown.
Prior to carrying out the 1H and 2H-Mims ENDOR measurements, X-band EPR spectra were recorded on samples quenched 4 s after mixing solutions of ToMOHred I100W:3ToMOD with dioxygen-saturated buffer. The g = 2.0 signal has two contributions, as previously mentioned, one from the diiron(III,IV) cluster and the other from the tryptophan radical. The radical signal is visible at temperatures up to ~ 77 K, whereas the diiron signal is visible only below ~ 40 K (Figure S4). The peak width of the mixed-valent diiron(III,IV)–W° transient is smaller than that reported elsewhere for tryptophan cation and neutral radicals (33).
Kinetic Isotope Effects for Formation and Decay of the Diiron(III,IV)–W• Species
Both the rates of formation, kf, and decay, kd, were sensitive to the hydrogen isotope concentration with normal isotope effects, kH > kD, observed over the examined temperature range (Table S4). The ratio of kH/kD decreased with increasing temperature from 2.51 to 1.97 for kf and from 3.2 to 1.72 for kd. From Arrhenius plots, the formation activation energy is greater in deuterated, 15.6 ± 0.5 kcal/mol, than in protic, 13.5 ± 0.1 kcal/mol, solvent as expected from the ratio of kH/kD (Figure S2). The solvent kinetic isotope effect (SKIE) for the formation process is weakly dependent on temperature and contrasts with the stronger, non-linear dependence for the decay process. The magnitude and sensitivity to temperature of the SKIEs indicate that hydrogen atom transfer or tunneling does not occur in the transition state during the reaction of the diiron(III) intermediate with W100, or for subsequent decay of the mixed-valent diiron(III,IV)–W• species.
Effect of pH on Reaction of ToMOH I100W with O2-Saturated Buffer
As the pH increased from 6.5 and 7.5, the rates of oxidation of W100 by the diiron(III) intermediate and decay of the diiron(III,IV)–W• species also increased (Table S5). From data collected in diode array mode, the absorption maximum of the tryptophanyl radical was unchanged over the examined pH range. To explain the pH dependence, we propose a model in which a rate-limiting deprotonation precedes a fast oxidation reaction. The pH dependence data agree reasonably well with this model to give a calculated proton-independent electron transfer rate of 1.22 s−1 (Figure 8). The decay rate of the transient is also sensitive to pH, following a similar trend as observed for the formation rate. Values of kd increase almost two-fold with an order of magnitude decrease in proton concentration. The data were adequately fit with the model applied to the formation rate, rate-limiting deprotonation followed by oxidation, yielding a rate constant of 0.12 s−1 for the rapid second step of the reaction (Figure 8).
Figure 8.
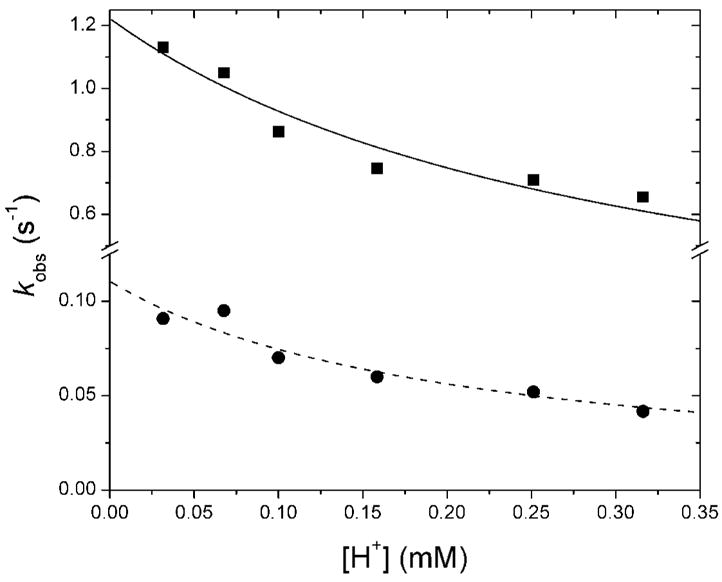
Effect of proton concentration on formation and decay of the diiron(III,IV)–W• species. Increasing pH increases kf and kd of the ToMOH I100W transient. The rate constants both increase with decreasing [H+], with kf (▲) and kd (●) increasing ~ 2-fold between pH 6.5 and 7.5. The y-axis scales differ above and below the break.
Effect of Substrates on the Decay Rate of the ToMOH I100W Transient
The presence of phenol increased the decay rate constant of the tryptophanyl radical from 0.054 s−1 to 13.7 ± 0.4 s−1. The concentration dependence of kd was modeled with a saturation binding model applied to the reaction of oxygenated intermediates in MMOH with alternative substrates, where a substrate-enzyme complex forms prior to reaction (eq 1) (34). From this analysis, the rate constant for reaction with phenol, krxn, and the substrate-enzyme dissociation constant, Kd, are 20 ± 2 s−1 and 2.3 ± 0.5 mM, respectively (Figure 9). In double-mixing experiments employing phenol, the absorbance over all wavelengths increased after decay of the transient. This growth in absorption was modeled as an independent exponential function in all data for which phenol was the substrate. The decay rate of W• is also accelerated in the presence of propylene. The observed decay rate constants are 0.239 ± 0.003 s−1 and 2.8 ± 0.1 s−1 for propylene concentrations of 19 μM and 3.8 mM, respectively. Addition of acetylene to the tryptophanyl radical had little influence on the decay process, increasing the rate constant from 0.079 ± 0.002 s−1 to 0.114 ± 0.006 s−1 (Figure S3).
Figure 9.

Phenol increases the decay rate of the tryptophanyl radical. The dependence of the observed rate on phenol concentration is well modeled by saturation kinetics. The determined reaction rate of the transient, krxn, with phenol and the substrate binding constant, Kd, are 20 ± 2 s−1 and 2.3 ± 0.5 mM, respectively.
1H- and 2H-ENDOR Spectra of the Mixed Valent Diiron(III,IV)–W• Couple
To confirm that the protein-based radical resides on a tryptophan residue, 1,2H-ENDOR spectra were recorded of this transient in H2O/D2O buffers using enzymes that contained either natural abundance tryptophan or the tryptophan selectively deuterated on the indole ring. Although the radical EPR signal overlaps that of the diiron center, the spectrum of the latter is substantially broader. Signals from the two can therefore be distinguished by their field dependence as well as by their different relaxation properties.
The 1H–CW ENDOR spectra collected at fields associated with the radical for samples containing natural abundance tryptophan residues are dominated by signals from the Cβ protons of the residue, with strong hyperfine couplings of ~ 20 MHz, and by those from the indole ring, with smaller couplings (Figure S5). Additional features near νH have been assigned to protein matrix protons in other systems (35). To determine that the radical is centered at tryptophan, we collected 2H-Mims ENDOR spectra from natural-abundance and selectively deuterated, tryptophan(d5-indole), hydroxylase.
The sample prepared with isotopically enriched tryptophan shows 2H-Mims ENDOR signals with hyperfine couplings corresponding to AH > 10 MHz when the field of observation is where the radical signal is strongest (Figure 10). These signals arise from the aromatic protons of the indole ring because they are absent at fields outside the EPR envelope of the radical signal in the natural-abundance sample. The signals unequivocally confirm that the protein-based radical resides on a tryptophan residue.
Figure 10.

2H-Mims ENDOR spectra of the tryptophanyl radical. Protein in this sample was expressed in media containing isotopically enriched tryptophan(d5-indole). Saturation of the transition corresponding to the radical signal at g ~ 2.00 (middle) yields 2H signals from the labeled indole ring. These peaks disappear upon saturation at higher (top) or lower (bottom) g-values. Magnetic fields scanned are reported as g-values in the figure.
