Abstract
Background
A genetic study was performed to identify candidate genes associated with day blindness in the standard wire haired dachshund. Based on a literature review of diseases in dogs and human with phenotypes similar to day blindness, ten genes were selected and evaluated as potential candidate genes associated with day blindness in the breed.
Results
Three of the genes, CNGB3, CNGA3 and GNAT2, involved in cone degeneration and seven genes and loci, ABCA4, RDH5, CORD8, CORD9, RPGRIP1, GUCY2D and CRX, reported to be involved in cone-rod dystrophies were studied. Polymorphic markers at each of the candidate loci were studied in a family with 36 informative offspring. The study revealed a high frequency of recombinations between the candidate marker alleles and the disease.
Conclusion
Since all of the markers were at the exact position of the candidate loci, and several recombinations were detected for each of the loci, all ten genes were excluded as causal for this canine, early onset cone-rod dystrophy. The described markers may, however, be useful to screen other canine resource families segregating eye diseases for association to the ten genes.
Background
Inherited retinal degenerations form a diverse spectrum of blinding disorders in humans and other mammals, including a large number of dog breeds [1]. Retinal diseases are inherited as monogenic or complex traits. In the last two decades over 130 genes that cause retinal diseases have been identified [2].
Progressive retinal atrophies (PRA) are the most common retinopathies in dogs and constitute a heterogeneous group of phenotypically similar disorders equivalent to retinitis pigmentosa (RP) in man. PRA, like RP, is primarily a disease of rod photoreceptors, while cone function and structure degenerate secondarily. PRA has been identified and studied in more than 100 breeds and distinct loci primarily responsible for each disorder have been identified in at least 22 different breeds [1,3].
The cone dystrophies comprise a phenotypically heterogeneous group of hereditary retinal degenerations characterized by progressive dysfunction of the photopic (cone-mediated) system. The presenting signs include day blindness, loss of colour vision, reduced central visual acuity and preserved peripheral vision [4]. The cone dystrophies are genetically heterogenous and may be sporadic, autosomal dominant, autosomal recessive or X-linked recessive [2].
Canine cone degeneration is phenotypically similar to human achromatopsia [5]. Identified genes associated with human achromatopsia are the phototransducin genes GNAT2 (HSA1) [6], the CNGA3 (HSA2) [7] and the CNGB3 (HSA8) [8] genes.
Autosomal recessive cone degeneration occurs naturally in Alaskan malamute [9] and German shorthaired pointer (GSP) and is due to different mutations in CNGB3 [5,10]. Sporadic cases is seen in a number of other breeds [11,12].
The cone-rod dystrophies (crd) are characterized by a predominant loss of cone function, with relative preservation of rod function [13,14]. The number of genes or loci currently identified for crd in humans is low, compared to those for RP [2]. Identified genes associated with autosomal recessive inherited crd in man include ABCA4 [15,16] and RDH5 [17] in addition to the mapped loci CORD8 [18,19] and CORD9 [20]. ABCA4 and RDH5 are genes involved in the retinoid cycle, the CORD8 and CORD9 loci are yet to be identified. The standard wire haired dachshund (SWHD), the miniature long haired dachshund (MLHD) and the pit bull terrier (PBT) are the only dog breeds to date known to be affected by crd [21-23].
Canine ABCA4 has been studied as a candidate gene for cone-rod degeneration in the pit bull terrier, although no mutations were found [21]. Mutations in the RPGRIP1 gene have been reported to be a cause of Leber congenital amaurosis (LCA) [24,25]. LCA is the earliest and most severe form of all retinal dystrophies responsible for congenital blindness [26]. Mutations causing residual RPGRIP1 activity may lead to phenotypes such as RP or crd, which are less severe than LCA [27]. In 2003, Hameed et al. (2003) [28] showed evidence that some RPGRIP1 gene mutations are associated with recessive cone-rod dystrophy. Recently Mellersh et al. (2006) [22] found a mutation in canine RPGRIP1 associated with autosomal recessive crd in miniature longhaired dachshunds.
A high level of genetic heterogeneity of the disease group is observed and a range of mutations in RPGRIP1, CRX and GUCY2D cause different retinal diseases with different modes of inheritance [14,27-34].
A resource strain of standard wire haired dachshund displaying day blindness was developed to allow characterization of the phenotype and identification of the causal mutation. Electroretinography (ERG) and clinical studies showed that secondary degeneration of the rods appeared during the progression of the disease of the dogs of the colony, indicating a progressive cone-rod dystrophy (crd) [23]. The disease seems to be inherited in an autosomal recessive manner, and diversity in the age of onset and progression of the retinal degeneration within the group of affected dachshunds is observed.
This study was conducted in parallel with clinical studies [35] and genes known to be involved in human cone degeneration, cone dystrophy and cone-rod dystrophy were evaluated as candidate genes for the day blindness in the SWHD (See Table 1).
Table 1.
Overview of the candidate genes
| Candidate | Canine chr | Location | Annotation | Coding for | Disease* |
| CNGB3 | CFA29 | 35896147-35752878 | NC_006611 | cyclic nucleotide gated channel beta 3 | cd |
| CNGA3 | CFA10 | 47377091-47357441 | NC_006592 | cyclic nucleotide gated channel alpha 3 | cd |
| GNAT2 | CFA6 | 45319301-45327681 | NC_006588 | guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 | cd |
| ABCA4 | CFA6 | 58112924-58240779 | NC_006588 | ATP-binding cassette, sub-family A (ABC1), member 4 | crd |
| RDH5 | CFA10 | 3102451-3106432 | NC_006592 | retinol dehydrogenase 5 (11-cis/9-cis) | crd |
| CORD8 | CFA9 | HSA1q12–24 | unknown | crd | |
| CORD9 | CFA29 | HSA8p11 | unknown | crd | |
| RPGRIP1 | CFA15 | 21355638-21394395 | NC_006597 | retinitis pigmentosa GTPase regulator interacting protein 1 | crd |
| GUCY2D | CFA5 | 35838279-35853509 | NC_006587 | guanylate cyclase 2D, membrane (retina-specific) | crd |
| CRX | CFA1 | 111135799-111146903 | NC_006583 | cone-rod homeobox | crd |
*cd = cone degeneration and crd = cone-rod-dystrophy
Results
An overview of the studied genes and their location is shown in Table 1. Studies of segregation of each of the 10 candidate loci and clinical disease in informative parents and offspring revealed a number of new candidate gene allele-disease phenotype combinations ("recombinants") in the offspring. The segregation of potential candidate gene alleles was studied in inbred offspring from parents with known disease genotypes. Females where either homozygous affected (= clinical diseased) or carriers (= clinical healthy offspring of affected male). Since the father in these litters usually was homozygous affected, the theoretical phenotype of the offspring would depend on which of the two potential alleles (affected or non-affected) it received from its carrier mother. The most frequently occurring genotype-phenotype combination was counted as the non-recombinants, while any new genotype-phenotype combination, with regard to their parents, was counted as recombinant. Each litter was counted as a separate unit and the recombination frequency were summed over all families. A minimum of 6 recombinants were identified for each of the 10 loci (See Table 2). This excludes all the candidate genes as the cause of mutation causing the disease. The family is illustrated in Figure 1. The genotypes of ABCA4-markers are shown below each individual and show free recombination of ABCA4-variants between affected and unaffected offspring.
Table 2.
Recombination frequencies between candidate gene loci (microsatellites at locus) and disease
| Locus | no. of informative offspring | no. of recombinant offspring | Recombination frequency | LOD score |
| CNGB3 | 34 | 11 | 0.32 | 0.940 |
| CNGA3 | 20 | 7 | 0.35 | 0.397 |
| GNAT2 | 31 | 6 | 0.19 | 2.717 |
| ABCA4 | 34 | 11 | 0.32 | 0.940 |
| RDH5 | 34 | 11 | 0.32 | 0.940 |
| CORD8 | 25 | 10 | 0.40 | 0.219 |
| CORD9 | 18 | 5 | 0.28 | 0.800 |
| CRX | 24 | 8 | 0.33 | 0.590 |
| GUCY2D | 23 | 7 | 0.30 | 0.786 |
Figure 1.
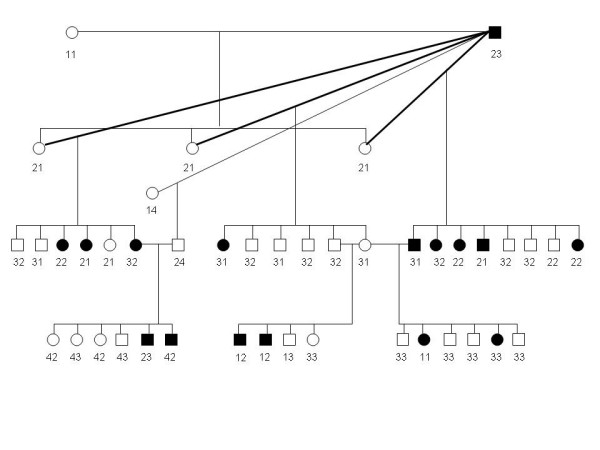
The informative resource family, consisting of offspring from a single affected male. The numbers below each individual represent ABCA4 genotypes, and clearly shows the free recombination of marker-alleles compared to disease in the family.
Discussion
The large number of retinal genes or loci have been identified and presented at the RetNet™ web page [2], facilitates a candidate gene approach in the study of canine retinal diseases. Human genes with mutations giving similar phenotypes and inheritance patterns to the one found in the affected SWHDs were selected as candidate genes. The preliminary clinical diagnoses for the affected SWHDs suggested that the disease was comparable with human cone degeneration, hence the inclusion of CNGB3, CNGA3 and GNAT2, which are associated primarily with cone degeneration. In the initial phase of the study the family material was not sufficient for segregation studies and we therefore sequenced the coding parts of these three genes. No mutations where identified in any of the three genes (methods and results not shown). Continued electrophysiological and clinical studies of the affected dachshunds were conducted in parallel with the genetic studies and revealed secondary continuing degeneration of the rods, indicating a progressive cone-rod dystrophy (crd) [23]. Seven genes reported to be associated with autosomal recessive cone-rod dystrophies (crd) [2], ABCA4, RDH5, CORD8, CORD9, RPGRIP1, CRX and GUCY2D were therefore included in the study.