1H–CW ENDOR spectra collected at the field corresponding to the maximum intensity of the radical are similar for the diiron(III,IV)–W• transient generated with unlabelled protein in deuterated and protic buffers, but such comparisons are difficult because of strong signals from non-exchangeable protons (Figure S5). As a result, the presence of potentially exchangeable protons was investigated by 2H Mims ENDOR measurements. 2H-Mims ENDOR signals corresponding to 1H coupling of AH ~ 4–8 MHz were observed in samples generated in deuterated buffer (Figure 11). Because these signals are observed at fields across the EPR envelope of the diiron center, and outside that of the tryptophanyl radical, they can be assigned to the diiron center (compare Figures 10 and 11). By analogy to the diiron centers of intermediate X and MMOHmv, for which ENDOR signals from terminal water molecules on the iron(III) ion correspond to species with AH ~ 7–9 MHz, the signals observed in the diiron(III,IV)–W• transient can be assigned to a terminal water molecule or hydroxide ion on the ferric ion (35, 36).
Figure 11.

2H-Mims ENDOR spectra of the diiron(III,IV)–W• species generated in deuterated buffers. Signals arising from protonated species are observed over the swept magnetic fields. The signal shows minor changes upon saturation at g = 1.9875 to 2.0 and does not correlate with the tryptophan radical. The corresponding proton scale is shown for comparison.
If the tryptophanyl radical were protonated at the indole nitrogen atom, one would expect an additional 2H signal, with AH > 10 MHz, at fields associated with this radical (37–41). No such 2H signals are observed, however (Figure 11). The absence of an exchangeable proton associated with the protein radical is consistent with our interpretation of the stopped-flow optical data, where a band with λmax of 500 nm indicated a deprotonated tryptophanyl radical.
Tryptic Digestion and Mass Spectrometric Analyses of ToMOH I100W and its Oxidation Product
The most intense ion envelopes in the MALDI-TOF(+) spectra are between 700 and 2500 m/z for the in-gel tryptic digested α-subunit of as-isolated and O2-reacted ToMOHred I100W. The expected tryptic peptide containing W100, 85ADPGWVSTMQLHFGAWALEEYAASTAEAR113, has a predicted monoisotopic mass of 3165.5 Da for the [M+H]+ parent ion. The ion envelope at m/z = 3166 in the as-isolated sample is well separated from other ions in the spectrum and is assigned to this 29mer peptide containing W100 (Figure 12). Two additional envelopes of lower intensity are present at m/z values of 3182 and 3198. The relative intensities of these latter two envelopes in the reacted sample increase markedly, with that at m/z = 3182 becoming the most intense. The envelopes at m/z = 3182 and 3198 were unchanged in MALDI-TOF spectra of digestion products of reduced ToMOH I100W reacted with 18O2-saturated buffer.
Figure 12.
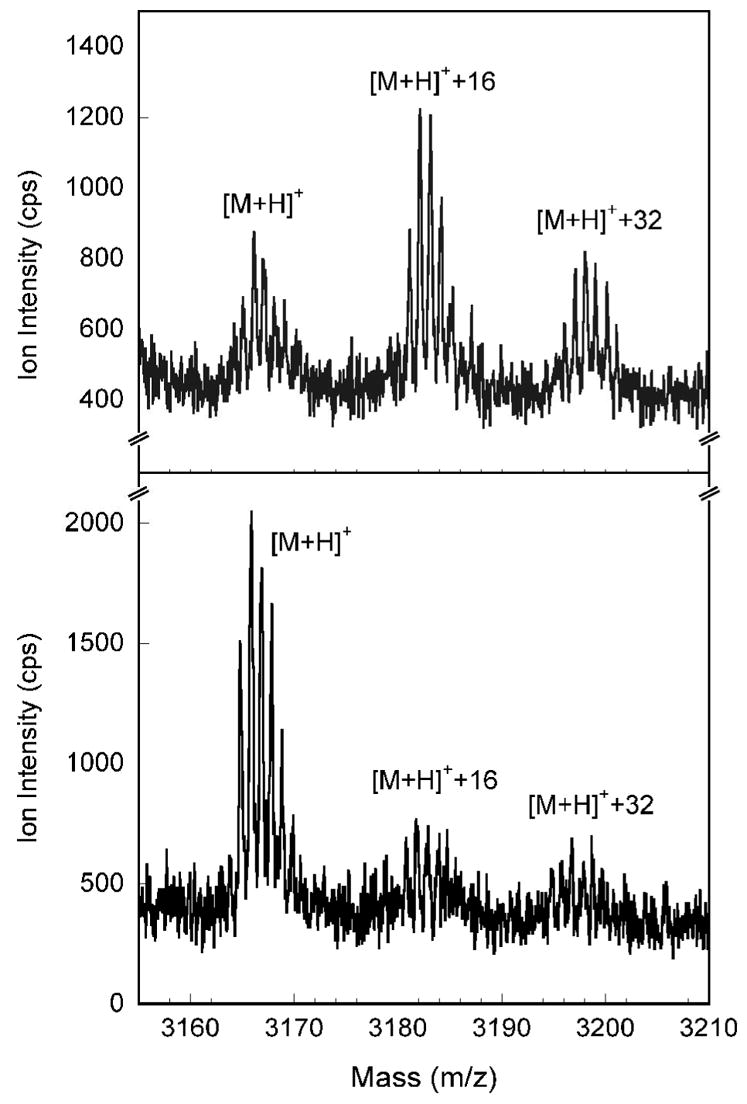
MALDI-TOF spectra of tryptic peptides for the α-subunit of O2-reacted (top) and as-isolated (bottom) ToMOH I100W. The mass of the [M+H]+ ion at 3166 m/z agrees with the predicted mass for the 29mer peptide. Two additional envelopes of 16 and 32 Da higher mass are also present. The relative intensity of these three envelopes changes in the reacted sample, with [M+H]+ less abundant than [M+H]++16.
The tryptic peptide of interest also contains other residues sensitive to oxidation, such as methionine and histidine. To determine the decay pathway chemistry for the W• species, we attempted to identify the specific residue that is oxidized. Fragmention of the tryptic peptide was carried out by ESI(+)-MS/MS. Ions resulting from the [M+H]3+ ion were assigned to respective b and y peptides by considering the fragment ion mass and the peak-to-peak separation within the ion envelope (Figure S6) (42). We could not identify the ions corresponding to every possible fragment because the peak intensities were below our detection threshold. From the ions that were isolated, the b ions limit the site of modification to lie between S91 and E104, a region that includes two other possible sites of oxidation, M93 and H96. The y fragment ions further narrowed the possible region of oxidation, excluding M93. Probable sites of oxidation are therefore limited to H96 and W100.
Discussion
Mixed-Valent Diiron(III,IV)–W• Species as an Entry to Identifying Oxygenated Intermediates in ToMOH
Dioxygen reacts rapidly, kobs ~ 18 s−1, with reduced diiron(II) ToMOH I100W preincubated with ToMOD to yield a diiron(III) intermediate that was only disclosed because it generated the tryptophan neutral radical chromophore. One-electron reduction of this diiron(III) transient by W100 gives rise to a mixed-valent diiron(III,IV) center. The peroxodiiron(III) species in RNR-R2 reacts in a similar manner. Oxidation of W48 and protonation of one of the oxygen atoms in the peroxo-adduct of RNR-R2 facilitates O–O bond cleavage to form intermediate X (43, 44). The proposed mechanism for dioxygen activation in cytochrome P450 enzymes requires protonation of the distal oxygen atoms in the peroxoiron(III) intermediate by a conserved threonine-aspartic acid pair (6, 31). Residue T201 in the active site cavity of ToMOH, which is strongly conserved among the BMMs, could function in a proton shuttle pathway to the diiron core. In MMOH, this threonine is proposed either to deliver protons to the peroxodiiron(III) species directly or, more plausibly, by strategically holding a hydronium ion for proton transfer during reduction of the oxidized diiron(III) center (45, 46). Conversion of the diiron(III) intermediate in ToMOH to the mixed-valent diiron center could proceed by an analogous pathway as that in the heme systems and RNR-R2. In the I100W variant, T201 could help to provide a crucial proton to facilitate cleavage of the O–O bond to form the mixed-valent transient. Protons would be consumed during the oxidative phase of the ToMOH catalytic cycle, which would contrast with MMOH where protons are proposed to be required during reduction of the oxidized diiron(III) core (47). In the native system, T201 may be important for steady-state catalysis if substrate radical generation is a pathway for arene hydroxylation. Pre-steady-state studies of a series of variants at this position would be valuable in discerning its possible role in the mechanism of substrate oxidation.
An investigation of T201 variants of T4MOH demonstrated that this residue does not affect steady-state catalysis (48). For an observable effect under steady-state conditions, however, T201 must be involved in the rate-determining step. Product release is believed to be rate-limiting for hydroxylation by MMOH (49). The products catechol and phenol can bind to the diiron center in ToMOHox isolated after purification or following single-turnover experiments (50). Lack of knowledge of the rate-determining step under steady-state catalysis prevents us from making a meaningful comparison between the earlier steady-state and current pre-steady-state analyses.
Values of δ and ΔEQ for the mixed-valent diiron(III,IV) transient in ToMOH I100W are comparable to those of the Fe(III)Fe(IV) centers of intermediate X in RNR-R2 and QX in MMOH (43, 51). In addition, the 2H-ENDOR spectra of the ToMOH diiron(III,IV) species prepared in deuterated buffer suggest that an exchangeable proton-containing species is coordinated to the iron(III) ion. The observed hyperfine couplings for this protonated ligand are within the range of those reported for terminal water molecules on the ferric centers in MMOHmv and intermediate X (36, 44). This terminal hydroxide ion or water molecule may arise by a mechanism similar to that proposed for RNR-R2 (44). Protonation-aided cleavage of the O–O bond in the diiron(III) species would yield a water molecule that coordinates to the ferric ion. Despite the differences between the diiron(III) intermediate in ToMOH and those of other CBDI enzymes, the ability to form high-valent transients with similar spectroscopic parameters might arise from the homologous primary coordination spheres that these enzymes share.
Our preliminary assignment of the protein-based radical as W• was based on stopped-flow optical experiments. In these studies, a λmax was measured to be 500 nm, within the previously reported range for tryptophanyl radicals (52–54). Since the species (λmax 500 nm) was observed only in the tryptophan variant, this specific residue seemed to be critical for transient formation. 1H- and 2H-Mims ENDOR spectra of this species containing unlabeled and selectively deuterated tryptophan residues firmly establish the presence of a tryptophanyl radical. Samples made in protic and deuterated buffers displayed similar 1H-Mims ENDOR spectra, providing evidence that the tryptophanyl radical is deprotonated, confirming unequivocally our initial assignment that the optically active intermediate observed in stopped-flow studies is a neutral tryptophanyl radical.
The redox potentials of the aromatic side chains of tyrosine and tryptophan are pH dependent with lower proton concentrations favoring oxidation (52, 54). In the present study, we observed the deprotonated form of the radical, the λmax value of which was invariant across the examined pH range of 6.5 to 7.5. We were unable to access pH values near the reported pKa values for the N–H group of the indole ring and the cationic indolyl radical because of protein instability (54, 55). Nonetheless, deprotonation of the indole nitrogen is tightly coupled to electron abstraction because the tryptophan cation radical is not observed. We exclude mechanisms such as hydrogen atom transfer from W100 to the diiron center or a proton tunneling event during the rate-determing step for formation and decay of the diiron(III,IV)–W• transient because of the small magnitude and temperature dependence of the kinetic isotope effects. In enzymes where hydrogen-atom transfer or proton tunneling are proposed to occur, the isotope effects range from 3 to > 100 and are temperature independent (56, 57). The isotope effects observed here are < 3.2 for either rate constant and temperature dependent, both of which are atypical of hydrogen-atom transfer or tunneling.
Disruption of electron transfer pathways from W48 and Y122 to oxygenated diiron intermediates in RNR-R2 results in oxidation of phenylalanine and tyrosine residues near the dimetallic center (58–60). Models of the I100Y mutation estimate the distance between the diiron center and this tyrosine at 7 to 10Å, positioning this residue for possible oxidation (61). Absorption bands corresponding to tyrosyl radicals or iron-catecholate species were not observed during or after the reaction of ToMOHred I100Y with dioxygen. Absence of these bands suggests that, if oxidation of the tyrosine residue were to occur, a stable radical species is not formed and the L-dopa product is unable to coordinate to the diiron center. Alternatively, the diiron(III) intermediate may not be a strong enough oxidant to abstract an electron from Y100. This limitation may arise from the redox potential of tyrosine or from other parameters that affect ET, such as the distance.
As mentioned in the Results Section, a model whereby a slow reversible equilibrium precedes fast oxidation provides the most satisfactory explanation for the pH dependence data. Deprotonation of the tryptophan residue, the diiron(III) intermediate, or an amino acid side-chain required to accept the proton from the tryptophan residue could limit the oxidation rate. The tryptophan cation radical is not observed in our optical studies, implying that proton loss must occur upon oxidation of this residue. Deprotonation of the indole nitrogen before electron abstraction by the diiron(III) intermediate is expected to be unfavorable because the pKa value for this proton is estimated to be 17 (55). Oxidation of W100 prior to proton loss would yield a transient tryptophan cation radical, the pKa of which is 3.7 (54). Deprotonation of the radical cation is therefore predicted to be facile within the examined pH range. Proton loss from the indole ring could limit the oxidation rate if it is required to occur prior to this reaction. Alternatively, if loss occurs after oxidation, the rate limiting deprotonation event might involve either the diiron(III) intermediate or nearby amino acid residues.
A Novel Diiron(III) Intermediate Oxidizes W100
The mechanism for arene hydroxylation by the native hydroxylase could proceed by either one- or two-electron oxidation pathways. If the reaction occurs by initial electron abstraction from substrate, the hydroxylation mechanism could involve generation of a species similar to the mixed-valent diiron(III,IV)–W• center in ToMOH I100W. However, oxidation of the aromatic substrate by electrophilic attack on the π-system would be comparable to the reactions of MMOHperoxo with electron-rich substrates, bypassing stable radical intermediates (62).