It is important to be aware of the fact that the disease-allele/haplotype present in all dogs in the family arises from inbreeding on one single affected founder (Figure 1). The informative markers were selected at the exact position of the candidate genes, most of them less than approximately 0.1 Mb distance. A distance of 0.1 Mb compares to a genetic distance of ~0.1 cM, or 1 recombination per 1000 offspring. The minimum number of detected recombinations in our family (1 recombination per 36 informative offspring) would compare to a recombination frequency/genetic distance of ~2.8 cM or a physical distance of ~2.8 Mb. We would not expect any recombinants if a mutation in any of the candidate genes caused this disease with a simple recessive inheritance. Our results show a minimum of 6 recombinations (GNAT2; 31 informative offspring) or more than 19.3 Mb distance (Table 2), excluding all the candidate genes as the site of the mutation causing the disease. However, even though the specific candidate genes can be excluded as the cause of the disease because of the high number of recombinations, the chromosomal regions are not necessarily excluded. The clinical findings defining the diseases as a cone-rod dystrophy rather than cone degeneration also support the exclusion of CNGB3, CNGA3 and GNAT2. The candidate loci CORD8 and CORD9 are not yet mapped and the genes remain to be identified, but the markers typed for these loci are located within 2.8 Mb of the gene and several recombinations exclude these loci as well.
It might be considered likely that crd in MLDH and SWDH was caused by the same mutation, since these two breeds have in part the same genetic background. There are, however, obvious differences in the clinical phenotypes of the disease in the two breeds. One of the most characteristic clinical findings in the SWDH is pinpoint-sized pupils, observed in 60% of the 8-week old, crd-affected puppies. Older crd-affected dogs displayed dilated pupils and delayed pupillary light reflexes (PLRs) when stimulated with a focal light source [23]. This is in contrast to the clinical signs of PBT and MLHD, which include dilated pupils at 7–8 weeks of age for PBT and normal PLRs at 25 weeks of age for MLHD, respectively [21,34]. The differences in clinical signs between the MLHD and the SWHD, support the finding of free recombination between RPGRIP1 and the disease in the SWHD. This shows that this disease in the SWHDs is a unique animal model for cone-rod dystrophy.
The development of animal models of ocular disease represents an invaluable resource for testing and evaluating treatment strategies relevant to both human and canine disease. Attempts at restoring vision in dogs and human by gene therapy have been made, providing optimism regarding potential for recovery of functional vision [36-41]. Uncovering the genetic basis of the cone-rod dystrophy in the SWHD may contribute to finding new gene therapy treatments.
The present work has excluded a number of genes as a cause of crd in the SWHD. Research in human and animals continuously identifies new genes that may be associated with crd. In addition to continued studies of a few specific candidate genes, future research will focus on a whole genome scan by linkage studies with polymorphic markers [42] or by SNP-array typing [43]. The development of the canine genotyping array of ~27000 SNPs show that genome-wide association mapping of mendelian traits in dog breeds can be achieved with only ~20 dogs [44]. The use of SNP array technology, followed by fine mapping based on microsatellites and regional gene studies may be the best method to elucidate which gene is involved in this disease.
Conclusion
The genes involved in cone degeneration, CNGB3, CNGA3 and GNAT2, and the genes involved in cone-rod dystrophy, ABCA4, RDH5, Cord8, Cord9, RPGRIP1, GUCY2D and CRX, were all excluded from being involved in the cone-rod dystrophy described in this family of SWHD. The study provides a number of genetic markers for use in other studies. Further work, based on a whole genome association study will be performed to identify the mutation involved in the disease.
Methods
Animals
Day blindness was diagnosed in two wire haired dachshund littermates, one male and one female, in a litter of four. The parents were phenotypically normal. The male was bred to two unrelated crossbred dogs with no history of previous eye disease and the dogs in the F1 generation were unaffected. A purpose-bred colony was established through back-crossing the male with his daughters, producing both affected and unaffected offspring. The retinal changes were always bilaterally symmetrical and the initial onset was observed from 10 months to 3 years of age. A complete retinal atrophy was evident within the age of 5–6 years.
Clinical diagnosis
To evaluate vision, all dogs were subjected to behavioral testing (maze test), examination of pupillary light reflexes (PLRs), indirect ophthalmoscopy and bilateral full-field electroretinography (ERG). Light microscopy, electron microscopy and immunohistochemistry were carried out in 22 selected cases at ages 5–304 weeks, in order to confirm the diagnoses [23].
All procedures used in this study adhered to the guidelines of the Norwegian Animal Research Authority (Forsøksdyrutvalget) and to the Association for Research in Vision and Ophthalmology's "Statement for the use of Animals in Ophthalmic Vision and Research".
Samples
Blood samples were collected into EDTA-coated tubes from all the related dogs in the day blind colony and from two non-affected, unrelated dogs. Genomic DNA was isolated from the blood samples using DNeasy Tissue kit (Qiagen, GmbH, Germany).
The collected DNA samples were subjected to candidate gene screening. All the ten loci were typed by polymorphic markers at these loci in five informative dachshund families comprising 43 dogs -7 parents and 36 offspring (Figure 1), to detect recombinants between each of the loci and the disease.