Both radical- and cation-derived products were observed for oxidation of RCS probes by T4MO, implying that one- and two-electron oxidation mechanisms might occur in this system (63). Formation of both of these products could reflect two competing mechanisms for substrate oxidation depending upon the ease of approach of these unnatural substrates to the diiron site. If substrate is bound close to the reactive intermediate, then oxidation might proceed by hydride abstraction to generate a substrate cation. Ring-opening of the substrate would give rise to the observed cation-derived product. On the other hand, if the hydrocarbon were not able to approach the oxidizing intermediate, electron abstraction may predominate to generate substrate-radical intermediates. If the diiron(III) species is the only iron-based transient formed during dioxygen activation in the native hydroxylase, the preference for electron-rich substrates, such as the indole ring of W100 or aromatic compounds, by this transient in ToMOH would be similar to the observed reactivity of MMOHperoxo and the peroxodiiron(III) species in RNR-R2. The formation of both radical- and cation-derived RCS products could arise from two distinct intermediates that react with these substrates. In MMOH, Q is proposed to carry out one-electron oxidations and MMOHperoxo by hydride abstraction and epoxidation mechanisms (62). The diiron(III) intermediate observed in ToMOH I100W is the only observed precursor to the mixed-valent diiron(III,IV) center, yet we cannot exclude formation of a short-lived oxo-bridged diiron(IV) intermediate. Formation of such a Q-type intermediate would facilitate electron abstraction pathways and the diiron(III) intermediate would allow for two-electron oxidation mechanisms. We recently reported a detailed examination of dioxygen activation in the native system where a similar diiron(III) intermediate forms and is kinetically competent for substrate hydroxylation (50).
The Mössbauer parameters and lack of optical absorption features in the near-IR region for the diiron(III) transient species are unique among intermediates at this oxidation level in the CBDI enzyme family (4). Specifically, values for λmax near 700 nm, δ > 0.6 mm/s, and ΔEQ > 1 mm/s are characteristic parameters of μ-1,2-peroxodiiron(III) clusters in synthetic and enzyme systems (12). This intermediate is EPR–silent, like other peroxo-bridged diiron(III) clusters. We tentatively assign this intermediate as a peroxodiiron(III) species based on the similarities of the proposed reactivity to other peroxo-intermediates and by analogy to the mechanism of dioxygen activation at synthetic and enzymatic CBDI centers.
The differences between the Mössbauer parameters for this intermediate and other peroxodiiron(III) species might arise from an alternate binding geometry of the peroxide moiety to the diiron core. Binding modes, such as μ-η 1:η 2- and μ-1,1-, of peroxide fragments at diiron(III) centers are proposed to occur during formation of MMOHperoxo and X (64–66). The peroxide fragment here might adopt such a conformation. The protonation state of the peroxide ion may also differ. Hydroperoxoiron(III) intermediates reported in heme systems are proposed to be electrophilic oxidants, similar to the observed reactivity of peroxodiiron(III) centers (6, 67). Structural investigations of this intermediate in the native system by ENDOR and XAS would provide insight into the structure of this species.
The redox potential of the diiron(III) intermediate can be estimated by Marcus theory (eq 2). The variable kET is the electron transfer rate, k0 is the characteristic frequency of the nuclei, usually assigned a value of 1013, R is the gas constant, and T is the temperature in K (68). The term β, fixed at 1.1 Å−1, is related to the nature of the intervening medium between the redox partners. The distance between the tryptophan residue and the diiron center, r, was determined from the crystal structure to be 6.5 Å, and r0 is the contact distance, which is generally set to 3 Å. Reorganization energies, λ, of 1.0 and 0.1 V were used to calculate a range for the reduction potential of the diiron(III) intermediate. The driving force, ΔG, is the sum of the reduction potentials for the forward reaction. Since we did not observe a transient tyrosyl radical in the I100Y variant, the reduction potential of the diiron(III) intermediate might lie between that of Y and W. Depending on whether deprotonation of the indole ring occurs before or after electron abstraction, the potential may be further limited to between those of the W•/W− and the W•+/W half reactions. To obtain a conservative estimate of the oxidizing power of the diiron(III) intermediate, we assumed that proton loss must occur prior to electron transfer because the energetic cost is less for oxidation of the deprotonated versus neutral indole ring. The foregoing analysis was used to estimate the potential for the Fe2III/Fe2III,IV half reaction, assuming that the W•/W− couple is 0.73 V, which corresponds to deprotonation of 50% of the tryptophan residues (54). To negate the inhibitory effect of protons on the oxidation rate, the calculated proton-independent oxidation rate of 1.22 s−1 determined from the variable pH data was used for kET. With these assumptions, we estimate the reduction potential of the intermediate to lie between 1.1 and 1.3 V versus NHE. This range is close to the reduction potentials of oxidants such as hydrogen peroxide (0.878 V), manganese dioxide (1.224 V), and chromate (1.35 V). The latter two oxidants, MnO2 and CrO4−, are commonly employed for the conversion of hydroxyl groups to either carbonyl or carboxylate functionalities. The peroxodiiron(III) intermediate in MMOH is proposed to carry out similar oxidation reactions, albeit via hydride abstraction (62).
Mechanism of Dioxygen Activation and W100 Oxidation
Scheme 1 depicts our proposed mechanism of formation for the mixed-valent diiron(III,IV)–W• transient. Dioxygen binding to the reduced diiron(II) enzyme would yield a peroxodiiron(III) intermediate, two possible geometries for which are shown. Electron abstraction from W100 by the diiron(III) center is accompanied by protonation-aided cleavage of the O–O bond to form the mixed-valent diiron(III,IV)–W• transient. The structure of the mixed-valent diiron core is similar to that proposed for intermediate X, where the oxygen atoms derived from dioxygen (filled circles in Scheme 1) become an oxo-bridge and either a terminal hydroxide ion or water molecule.
Scheme 1.
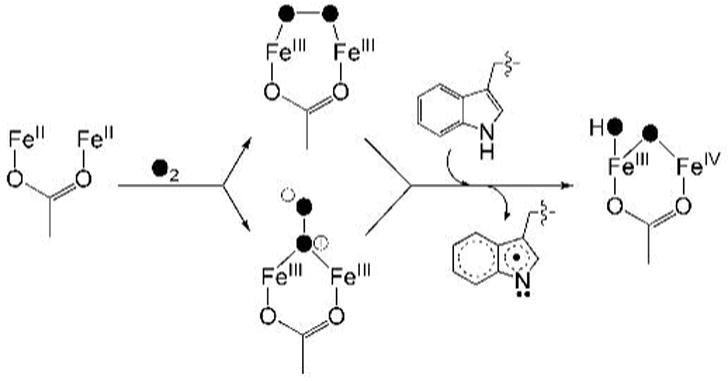
Proposed Mechanism for Formation of the Diiron(III,IV)–W• Transient.
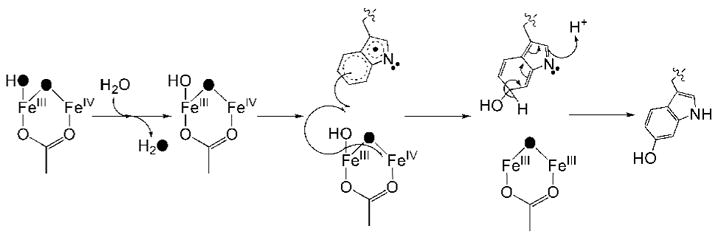
The diiron(III,IV) cluster and W• radical decay at the same rate as measured by Mössbauer, EPR, and optical spectroscopy. The two S = ½ centers are therefore likely to share a common mechanism to restore the oxidized diiron(III) cluster and quench the protein radical. In reacted ToMOH I100W, the installed tryptophan appears to be oxidized, as evidence by the 16 Da increase in mass of the tryptic peptide containing W100 and the fragmentation peptides thereof. In the crystal structure of ToMOH I100W, the indole ring adopts conformations in which the C4–C7 vector is oriented either away from or toward the active site. We predict either C5 or C6 to be the site of oxidation, based on the distances between these two carbon atoms and the diiron core in the oxidized crystal structure. Oxidation of W• arising from attack by dioxygen is unlikely, since the mass of the predicted products would be 32 Da greater than the parent [M+H]+ ion of the unmodified peptide, in disagreement with the experimental results. Dioxygen attack on W• gives rise to a transient tryptophan peroxyl radical, which results in cleavage of the pyrrole ring (69). The peak at m/z = 3198 in the MALDI-TOF spectra is of weak intensity and is attributed to further oxidation of the W100 from multiple turnovers of the hydroxylase, instead of oxidation of the indolyl radical by O2. In addition, such a pathway would not explain the simultaneous decay of the mixed-valent diiron(III,IV) center. Instead, we propose a radical recombination mechanism, where an oxygen atom species on the diiron(III,IV) cluster is transferred to the indolyl radical (Scheme 2).