Study of marker-disease segregation in the resource family
Genotyping of CNGB3 was done with two closely linked markers, FH2772 [45] and c29.002 [46], embracing the gene (530 Kb and 350 Kb distance respectively). Markers for genotyping of CNGA3, GNAT2 ABCA4, RDH5, CRX, GUCY2D and RPGRIP1 were identified by searching a sequence of about 200,000 bp, including the gene(s) (gene +/- ~100 000 bp), for various microsatellite motives. FH2370, a marker closely linked to ABCA4 [46] was also genotyped. For the loci CORD8 and CORD9, 200 000 basepairs surrounding genes at the site of the two loci, respectively FM05 and RP1, were used to identify polymorphic markers. Tetra- and dinucleotide repeat microsatellites with more than ten and nineteen repeats, respectively, were selected. Primers for amplification of selected microsatellites were designed using primer3 [47] (See Table 3). One primer in each pair was labelled with fluorescein for automated detection in an automated fragment analyzer, ABI3100 Genetic Analyser.
Table 3.
Primers for amplification of the microsatellites at the exact position of the candidate gene loci
| Microsatellite | Forward primer (5'-3') | Reverse primer (5'-3') | Size of PCR-product (bp) | Nucleotide repeat | Location (bp) | Accession number |
| CNGB3-FH2772 | CCCAAAGCACATCCTAATTC | GGAGTCTGCTTCTCCCTTTC | 168 | di | Chr29:35405042-35405217 | UniSTS 263646 |
| CNGB3-C29.002 | TATTAAATCCCAGTCACCACCC | AGGTCCCAGACCGAGTCC | 208 | di | Chr29:36423257-36423471 | UniSTS 262830 |
| CNGA3-GT19 | CCTCCCACTCTCCCCTCTAC | CCAGGGGAGCTTTTACAACA | 337 | di | Chr10:47307640-47307977 | BV729091 |
| CNGA3-GT20 | GCAAGCAGTCCCGATTTTTA | TCAGCTTTGGTCATGCACTC | 304 | di | Chr10:47283009-47283313 | BV729076 |
| GNAT2-GAAA16 | CCCATGCTTGGTTTAATGCT | GACTGTCCTGCCTTCCATGT | 350 | tetra | Chr6:45194834-45195184 | BV729077 |
| GNAT2-CTT22 | CAGCTGGATTCTTCCCATGA | GCCCAAATTGCAAATCCTTA | 273 | tri | Chr6:45461591-45461864 | BV729078 |
| ABCA4-FH2370 | CCTGAAAAATAGCTAGATGATGG | GTCTTTACCTGCCTATATAGCTGC | 380 | tetra | Chr6:58367514-58367919 | UniSTS 263571 |
| ABCA4-TTTC16m | GGAATCAGTGGACTCATCCAA | GGGGATTGGACAGTGGTAGA | 238 | tetra | Chr6:58046419-58046656 | BV729079 |
| RDH5-TAGA10 | GAAGATGACGATGATGATGAAGA | GCTGAAGGTAGACGCTGGAC | 286 | tetra | Chr10:3068645-3068931 | BV729080 |
| RDH5-GAAA16 | CCCTGCTGTGAGGAGTCAGT | AGCCAGATGCAGGACTTGAT | 209 | tetra | Chr10:3002063-3002276 | BV729081 |
| CORD8-FMO5-GAAA12 | CCACAAGTTGGGGTTTCAAG | CCCCTCCTCTCTCTCTTTCC | 204 | tetra | Chr9:60070458-60070662 | BV729082 |
| CORD8-FMO5-TTTA10 | CCCAGGTGTCCCTATTTTCC | GCTCACTGGGGAGTCTGCT | 175 | tetra | Chr9:60654305-60654480 | BV729083 |
| CORD9-RP1-ATm22 | CACTGGATGCACACAGATCC | GGTCCTTGAGAAGGAAGCTG | 296 | di | Chr29:9128657-9128953 | BV729084 |
| CORD9-RP1-TTTC21m | TCCAGTAGGCGTCCTCTGAC | AGTCAATGGAGCCTGCAACT | 410 | tetra | Chr29:9011174-9011583 | BV729085 |
| RPGRIP1-GAAA26 | TGTTACCTGTTCCAAAGTTGTTTT | AGTTACAGCCATGGGAATGC | 475 | tetra | Chr15:21287275-21287750 | BV729086 |
| GUCY2D-TTCC15 | ACAATGGGCACATCTGTTGA | TTCTCCCTCTGCCTGTGTCT | 405 | tetra | Chr5:35459311-35459716 | BV729089 |
| GUCY2D-GAAA17 | CCCCTCTTCTCCACTCTCCT | TCATATTCTTGCCCCAGTCC | 444 | tetra | Chr5:35514793-35515246 | BV729090 |
| CRX-GAAA15m | TGGTCTCACATTCCCACTGA | AGAAGTGGCAGAGCACAGGT | 438 | tetra | Chr1:111096908-111097346 | BV729087 |
| CRX-GT21 | ACCAGAACCAAAGGCAGATG | TCAGGGTTGGAGTTTTGAGC | 427 | di | Chr1:111076912-111077339 | BV729088 |
PCR amplification reactions were performed using 83 μM of each primer in a 15 μl reaction containing 1.5 μl DNA prepared as described above, 1× PCR buffer containing 1.5 mM MgCl2, 83 μM each of dATP, dCTP, dGTP and dTTP, and 0,33 units Taq DNA polymerase (Qiagen). After an initial denaturation at 95°C for 2 min and 30 sec, samples were amplified for 28 cycles at 95°C for 30 sec, 58°C for 40 sec and 72°C for 50 sec, followed by a final extension of 72°C for 5 min and 30 sec. The sizes of the alleles were estimated with an automated sequencer (ABI PRISM® 3100 Genetic Analyzer, Applied Biosystems, Foster City, CA, U.S.A.) with software for fragment analysis.