Scheme 2.
Proposed Mechanism for Decay of the Diiron(III,IV)–W• Transient.
The terminal hydroxide or water molecule must be exchangeable to explain the insensitivity of the mass of the tryptic peptide and its fragments containing W100 to the isotopic content of the dioxygen source. 2H-ENDOR spectra of the transient in deuterated buffers identified an exchangeable species with hyperfine coupling constants similar to those reported for terminal water ligands on the ferric ion in X and MMOHmv (35, 36). The exchange rate of this labile species must be significantly faster than the rate of the decay of the transient species to prevent incorporation of 18O-atoms into the tryptic peptide. Rapid exchange of oxygen atom ligands at high-valent iron centers has been reported for bridging and terminal ligands in intermediate X and terminal oxo-groups at mononuclear iron centers (44, 70, 71). Hydroxyl radical transfer from the diiron center to W• followed by rearomatization of the indole ring gives rise to an oxo-bridged diiron(III) cluster and a hydroxyindole side chain at W100. Addition of water to the diiron core reforms the di(μ-hydroxo)diiron(III) resting state.
The decay reaction is sensitive to proton concentration. The active site of ToMOH is charge neutral, and depending on the ligand type and binding geometries in the diiron(III,IV) transient, the terminal ligand could be either a bound water or hydroxide ion. If the terminal species is a bound water molecule, deprotonation might be required prior to hydroxyl radical transfer. Alternatively, the protonation state of residues within the active site cavity or channel near W100 may be important for stabilizing the diiron(III,IV) or W•. Deprotonation of these residues could favor decay of these transient species. More detailed structural information on the transient species formed during dioxygen activation in the native and I100W variant of the hydroxylase is required to determine the source of this proton dependence.
Potential substrates for BMMs, such as propylene and phenol, accelerate decay of the tryptophanyl radical in stopped-flow optical studies. In the steady state, the activity of ToMOH I100W is more than an order of magnitude lower than that of the wild type enzyme, possibly due to retardation of substrate access or product egress in the variant hydroxylase. The ability of phenol and propylene to accelerate the decay rate is interesting. Phenol could quench the tryptophanyl radical by hydrogen atom transfer from the O–H group to form a phenoxyl radical. Similar reactivity is observed in small molecule chemistry where analogues of phenol are used as radical scavengers. Neutral and cationic tryptophanyl radicals reportedly abstract hydrogen atoms from the hydroxyl group of p-methoxyphenol at rates exceeding 105 s−1 (72). No absorption bands were observed at 410 nm during our double-mixing optical experiments, which indicates that any phenoxyl radical formed by hydrogen-atom abstraction must be short-lived. To explain the sensitivity of the decay rate on propylene and acetylene, we consider the bond dissociation energies of the weakest C–H bonds in these molecules. The BDE of the phenolic O–H bond is similar to that of the methyl sp3 C–H bond in propylene, ~ 87 kcal/mol (73, 74). Acetylene, by comparison, has a C–H BDE almost 50 kcal/mol greater than that of phenol or propylene. The tryptophanyl radical is capable of abstracting a hydrogen atom from the weak O–H bond in phenol and C–H bond in propylene, accelerating the decay of this transient. Because the same reaction with acetylene requires more energy, the decay rate is not appreciably perturbed upon mixing the radical with this substrate.
The reaction of phenol with the neutral tryptophan radical is a pathway distinct from hydroxylation of to yield catechol under steady state conditions where the diiron centers are distributed into populations of reduced, diiron(III) intermediate, diiron(III,IV)–W• transient, and diiron(III) product species. The presence of W100 effectively out-competes the phenolic substrate for the diiron(III) intermediate, if substrate binding occurs after dioxygen activation. Substrate could conceivably bind to the active site in either the reduced state, prior to dioxygen binding and activation, or in the oxidized state after oxidation of W100 or phenol. The reduced capacity of the I100W hydroxylase to oxidize phenol reflects the retarded access of substrate into the active site pocket afforded by the mutation. Considering the location of residue 100 in the ToMOH α-subunit, separating the active site pocket from the rest of the channel and cavity 2, it is reasonable to propose that it may serve to gate substrate entry, product egress, or solvent access to the diiron center as proposed for L110 in MMOH and L98 in hemerythrin (16, 32). Evidence supporting this proposal is illustrated by the two conformations of the indole side chain in the crystal structure of I100W, which allow different levels of access to the diiron center of ToMOH.
Conclusions
The reaction of diiron(II) ToMOH I100W with dioxygen yields a diiron(III) intermediate, which subsequently abstracts an electron from W100 to form a chromophoric mixed-valent diiron(III,IV)–W• species. This coupled species could decay by transfer of an O-atom from the diiron core to the protein-based radical. The one-electron redox chemistry afforded by the diiron(III) intermediate resembles that of the peroxodiiron(III) intermediate in RNR-R2. No other high-valent diiron species were observed, suggesting that oxidation of hydrocarbons in this system occurs at the diiron(III) level. This diiron(III) intermediate is spectroscopically different from that of μ-1,2-peroxodiiron(III) clusters in CBDI enzymes and model compounds. These differences suggest that this intermediate in ToMOH may have an alternate binding geometry or protonation state of the dioxygen-derived fragment. The oxidation of W100 to form deprotonated tryptophanyl radical has allowed us to estimate the reduction potential of the diiron(III) intermediate as 1.1–1.3 V. The mixed-valent diiron(III,IV) center has spectroscopic parameters similar to those of intermediates X and QX, although the diiron(III) precursors differ spectroscopically.
Supplementary Material
SUPPORTING INFORMATION. Distances between the atoms of the indole ring of W100 and the diiron center, HPLC traces from the steady-state product determination for phenol, data for the effect of pH on kf and kd, the effect of propylene and acetylene on kd, selected ENDOR spectra, and ESI-MS/MS data. This information is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
X-ray data were collected at the Stanford Synchrotron Radiation Laboratory (SSRL), which is funded by the Department of Energy (BES, BER) and the National Institutes of Health (NCRR, NIGMS). Crystallographic coordinates for ToMOH I100W have been deposited into the RCSB Protein Databank with accession code 2RDB. Crystallographic figures were generated using PyMOL (75). We thank Dr. Ioannis Papayannopoulos (Center for Cancer Research, M.I.T.) for collection, acquisition, and analysis of the MS data.
ABBREVIATIONS
- CBDI
carboxylate-bridged diiron
- Δ9D
stearoyl-ACP Δ9 desaturase
- Ft
ferritin
- MMOH
hydroxylase component of methane monooxygenase
- ToMO
toluene/o-xylene monooxygenase
- ToMOC
Rieske component of ToMO
- ToMOD
regulatory component of ToMO
- ToMOF
NADH oxidoreductase component of ToMO
- ToMOH
hydroxylase component of ToMO
- Hmv
diiron(II,III) form of the hydroxylase
- Hperoxo
peroxodiiron(III) intermediate of the hydroxylase
- Hred
diiron(II) form of the hydroxylase
- Q
di(μ-oxo)diiron(IV) transient of MMOH
- RNR-R2
ribonucleotide reductase R2 subunit (Class I)
- SSRL
Stanford Synchrotron Radiation Laboratories
- X
mixed-valent diiron(III,IV) intermediate formed in RNR-R2
Footnotes
This study was supported by NIH Grants GM32134 (to SJL), GM47295 (to BHH), and HL13531 (to BMH). LJM was supported in part by a fellowship from the Martin Society (M.I.T.).
References
- 1.Song WJ, Seo MS, George SD, Ohta T, Song R, Kang MJ, Tosha T, Kitagawa T, Solomon EI, Nam W. Synthesis, characterization, and reactivities of manganese(V)-oxo porphyrin complexes. J Am Chem Soc. 2007;129:1268–1277. doi: 10.1021/ja066460v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nam W. High-valent iron(IV)–oxo complexes of heme and non-heme ligands in oxygenation reactions. Acc Chem Res. 2007;40:522–531. doi: 10.1021/ar700027f. [DOI] [PubMed] [Google Scholar]
- 3.Mirica LM, Vance M, Jackson Rudd D, Hedman B, Hodgson KO, Solomon EI, Stack DP. Tyrosinase reactivity in a model complex: an alternative hydroxylation mechanism. Science. 2005;308:1890–1892. doi: 10.1126/science.1112081. [DOI] [PubMed] [Google Scholar]
- 4.Lippard SJ. Hydroxylation of C-H bonds at carboxylate-bridged diiron centres. Philos Trans R Soc A. 2005;363:861–877. doi: 10.1098/rsta.2004.1532. [DOI] [PubMed] [Google Scholar]
- 5.Sinnecker S, Svensen N, Barr EW, Ye S, Bollinger JM, Jr, Neese F, Krebs C. Spectroscopic and computational evaluation of the structure of the high-spin Fe(IV)-oxo intermediates in taurine: α-ketoglutarate dioxygenase from Escherichia coli and its His99Ala ligand variant. J Am Chem Soc. 2007;129:6168–6179. doi: 10.1021/ja067899q. [DOI] [PubMed] [Google Scholar]
- 6.Denisov IG, Makris TM, Sligar SG, Schlichting I. Structure and chemistry of cytochrome P450. Chem Rev. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 7.Rosenzweig AC, Brandstetter H, Whittington DA, Nordlund P, Lippard SJ, Frederick CA. Crystal structures of the methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): implications for substrate gating and component interactions. Proteins. 1997;29:141–152. [PubMed] [Google Scholar]
- 8.Logan DT, Su X-D, Åberg A, Regnström K, Hajdu J, Eklund H, Nordlund P. Crystal structure of reduced protein R2 of ribonucleotide reductase: the structural basis for oxygen activation at a dinuclear iron site. Structure. 1996;4:1053–1064. doi: 10.1016/s0969-2126(96)00112-8. [DOI] [PubMed] [Google Scholar]
- 9.Lindqvist Y, Huang W, Schneider G, Shanklin J. Crystal structure of Δ9 stearoyl-acyl carrier protein desaturase from castor seed and its relationship to other di-iron proteins. EMBO J. 1996;15:4081–4092. [PMC free article] [PubMed] [Google Scholar]
- 10.Liu KE, Wang D, Huynh BH, Edmondson DE, Salifoglou A, Lippard SJ. Spectroscopic detection of intermediates in the reaction of dioxygen with the reduced methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath) J Am Chem Soc. 1994;116:7465–7466. [Google Scholar]
- 11.Sturgeon BE, Burdi D, Chen S, Huynh BH, Edmondson DE, Stubbe J, Hoffman BM. Reconsideration of X, the diiron intermediate formed during cofactor assembly in E. coli ribonucleotide reductase. J Am Chem Soc. 1996;118:7551–7557. [Google Scholar]
- 12.Merkx M, Kopp DA, Sazinsky MH, Blazyk JL, Müller J, Lippard SJ. Dioxygen activation and methane hydroxylation by soluble methane monooxygenase: a tale of two irons and three proteins. Angew Chem, Int Ed. 2001;40:2782–2807. doi: 10.1002/1521-3773(20010803)40:15<2782::AID-ANIE2782>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 13.Broadwater JA, Ai J, Loehr TM, Sanders-Loehr J, Fox BG. Peroxodiferric intermediate of stearoyl-acyl carrier protein Δ9 desaturase: oxidase reactivity during single turnover and implications for the mechanism of desaturation. Biochemistry. 1998;37:14664–14671. doi: 10.1021/bi981839i. [DOI] [PubMed] [Google Scholar]
- 14.Jin S, Kurtz DJ, Jr, Liu ZJ, Rose J, Wang BC. X-ray crystal structures of reduced rubrerythrin and its azide adduct: a structure-based mechanism for a non-heme diiron peroxidase. J Am Chem Soc. 2002;124:9845–9855. doi: 10.1021/ja026587u. [DOI] [PubMed] [Google Scholar]
- 15.Sazinsky MH, Bard J, DiDonato A, Lippard SJ. Crystal structure of the toluene/o-Xylene monooxygenase hydroxylase from Pseudomonas stutzeri OX1: insight into the substrate specificity, substrate channeling, and active site tuning of multicomponent monooxygenases. J Biol Chem. 2004;279:30600–30610. doi: 10.1074/jbc.M400710200. [DOI] [PubMed] [Google Scholar]
- 16.Farmer CS, Kurtz DJ, Jr, Phillips RS, Ai J, Sanders-Loehr J. A leucine residue “gates” solvent but not O2 access to the binding pocket of Phascolopsis gouldii hemerythrin. J Biol Chem. 2000;275:17043–17050. doi: 10.1074/jbc.M001289200. [DOI] [PubMed] [Google Scholar]
- 17.Murray LJ, García-Serres R, Naik S, Huynh BH, Lippard SJ. Dioxygen activation at non-heme diiron centers: characterization of intermediates in a mutant form of toluene/o-xylene monooxygenase hydroxylase. J Am Chem Soc. 2006;128:7458–7459. doi: 10.1021/ja062762l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeMaster DM, Richards FM. 1H-15N heteronuclear NMR studies of Escherichia coli thioredoxin in samples isotopically labeled by residue type. Biochemistry. 1985;24:7263–7268. doi: 10.1021/bi00346a036. [DOI] [PubMed] [Google Scholar]