Authors' contributions
FL and EB jointly conceived of the study. ACW carried out most of the practical molecular part of the study supervised by FL. ACW drafted the manuscript in collaboration with FL, EOR and EB. All authors read and approved the final manuscript.
Contributor Information
Anne Caroline Wiik, Email: annecaroline.wiik@veths.no.
Ernst-Otto Ropstad, Email: Ernst-Otto.Ropstad@veths.no.
Ellen Bjerkås, Email: ellen.bjerkaas@veths.no.
Frode Lingaas, Email: frode.lingaas@veths.no.
References
- Kijas JW, Miller BJ, Pearce-Kelling SE, Aguirre GD, Acland GM. Canine models of ocular disease: outcross breedings define a dominant disorder present in the English mastiff and bull mastiff dog breeds. J Hered. 2003;94:27–30. doi: 10.1093/jhered/esg007. [DOI] [PubMed] [Google Scholar]
- RetNet™ Retinal Information Network. 2007. http://www.sph.uth.tmc.edu/Retnet/
- Narfstrom K, Petersen-Jones S. Diseases of the canine ocular fundus. Blackwell publishing; 2006. [Google Scholar]
- Sharpe LT, Stockman A, Jägle H, Nathans J. In: Color vision: from genes to perception. edited by Karl R.Gegenfurtner LTS, editor. Cambridge, Cambridge University Press; 1999. pp. 3–52. (Opsin genes, cone photopigments and colorblindness.). [Google Scholar]
- Sidjanin DJ, Lowe JK, McElwee JL, Milne BS, Phippen TM, Sargan DR, Aguirre GD, Acland GM, Ostrander EA. Canine CNGB3 mutations establish cone degeneration as orthologous to the human achromatopsia locus ACHM3. Hum Mol Genet. 2002;11:1823–1833. doi: 10.1093/hmg/11.16.1823. [DOI] [PubMed] [Google Scholar]
- Aligianis IA, Forshew T, Johnson S, Michaelides M, Johnson CA, Trembath RC, Hunt DM, Moore AT, Maher ER. Mapping of a novel locus for achromatopsia (ACHM4) to 1p and identification of a germline mutation in the alpha subunit of cone transducin (GNAT2) J Med Genet. 2002;39:656–660. doi: 10.1136/jmg.39.9.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl S, Marx T, Giddings I, Jagle H, Jacobson SG, Apfelstedt-Sylla E, Zrenner E, Sharpe LT, Wissinger B. Total colourblindness is caused by mutations in the gene encoding the alpha-subunit of the cone photoreceptor cGMP-gated cation channel. Nat Genet. 1998;19:257–259. doi: 10.1038/935. [DOI] [PubMed] [Google Scholar]
- Kohl S, Baumann B, Broghammer M, Jagle H, Sieving P, Kellner U, Spegal R, Anastasi M, Zrenner E, Sharpe LT, Wissinger B. Mutations in the CNGB3 gene encoding the beta-subunit of the cone photoreceptor cGMP-gated channel are responsible for achromatopsia (ACHM3) linked to chromosome 8q21. Hum Mol Genet. 2000;9:2107–2116. doi: 10.1093/hmg/9.14.2107. [DOI] [PubMed] [Google Scholar]
- Rubin LF, Bourns TK, Lord LH. Hemeralopia in dogs: heredity of hemeralopia in Alaskan Malamutes. Am J Vet Res. 1967;28:355–357. [PubMed] [Google Scholar]
- Seddon JM, Hampson EC, Smith RI, Hughes IP. Genetic heterogeneity of day blindness in Alaskan Malamutes. Anim Genet. 2006;37:407–410. doi: 10.1111/j.1365-2052.2006.01484.x. [DOI] [PubMed] [Google Scholar]
- Hurn SD, Hardman C, Stanley RG. Day-blindness in three dogs: clinical and electroretinographic findings. Vet Ophthalmol. 2003;6:127–130. doi: 10.1046/j.1463-5224.2003.00278.x. [DOI] [PubMed] [Google Scholar]
- Rubin LF. Inherited eye diseases in purebred dogs. Baltimore, Williams & Wilkins; 1989. [Google Scholar]
- Michaelides M, Holder GE, Hunt DM, Fitzke FW, Bird AC, Moore AT. A detailed study of the phenotype of an autosomal dominant cone-rod dystrophy (CORD7) associated with mutation in the gene for RIM1. Br J Ophthalmol. 2005;89:198–206. doi: 10.1136/bjo.2004.050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udar N, Yelchits S, Chalukya M, Yellore V, Nusinowitz S, Silva-Garcia R, Vrabec T, Hussles M, I, Donoso L, Small KW. Identification of GUCY2D gene mutations in CORD5 families and evidence of incomplete penetrance. Hum Mutat. 2003;21:170–171. doi: 10.1002/humu.9109. [DOI] [PubMed] [Google Scholar]