- 19.McCormick MS, Sazinsky MH, Condon KL, Lippard SJ. X-ray crystal structures of manganese(II)-reconstituted and native toluene/o-xylene monooxygenase hydroxylase reveal rotamer shifts in conserved residues and an enhanced view of the protein interior. J Am Chem Soc. 2006;128:15108–15110. doi: 10.1021/ja064837r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McPhillips TM, McPhillips SE, Chiu HJ, Cohen AE, Deacon AM, Ellis PJ, Garman E, Gonzalez A, Sauter NK, Phizackerley RP, Soltis SM, Kuhn PJ. Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. J Synchrotron Rad. 2002;9:401–406. doi: 10.1107/s0909049502015170. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Kissinger CR, Gehlhaar DK, Fogel DB. Rapid automated molecular replacement by evolutionary search. Acta Crystallogr D. 1999;55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 24.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 27.Kleywegt G, Jones TA. Databases in protein crystallography. Acta Crystallogr D. 1998;54:1119–1131. doi: 10.1107/s0907444998007100. [DOI] [PubMed] [Google Scholar]
- 28.Cafaro V, Scognamiglio R, Viggiani A, Izzo V, Passaro I, Notomista E, Dal Piaz F, Amoresano A, Casbarra A, Pucci P, DiDonato A. Expression and purification of the recombinant subunits of toluene/o-xylene monooxygenase and reconstitution of the active complex. Eur J Biochem. 2002;269:5689–5699. doi: 10.1046/j.1432-1033.2002.03281.x. [DOI] [PubMed] [Google Scholar]
- 29.Wilhelm E, Battino R, Wilcock RJ. Low-pressure solubility of gases in liquid water. Chem Rev. 1977;77:219–262. [Google Scholar]
- 30.Ravi N, Bollinger JM, Jr, Huynh BH, Edmondson DE, Stubbe J. Mechanism of assembly of the tyrosyl radical-diiron(III) cofactor of E. coli ribonucleotide reductase. 1 Mössbauer characterization of the diferric radical precursor. J Am Chem Soc. 1994;116:8007–8014. [Google Scholar]
- 31.Davydov R, Perera R, Jin S, Yang TC, Bryson TA, Sono M, Dawson JH, Hoffman BM. Substrate modulation of the properties and reactivity of the oxy-ferrous and hydroperoxo-ferric intermediates of cytochrome P450cam as shown by cryoreduction-EPR/ENDOR spectroscopy. J Am Chem Soc. 2005;127:1403–1413. doi: 10.1021/ja045351i. [DOI] [PubMed] [Google Scholar]
- 32.Whittington DA, Sazinsky MH, Lippard SJ. X-ray crystal structure of alcohol products bound at the active site of soluble methane monooxygenase hydroxylase. J Am Chem Soc. 2001;123:1794–1795. doi: 10.1021/ja0031725. [DOI] [PubMed] [Google Scholar]
- 33.Lendzian F. Structure and interactions of amino acid radicals in class I ribonucleotide reductase studied by ENDOR and high-field EPR spectroscopy. Biochim Biophys Acta. 2005;1707:67–90. doi: 10.1016/j.bbabio.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 34.Ambundo EA, Friesner RA, Lippard SJ. Reactions of methane monooxygenase intermediate Q with derivatized methanes. J Am Chem Soc. 2002;124:8770–8771. doi: 10.1021/ja0265759. [DOI] [PubMed] [Google Scholar]
- 35.Willems JP, Lee HI, Burdi D, Doan PE, Stubbe J, Hoffman BM. Identification of the protonated oxygenic ligands of ribonucleotide reductase intermediate X by Q-Band 1,2H CW and pulsed ENDOR. J Am Chem Soc. 1997;119:9816–9824. [Google Scholar]
- 36.Smoukov SK, Kopp DA, Valentine AM, Davydov R, Lippard SJ, Hoffman BM. Product binding to the diiron(III) and mixed-valence diiron centers of methane monooxygenase hydroxylase studied by 1,2H and 19F ENDOR spectroscopy. J Am Chem Soc. 2002;124:2657–2663. doi: 10.1021/ja010123z. [DOI] [PubMed] [Google Scholar]
- 37.Himo F, Eriksson LA. Theoretical study of model tryptophan radicals and radical cations: comparison with experimental data of DNA photolyase, cytochrome c peroxidase, and ribonucleotide reductase. J Phys Chem B. 1997;101:9811–9819. [Google Scholar]
- 38.Huyett JE, Doan PE, Gurbiel R, Houseman ALP, Sivaraja M, Goodin DB, Hoffman BM. Compound ES of cytochrome c peroxidase contains a Trp π-cation radical: characterization by CW and pulsed Q-band ENDOR spectroscopy. J Am Chem Soc. 1995;117:9033–9041. [Google Scholar]
- 39.Walden SE, Wheeler RA. Distinguishing features of indolyl radical and radical cation: implications for tryptophan radical studies. J Phys Chem. 1996;100:1530–1535. [Google Scholar]
- 40.Rigby SEJ, Jünemann S, Rich PR, Heathcote P. Reaction of bovine cytochrome c oxidase with hydrogen peroxide produces a tryptophan cation radical and a porphyrin cation radical. Biochemistry. 2000;39:5921–5928. doi: 10.1021/bi992614q. [DOI] [PubMed] [Google Scholar]
- 41.O’Malley PJ, Ellson DA. The calculation of 1H, 13C, 14N isotropic and anisotropic hyperfine interactions for the 3-methyl indole cation and neutral radicals using hybrid density functional methods models for in vivo tryptophan-based radicals. Chem Phys Lett. 1996;260:492–498. [Google Scholar]
- 42.Papayannopoulos IA. The interpretation of collision-induced dissociation tandem mass spectra of peptides. Mass Spectrom Rev. 1995;14:49–73. [Google Scholar]
- 43.Krebs C, Chen S, Baldwin J, Ley BA, Patel U, Edmondson DE, Huynh BH, Bollinger JM., Jr Mechanism of rapid electron transfer during oxygen activation in the R2 subunit of Escherichia coli ribonucleotide reductase 2 Evidence for and consequences of blocked electron transfer in the W48F variant. J Am Chem Soc. 2000;122:12207–12219. [Google Scholar]
- 44.Burdi D, Willems JP, Riggs-Gelasco P, Antholine WE, Stubbe J, Hoffman BM. The core structure of X generated in the assembly of the diiron cluster of ribonucleotide reductase: 17O2 and H217O. J Am Chem Soc. 1998;120:12910–12919. [Google Scholar]
- 45.Lee S-K, Lipscomb JD. Oxygen activation catalyzed by methane monooxygenase hydroxylase component: proton delivery during the O-O bond cleavage steps. Biochemistry. 1999;38:4423–4432. doi: 10.1021/bi982712w. [DOI] [PubMed] [Google Scholar]
- 46.Lovell T, Li J, Noodleman L. Energetics of oxidized and reduced methane monooxygenase active site clusters in the protein environment. Inorg Chem. 2001;40:5267–5278. doi: 10.1021/ic010116b. [DOI] [PubMed] [Google Scholar]
- 47.Murray LJ, Lippard SJ. Substrate trafficking and dioxygen activation in bacterial multicomponent monooxygenases. Acc Chem Res. 2007;40:466–474. doi: 10.1021/ar600040e. [DOI] [PubMed] [Google Scholar]
- 48.Pikus JD, Mitchell KH, Studts JM, McClay K, Steffan RJ, Fox BG. Threonine 201 in the diiron enzyme toluene 4-monooxygenase is not required for catalysis. Biochemistry. 2000;39:791–799. doi: 10.1021/bi992187g. [DOI] [PubMed] [Google Scholar]
- 49.Lipscomb JD. Biochemistry of soluble methane monooxygenase. Annu Rev Microbiol. 1994;48:371–399. doi: 10.1146/annurev.mi.48.100194.002103. [DOI] [PubMed] [Google Scholar]