- Allikmets R, Shroyer NF, Singh N, Seddon JM, Lewis RA, Bernstein PS, Peiffer A, Zabriskie NA, Li Y, Hutchinson A, Dean M, Lupski JR, Leppert M. Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science. 1997;277:1805–1807. doi: 10.1126/science.277.5333.1805. [DOI] [PubMed] [Google Scholar]
- Allikmets R, Singh N, Sun H, Shroyer NF, Hutchinson A, Chidambaram A, Gerrard B, Baird L, Stauffer D, Peiffer A, Rattner A, Smallwood P, Li Y, Anderson KL, Lewis RA, Nathans J, Leppert M, Dean M, Lupski JR. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet. 1997;15:236–246. doi: 10.1038/ng0397-236. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Simon A, Eriksson U, Harris E, Berson EL, Dryja TP. Mutations in the gene encoding 11-cis retinol dehydrogenase cause delayed dark adaptation and fundus albipunctatus. Nat Genet. 1999;22:188–191. doi: 10.1038/9707. [DOI] [PubMed] [Google Scholar]
- Ismail M, Abid A, Anwar K, Qasim MS, Khaliq S. Refinement of the locus for autosomal recessive cone-rod dystrophy (CORD8) linked to chromosome 1q23-q24 in a Pakistani family and exclusion of candidate genes. J Hum Genet. 2006;51:827–831. doi: 10.1007/s10038-006-0028-y. [DOI] [PubMed] [Google Scholar]
- Khaliq S, Hameed A, Ismail M, Anwar K, Leroy BP, Mehdi SQ, Payne AM, Bhattacharya SS. Novel locus for autosomal recessive cone-rod dystrophy CORD8 mapping to chromosome 1q12-Q24. Invest Ophthalmol Vis Sci. 2000;41:3709–3712. [PubMed] [Google Scholar]
- Danciger M, Hendrickson J, Lyon J, Toomes C, McHale JC, Fishman GA, Inglehearn CF, Jacobson SG, Farber DB. CORD9 a new locus for arCRD: mapping to 8p11, estimation of frequency, evaluation of a candidate gene. Invest Ophthalmol Vis Sci. 2001;42:2458–2465. [PubMed] [Google Scholar]
- Kijas JW, Zangerl B, Miller B, Nelson J, Kirkness EF, Aguirre GD, Acland GM. Cloning of the canine ABCA4 gene and evaluation in canine cone-rod dystrophies and progressive retinal atrophies. Mol Vis. 2004;10:223–232. [PubMed] [Google Scholar]
- Mellersh CS, Boursnell ME, Pettitt L, Ryder EJ, Holmes NG, Grafham D, Forman OP, Sampson J, Barnett KC, Blanton S, Binns MM, Vaudin M. Canine RPGRIP1 mutation establishes cone-rod dystrophy in miniature longhaired dachshunds as a homologue of human Leber congenital amaurosis. Genomics. 2006;88:293–301. doi: 10.1016/j.ygeno.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Ropstad EO, Bjerkas E, Narfstrom K. Clinical findings in early onset cone-rod dystrophy in the Standard Wire-haired Dachshund. Vet Ophthalmol. 2007;10:69–75. doi: 10.1111/j.1463-5224.2007.00503.x. [DOI] [PubMed] [Google Scholar]
- Dryja TP, Adams SM, Grimsby JL, McGee TL, Hong DH, Li T, Andreasson S, Berson EL. Null RPGRIP1 alleles in patients with Leber congenital amaurosis. Am J Hum Genet. 2001;68:1295–1298. doi: 10.1086/320113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber S, Perrault I, Hanein S, Barbet F, Ducroq D, Ghazi I, Martin-Coignard D, Leowski C, Homfray T, Dufier JL, Munnich A, Kaplan J, Rozet JM. Complete exon-intron structure of the RPGR-interacting protein (RPGRIP1) gene allows the identification of mutations underlying Leber congenital amaurosis. Eur J Hum Genet. 2001;9:561–571. doi: 10.1038/sj.ejhg.5200689. [DOI] [PubMed] [Google Scholar]
- Hanein S, Perrault I, Gerber S, Tanguy G, Barbet F, Ducroq D, Calvas P, Dollfus H, Hamel C, Lopponen T, Munier F, Santos L, Shalev S, Zafeiriou D, Dufier JL, Munnich A, Rozet JM, Kaplan J. Leber congenital amaurosis: comprehensive survey of the genetic heterogeneity, refinement of the clinical definition, and genotype-phenotype correlations as a strategy for molecular diagnosis. Hum Mutat. 2004;23:306–317. doi: 10.1002/humu.20010. [DOI] [PubMed] [Google Scholar]
- Cremers FP, van den Hurk JA, den Hollander AI. Molecular genetics of Leber congenital amaurosis. Hum Mol Genet. 2002;11:1169–1176. doi: 10.1093/hmg/11.10.1169. [DOI] [PubMed] [Google Scholar]
- Hameed A, Abid A, Aziz A, Ismail M, Mehdi SQ, Khaliq S. Evidence of RPGRIP1 gene mutations associated with recessive cone-rod dystrophy. J Med Genet. 2003;40:616–619. doi: 10.1136/jmg.40.8.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund CL, Gregory-Evans CY, Furukawa T, Papaioannou M, Looser J, Ploder L, Bellingham J, Ng D, Herbrick JA, Duncan A, Scherer SW, Tsui LC, Loutradis-Anagnostou A, Jacobson SG, Cepko CL, Bhattacharya SS, McInnes RR. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell. 1997;91:543–553. doi: 10.1016/S0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- Freund CL, Wang QL, Chen S, Muskat BL, Wiles CD, Sheffield VC, Jacobson SG, McInnes RR, Zack DJ, Stone EM. De novo mutations in the CRX homeobox gene associated with Leber congenital amaurosis. Nat Genet. 1998;18:311–312. doi: 10.1038/ng0498-311. [DOI] [PubMed] [Google Scholar]
- Kelsell RE, Evans K, Gregory CY, Moore AT, Bird AC, Hunt DM. Localisation of a gene for dominant cone-rod dystrophy (CORD6) to chromosome 17p. Hum Mol Genet. 1997;6:597–600. doi: 10.1093/hmg/6.4.597. [DOI] [PubMed] [Google Scholar]
- Sohocki MM, Sullivan LS, Mintz-Hittner HA, Birch D, Heckenlively JR, Freund CL, McInnes RR, Daiger SP. A range of clinical phenotypes associated with mutations in CRX, a photoreceptor transcription-factor gene. Am J Hum Genet. 1998;63:1307–1315. doi: 10.1086/302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain PK, Chen S, Wang QL, Affatigato LM, Coats CL, Brady KD, Fishman GA, Jacobson SG, Swaroop A, Stone E, Sieving PA, Zack DJ. Mutations in the cone-rod homeobox gene are associated with the cone-rod dystrophy photoreceptor degeneration. Neuron. 1997;19:1329–1336. doi: 10.1016/S0896-6273(00)80423-7. [DOI] [PubMed] [Google Scholar]
- Turney C, Chong NH, Alexander RA, Hogg CR, Fleming L, Flack D, Barnett KC, Bird AC, Holder GE, Luthert PJ. Pathological and electrophysiological features of a canine cone-rod dystrophy in the miniature longhaired dachshund. Invest Ophthalmol Vis Sci. 2007;48:4240–4249. doi: 10.1167/iovs.04-0737. [DOI] [PubMed] [Google Scholar]
- Ropstad EO, Bjerkas E, Narfstrom K. Electroretinographic findings in the Standard Wire Haired Dachshund with inherited early onset cone-rod dystrophy. Doc Ophthalmol. 2007;114:27–36. doi: 10.1007/s10633-006-9035-8. [DOI] [PubMed] [Google Scholar]
- Narfstrom K, Vaegan. Katz M, Bragadottir R, Rakoczy EP, Seeliger M. Assessment of Structure and Function Over a 3-year Period after Gene Transfer in RPE65-/- dogs. Doc Ophthalmol. 2005;111:39–48. doi: 10.1007/s10633-005-3159-0. [DOI] [PubMed] [Google Scholar]
- Acland GM, Aguirre GD, Bennett J, Aleman TS, Cideciyan AV, Bennicelli J, Dejneka NS, Pearce-Kelling SE, Maguire AM, Palczewski K, Hauswirth WW, Jacobson SG. Long-term restoration of rod and cone vision by single dose rAAV-mediated gene transfer to the retina in a canine model of childhood blindness. Mol Ther. 2005;12:1072–1082. doi: 10.1016/j.ymthe.2005.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acland GM, Aguirre GD, Ray J, Zhang Q, Aleman TS, Cideciyan AV, Pearce-Kelling SE, Anand V, Zeng Y, Maguire AM, Jacobson SG, Hauswirth WW, Bennett J. Gene therapy restores vision in a canine model of childhood blindness. Nat Genet. 2001;28:92–95. doi: 10.1038/88327. [DOI] [PubMed] [Google Scholar]
- Aguirre GK, Komaromy AM, Cideciyan AV, Brainard DH, Aleman TS, Roman AJ, Avants BB, Gee JC, Korczykowski M, Hauswirth WW, Acland GM, Aguirre GD, Jacobson SG. Canine and human visual cortex intact and responsive despite early retinal blindness from RPE65 mutation. PLoS Med. 2007;4:e230. doi: 10.1371/journal.pmed.0040230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofri R, Narfstrom K. Light at the end of the tunnel? Advances in the understanding and treatment of glaucoma and inherited retinal degeneration. Vet J. 2007;174:10–22. doi: 10.1016/j.tvjl.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Bainbridge JW, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K, Viswanathan A, Holder GE, Stockman A, Tyler N, Petersen-Jones S, Bhattacharya SS, Thrasher AJ, Fitzke FW, Carter BJ, Rubin GS, Moore AT, Ali RR. Effect of gene therapy on visual function in Leber's congenital amaurosis. N Engl J Med. 2008;358:2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- Clark LA, Tsai KL, Steiner JM, Williams DA, Guerra T, Ostrander EA, Galibert F, Murphy KE. Chromosome-specific microsatellite multiplex sets for linkage studies in the domestic dog. Genomics. 