- 50.Murray LJ, Naik SG, Ortillo DO, García-Serres R, Lee JK, Huynh BH, Lippard SJ. Characterization of the arene-oxidizing intermediate in ToMOH as a diiron(III) species. 2007 doi: 10.1021/ja076121h. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valentine AM, Tavares P, Pereira AS, Davydov R, Krebs C, Hoffman BM, Huynh BH, Lippard SJ. Generation of a mixed-valent Fe(III)Fe(IV) form of intermediate Q in the reaction cycle of soluble methane monooxyenase, an analogue of intermediate X in ribonucleotide reductase R2 assembly. J Am Chem Soc. 1998;120:2190–2191. [Google Scholar]
- 52.Reece SY, Stubbe J, Nocera DG. pH dependence of charge transfer between tryptophan and tyrosine in dipeptides. Biochim Biophys Acta. 2005;1706:232–238. doi: 10.1016/j.bbabio.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 53.Aubert C, Vos MH, Mathis P, Eker APM, Brettel K. Intraprotein radical transfer during photoactivation of DNA photolyase. Nature. 2000;405:586–590. doi: 10.1038/35014644. [DOI] [PubMed] [Google Scholar]
- 54.Tommos C, Skalicky JJ, Pilloud DL, Wand AJ, Dutton PL. De novo proteins as models of radical enzymes. Biochemistry. 1999;38:9495–9507. doi: 10.1021/bi990609g. [DOI] [PubMed] [Google Scholar]
- 55.Yagil G. The proton dissociation constant of pyrrole, indole, and related compounds. Tetrahedron. 1967;23:2855–2861. doi: 10.1016/0040-4020(67)85151-2. [DOI] [PubMed] [Google Scholar]
- 56.Aikens J, Sligar SG. Kinetic solvent isotope effects during oxygen activation by cytochrome P-450cam. J Am Chem Soc. 1994;116:1143–1144. [Google Scholar]
- 57.Kohen A. Kinetic isotope effects as probes for hydrogen tunneling, coupled motion and dynamics contributions to enzyme catalysis. Prog React Kinet Mech. 2003;28:119–156. [Google Scholar]
- 58.Baldwin J, Voegtli WC, Khidekel N, Moënne-Loccoz P, Krebs C, Pereira AS, Ley BA, Huynh BH, Loehr TM, Riggs-Gelasco PJ, Rosenzweig AC, Bollinger JM., Jr Rational reprogramming of the R2 subunit of Escherichia coli ribonucleotide reductase into a self-hydroxylating monooxygenase. J Am Chem Soc. 2001;123:7017–7030. doi: 10.1021/ja002114g. [DOI] [PubMed] [Google Scholar]
- 59.Parkin SE, Chen S, Ley BA, Mangravite L, Edmondson DE, Huynh BH, Bollinger JM., Jr Electron injection through a specific pathway determines the outcome of oxygen activation at the diiron cluster in the F208Y mutant of Escherichia coli ribonucleotide reductase protein R2. Biochemistry. 1998;37:1124–1130. doi: 10.1021/bi9723717. [DOI] [PubMed] [Google Scholar]
- 60.Ormö M, deMaré F, Regnström K, Åberg A, Sahlin M, Ling J, Loehr TM, Sanders-Loehr J, Sjöberg BM. Engineering of the iron site in ribonucleotide reductase to a self-hydroxylating monooxygenase. J Biol Chem. 1992;267:8711–8714. [PubMed] [Google Scholar]
- 61.McCormick MS, Lippard SJ. unpublished results. [Google Scholar]
- 62.Beauvais LG, Lippard SJ. Reactions of the peroxo intermediate of soluble methane monooxygenase with ethers. J Am Chem Soc. 2005;127:7370–7378. doi: 10.1021/ja050865i. [DOI] [PubMed] [Google Scholar]
- 63.Moe LA, Hu Z, Deng D, Austin RN, Groves JT, Fox BG. Remarkable aliphatic hydroxylation by the diiron enzyme toluene 4-monooxygenase in reactions with radical or cation diagnostic probes norcarane, 1,1-dimethylcyclopropane, and 1,1-diethylcyclopropane. Biochemistry. 2004;43:15688–15701. doi: 10.1021/bi040033h. [DOI] [PubMed] [Google Scholar]
- 64.Gherman BF, Baik MH, Lippard SJ, Friesner RA. Dioxygen activation in methane monooxygenase: a theoretical study. J Am Chem Soc. 2004;126:2978–2990. doi: 10.1021/ja036506+. [DOI] [PubMed] [Google Scholar]
- 65.Baldwin J, Krebs C, Saleh L, Stelling M, Huynh BH, Bollinger JM, Jr, Riggs-Gelasco PJ. Structural characterization of the peroxodiiron(III) intermediate generated during oxygen activation by the W48A/D84E variant of ribonucleotide reductase. Biochemistry. 2003;42:13269–13279. doi: 10.1021/bi035198p. [DOI] [PubMed] [Google Scholar]
- 66.Brunold TC, Tamura N, Kitajima N, Moro-oka Y, Solomon EI. Spectroscopic study of [Fe2(O2)(OBz)2{HB(pz′)3}2]: nature of the μ-1,2 peroxide-Fe(III) bond and its possible relevance to O2 activation by non-heme iron enzymes. J Am Chem Soc. 1998;120:5674–5690. [Google Scholar]
- 67.Nam W, Ryu YO, Song WJ. Oxidizing intermediates in cytochrome P450 model reactions. J Biol Inorg Chem. 2004;9:654–660. doi: 10.1007/s00775-004-0577-5. [DOI] [PubMed] [Google Scholar]
- 68.Blazyk JL, Gassner GT, Lippard SJ. Intermolecular electron-transfer reactions in soluble methane monooxygenase: a role for hysteresis in protein function. J Am Chem Soc. 2005;127:17364–17376. doi: 10.1021/ja0554054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Candeias LP, Wardman P, Mason RP. The reaction of oxygen with radicals from oxidation of tryptophan and indole-3-acetic acid. Biophys Chem. 1997;67:229–237. doi: 10.1016/s0301-4622(97)00052-5. references cited therein. [DOI] [PubMed] [Google Scholar]
- 70.Dorovska-Taran V, Posthumus MA, Boeren S, Boersma MG, Teunis CJ, Rietjens IMCM, Veeger C. Oxygen exchange with water in heme-oxo intermediates during H2O2-driven oxygen incorporation in aromatic hydrocarbons catalyzed by microperoxidase-8. Eur J Biochem. 1998;253:659–668. doi: 10.1046/j.1432-1327.1998.2530659.x. [DOI] [PubMed] [Google Scholar]
- 71.Seo MS, In JH, Kim SO, Oh NY, Hong J, Kim J, Que L, Jr, Nam W. Direct evidence for oxygen-atom exchange between nonheme oxoiron(IV) complexes and isotopically labeled water. Angew Chem, Int Ed. 2004;43:2417–2420. doi: 10.1002/anie.200353497. [DOI] [PubMed] [Google Scholar]
- 72.Jovanovic SV, Steenken S, Simic MG. Kinetics and energetics of one-electron-transfer reactions involving tryptophan neutral and cation radicals. J Phys Chem. 1991;95:684–687. [Google Scholar]
- 73.Blanksby SJ, Ellison GB. Bond dissociation energies of organic molecules. Acc Chem Res. 2003;36:255–263. doi: 10.1021/ar020230d. [DOI] [PubMed] [Google Scholar]
- 74.Silva G, Chen CC, Bozzelli JW. Bond dissociation energy of the phenol O-H bond from ab initio calculations. Chem Phys Lett. 2006;424:42–45. [Google Scholar]
- 75.DeLano WL. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPORTING INFORMATION. Distances between the atoms of the indole ring of W100 and the diiron center, HPLC traces from the steady-state product determination for phenol, data for the effect of pH on kf and kd, the effect of propylene and acetylene on kd, selected ENDOR spectra, and ESI-MS/MS data. This information is available free of charge via the Internet at http://pubs.acs.org.