2004;84:550–554. doi: 10.1016/j.ygeno.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, III, Zody MC, Mauceli E, Xie X, Breen M, Wayne RK, Ostrander EA, Ponting CP, Galibert F, Smith DR, DeJong PJ, Kirkness E, Alvarez P, Biagi T, Brockman W, Butler J, Chin CW, Cook A, Cuff J, Daly MJ, DeCaprio D, Gnerre S, Grabherr M, Kellis M, Kleber M, Bardeleben C, Goodstadt L, Heger A, Hitte C, Kim L, Koepfli KP, Parker HG, Pollinger JP, Searle SM, Sutter NB, Thomas R, Webber C, Baldwin J, Abebe A, Abouelleil A, Aftuck L, Ait-Zahra M, Aldredge T, Allen N, An P, Anderson S, Antoine C, Arachchi H, Aslam A, Ayotte L, Bachantsang P, Barry A, Bayul T, Benamara M, Berlin A, Bessette D, Blitshteyn B, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Brown A, Cahill P, Calixte N, Camarata J, Cheshatsang Y, Chu J, Citroen M, Collymore A, Cooke P, Dawoe T, Daza R, Decktor K, DeGray S, Dhargay N, Dooley K, Dooley K, Dorje P, Dorjee K, Dorris L, Duffey N, Dupes A, Egbiremolen O, Elong R, Falk J, Farina A, Faro S, Ferguson D, Ferreira P, Fisher S, FitzGerald M, Foley K, Foley C, Franke A, Friedrich D, Gage D, Garber M, Gearin G, Giannoukos G, Goode T, Goyette A, Graham J, Grandbois E, Gyaltsen K, Hafez N, Hagopian D, Hagos B, Hall J, Healy C, Hegarty R, Honan T, Horn A, Houde N, Hughes L, Hunnicutt L, Husby M, Jester B, Jones C, Kamat A, Kanga B, Kells C, Khazanovich D, Kieu AC, Kisner P, Kumar M, Lance K, Landers T, Lara M, Lee W, Leger JP, Lennon N, Leuper L, LeVine S, Liu J, Liu X, Lokyitsang Y, Lokyitsang T, Lui A, Macdonald J, Major J, Marabella R, Maru K, Matthews C, McDonough S, Mehta T, Meldrim J, Melnikov A, Meneus L, Mihalev A, Mihova T, Miller K, Mittelman R, Mlenga V, Mulrain L, Munson G, Navidi A, Naylor J, Nguyen T, Nguyen N, Nguyen C, Nguyen T, Nicol R, Norbu N, Norbu C, Novod N, Nyima T, Olandt P, O'Neill B, O'Neill K, Osman S, Oyono L, Patti C, Perrin D, Phunkhang P, Pierre F, Priest M, Rachupka A, Raghuraman S, Rameau R, Ray V, Raymond C, Rege F, Rise C, Rogers J, Rogov P, Sahalie J, Settipalli S, Sharpe T, Shea T, Sheehan M, Sherpa N, Shi J, Shih D, Sloan J, Smith C, Sparrow T, Stalker J, Stange-Thomann N, Stavropoulos S, Stone C, Stone S, Sykes S, Tchuinga P, Tenzing P, Tesfaye S, Thoulutsang D, Thoulutsang Y, Topham K, Topping I, Tsamla T, Vassiliev H, Venkataraman V, Vo A, Wangchuk T, Wangdi T, Weiand M, Wilkinson J, Wilson A, Yadav S, Yang S, Yang X, Young G, Yu Q, Zainoun J, Zembek L, Zimmer A, Lander ES. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Karlsson EK, Baranowska I, Wade CM, Salmon Hillbertz NH, Zody MC, Anderson N, Biagi TM, Patterson N, Pielberg GR, Kulbokas EJ, III, Comstock KE, Keller ET, Mesirov JP, von EH, Kampe O, Hedhammar A, Lander ES, Andersson G, Andersson L, Lindblad-Toh K. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat Genet. 2007;39:1321–1328. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- Guyon R, Kirkness EF, Lorentzen TD, Hitte C, Comstock KE, Quignon P, Derrien T, Andre C, Fraser CM, Galibert F, Ostrander EA. Building comparative maps using 1.5x sequence coverage: human chromosome 1p and the canine genome. Cold Spring Harb Symp Quant Biol. 2003;68:171–177. doi: 10.1101/sqb.2003.68.171. [DOI] [PubMed] [Google Scholar]
- Breen M, Jouquand S, Renier C, Mellersh CS, Hitte C, Holmes NG, Cheron A, Suter N, Vignaux F, Bristow AE, Priat C, McCann E, Andre C, Boundy S, Gitsham P, Thomas R, Bridge WL, Spriggs HF, Ryder EJ, Curson A, Sampson J, Ostrander EA, Binns MM, Galibert F. Chromosome-specific single-locus FISH probes allow anchorage of an 1800-marker integrated radiation-hybrid/linkage map of the domestic dog genome to all chromosomes. Genome Res. 2001;11:1784–1795. doi: 10.1101/gr.189401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- primer3. 2007. http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi


