Abstract
The homeostasis of both cornea and hair follicles depends on a constant supply of progeny cells produced by populations of keratin (K) 14-expressing stem cells localized in specific niches. To investigate the potential role of Co-factors of LIM domains (Clims) in epithelial tissues, we generated transgenic mice expressing a dominant-negative Clim molecule (DN-Clim) under the control of the K14 promoter. As expected, the K14 promoter directed high level expression of the transgene to the basal cells of cornea and epidermis, as well as the outer root sheath of hair follicles. In corneal epithelium, the transgene expression causes decreased expression of adhesion molecule BP180 and defective hemidesmosomes, leading to detachment of corneal epithelium from the underlying stroma, which in turn causes blisters, wounds and an inflammatory response. After a period of epithelial thinning, the corneal epithelium undergoes differentiation to an epidermis-like structure. The K14-DN-Clim mice also develop progressive hair loss due to dysfunctional hair follicles that fail to generate hair shafts. The number of hair follicle stem cells is decreased by at least 60% in K14-DN-Clim mice, indicating that Clims are required for hair follicle stem cell maintenance. In addition, Clim2 interacts with Lhx2 in vivo, suggesting that Clim2 is an essential co-factor for the LIM homeodomain factor Lhx2, which was previously shown to play a role in hair follicle stem cell maintenance. Together, these data indicate that Clim proteins play important roles in the homeostasis of corneal epithelium and hair follicles.
Keywords: LIM domain transcription factors, Co-factors of LIM domains, Clim, Lhx2, hair follicle stem cells, cornea, adhesion, hemidesmosomes, BP180
Introduction
The LIM domain, a zinc finger motif that is conserved throughout evolution, is found in a diverse set of proteins, including transcription factors belonging to the LIM-homeodomain (Lhx) and LIM-only (LMO) protein families (Kadrmas and Beckerle, 2004). The Cofactor of LIM domains (Clim), which is also referred to as LIM domain binding protein (Ldb) and nuclear LIM interactor (NLI), was originally identified as a co-activator for Lhx and LMO proteins (Agulnick et al., 1996; Bach et al., 1997; Jurata et al., 1996; Visvader et al., 1997). Hence, one function of LIM domains in transcription factors is thought to be recruitment of Clim co-factors to enhance transcription of target genes.
In mice, there are two highly conserved genes in the Clim family: Clim2 (Ldb1) and Clim1 (Ldb2). The LIM-binding portion of Clims has been localized to the carboxy-terminus, while the amino-terminal region is involved in homodimer formation (Agulnick et al., 1996; Jurata and Gill, 1997; Matthews and Visvader, 2003). Through these interaction properties, Clims may act as specific protein-binding adapters, facilitating assembly of large complexes consisting of LIM-HD or LMO, as well as other classes of transcription factors with which LMOs can interacts, including GATA and bHLH DNA-binding proteins (Bach, 2000; Dawid et al., 1998).
Both Clim genes show widespread expression throughout mouse embryogenesis (Bach et al., 1997; Bach et al., 1999; Bulchand et al., 2003; Ostendorff et al., 2006; Visvader et al., 1997), and deletion of the Clim2 gene leads to early embryonic lethality around day 9.5. The Clim2-deleted mice exhibit a pleiotropic phenotype with no heart anlage and head structures that are truncated anterior to the hindbrain. In about 40% of the mutants, posterior axis duplication is observed. There are also severe defects in mesoderm-derived extraembryonic structures (Mukhopadhyay et al., 2003). Clim1 knockout mice do not exhibit an obvious phenotype (information from Mouse Genome Informatics website), most likely due to redundancy in function between Clim1 and Clim2. Expression of a dominant-negative Clim (DN-Clim) during early developmental stages of zebrafish embryos results in an impairment of eye and midbrain-hindbrain boundary development and interference with the formation of the anterior midline (Mukhopadhyay et al., 2003). While these studies have established a key role for Clim2 in early vertebrate development, the potential roles of Clims in late development and in the adult remain unknown.
We have previously shown that Clim2 is prominently expressed in several self-renewing stratified epithelial tissues where it is co-expressed with LMO4 (Sugihara et al., 1998). These tissues include epidermis, hair follicles and cornea – all epithelia that contain slow-cycling and keratin 14-expressing stem cell populations, which in the case of hair follicles and cornea, reside in specific niches. Throughout adult life, these adult stem cell populations maintain epithelial homeostasis by supplying new cells that undergo a series of specific differentiation steps and replace cells that have been shed (Cotsarelis, 2006; Tumbar et al., 2004). The potential roles of Clims in adult epithelial tissues remain unknown.
The early embryonic lethality of Clim2 knockout mice, as well as the redundancy between Clim2 and Clim1 proteins, creates a challenge for studies of Clim function in the late embryo and the adult. To avoid these difficulties, we investigated the role of Clims in cornea, epidermis and hair development by expressing a dominant-negative Clim molecule (DN-Clim) (Becker et al., 2002) under control of the keratin 14 (K14) promoter in mice. In these mice (K14-DN-Clim), the DN-Clim interferes with the function of both Clim2 and Clim1 in K14 expressing epithelia. The K14 promoter directs high level of expression to epidermis, hair follicles and cornea; there are several examples of such transgenic mice exhibiting phenotypes in both cornea and hair follicles (Kaya et al., 1997; Nicolas et al., 2003; Xie et al., 1999). The corneas of K14-DN-Clim mice develop edema and blisters caused by the detachment of corneal epithelium from the underlying stroma. Eventually the corneal epithelium in DN-Clim mice undergoes differentiation to epidermis-like epithelial structure. In hair follicles of K14-DN-Clim mice, stem cell maintenance is impaired, ultimately leading to hair follicle dysfunction and hair loss. Together, our data indicate that Clim proteins play an important role in the homeostasis of corneal epithelium and hair follicles.
Materials and methods
K14-DN-Clim mice
The Myc-tagged DN Clim fragment (Bach et al., 1999) was cloned downstream of the 1.5kb human K14 promoter (Leask et al., 1990; Sinha et al., 2000; Vassar et al., 1989; Wang et al., 1997). The linearized K14-DN-Clim fragment was then used to create CB6F1 transgenic mice according to standard protocols. All mice were genotyped by PCR analysis using the following primers: sense 5’-TGTATCACCATGGACCCTCAT-3’ and antisense 5’-AAGAAGAAGGCATGAACATGG-3’. Animal housing and experiments were performed according to the guidelines of the Institutional Animal Care and Use Committee.
Histology and Immunohistochemistry
Back skins and corneas were dissected and fixed in 10% Formalin in PBS or in 60% pure Ethanol, 30% water and 10% Formaldehyde. Samples were then embedded in paraffin blocks, and sectioned at 6 μm. For histological analysis, sections were stained with hemotoxylin and eosin (H&E). For immunohistochemistry, sections were deparaffinized, redehydrated, and washed in PBS. Antigen-retrieval was performed by incubating slides in 0.01M citrate buffer (pH 6.0) at 95 °C for 20 min. Sections were then immunostained with DakoCytomation LSAB+ System-HRP (DAKO) according to the manufacturer's instructions. For BrdU staining, mice were sacrificed 2 hr after BrdU injection for sample preparation. The dilutions of primary antibodies utilized are: 1:1000 rabbit anti-mouse K14 and K12 (Covance), 1:400 mouse anti-human K10 (DakoCytomation), 1:500 mouse monoclonal Myc-tag (Upstate), 1:200 goat anti-human Clim2 (Santa Cruz), 1:200 rat anti-mouse Clim (Ingolf Bach, University of Massachusetts Medical School), 1:1000 rabbit anti-mouse Filaggrin (Covance), 1:50 rat anti-mouse CD45 (BD Pharmingen), 1:200 mouse monoclonal anti-BrdU(Roche). For immunofluorescence, tissues were embedded in OCT and frozen sections were fixed in ice-cold acetone and subjected to immunofluorescence microscopy. Antibody and dilution used: 1:200 rabbit anti-mouse BP180 (Zhi Liu, University of North Carolina at Chapel Hill), Alexa488 conjugated goat anti-rabbit secondary antibody (1:1000, Molecular Probes). Nuclei were stained using 4’6’-diamidino-2-phenylindole (DAPI). For Oil Red O staining, frozen skin sections were stained with 0.5% Oil-Red O in 2-propanol at 37 °C for 30 min, followed by washes with 70% 2-propanol and H2O. Sections were counterstained with hematoxylin.
RNA Isolation and RNase Protection Assays
Back skin tissue samples were collected at different time points and after certain experimental manipulations. Total RNA was extracted using TrizolTM solution (Invitrogen) and polytron homogenization according to the manufacturer's instructions. RNA was precipitated with isopropanol and re-dissolved in diethyl pyrocarbonate (DEPC)-treated water. Yield and purity were assessed by UV spectrophotometry and denatured agarose gel electrophoresis. Antisense probes against murine cytoplasmic actin, Clim2 and LMO4 were synthesized by T7 polymerase (Promega) in the presence of 32P-labeled UTP (Amersham). The probes were purified through 5% (vol/vol) polyacrylamide/7 M urea gels, and eluted. Total RNA (20μg) was mixed with the probes and hybridized overnight at 65°C. After digestion with RNase One (Promega), the protected fragments were precipitated with ethanol, separated by electrophoresis through sequencing gels, and visualized by autoradiography.
Immunoprecipitations and Western Blot Analysis
Whole skins from newborn wild type and DN-Clim transgenic mice were dissected and treated overnight with 0.25% trypsin (Gibco) at 4°C to separate dermis from epidermis and hair follicles. The epidermal fraction was cut into pieces and pipetted up and down for 10 minutes, and neutralized cell suspensions were strained (70μM pores; BD Bioscience). Isolated cells were lysed with TNE lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 5 mM EDTA, 5% glycerol, 10 μg leupeptin per ml, 10 μg of aprotinin per ml, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 2 mM sodium vanadate). Nuclear extracts were precleared at 4°C using Protein G Sepharose beads and affinity-purified IgG (rabbit and goat, Santa Cruz). 5μg of LMO4 (Santa Cruz) and Lhx2 (Edwin Monuki, University of California, Irvine) antibodies were used. Immunoprecipitations were performed in PBS overnight at 4°C using protein-G Sepharose beads. Washes were carried out at room temperature in PBS. Bound material was eluted by boiling in 2X SDS sample buffer. Proteins were separated by SDS-PAGE and electrotransferred to polyvinylidene difluoride membrane (100 V for 1 h) in Tris-glycine buffer. Blots were probed with primary antibodies Clim2 (1: 100, Santa Cruz) and anti-Myc tag (1: 200, Upstate) and detected, after incubation with horseradish peroxidase-conjugated secondary antibody by SuperSignal West Pico chemiluminescent substrate (Pierce).
Hair Depilation
Depilation was performed on 8-week-old mice in telogen. The backs of mice were painted with a mixture of wax and rosin, and the embedded dorsal fur was gently removed.
Transfections and luciferase assays
Cotransfection assays were performed in HEK293 cells. Cells at 50% confluency were transiently transfected using Calcium Phosphate as previously described (Wang et al., 2004).
DNA microarray experiments
Microarray experiments using Affymetrix MU74Av2 chips were performed as previously described (Lin et al., 2004). For each time point, RNA was isolated and analyzed from 3 to 5 transgenic mice and same number of wild type littermates. The Cyber-T program (Baldi and Long, 2001) was used to determine statistically significant differentially expressed genes. Overrepresented gene ontology biological process categories for differentially expressed genes were determined using the DAVID (Database for Annotation, Visualization and Integrated Discovery) 2007 program (Dennis et al., 2003). PANTHER/X ontology was used to classify the significantly differentially expressed genes into biological process ontology categories (Thomas et al., 2003). Primary microarray data from this paper is accessible at NCBI Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo), GSE8227.
Fluorescence Activated Cell Sorting (FACS)
Back skins from 8-week-old wild type and DN-Clim transgenic mice were dissected and treated overnight with 0.25% trypsin (Gibco) at 4°C, which helps to separate the epidermis and hair follicles from the rest of the skin. This epidermal fraction was cut into pieces and pipetted up and down for 10 minutes, and neutralized cell suspensions were strained (70μM pores; BD Bioscience). Single cells were resuspended in PBS (Gibco) with 5% FBS, and incubated with primary antibodies for 30 minutes at room temperature. After washing with PBS, cells were stained with streptavidin and antibodies directly conjugated to specific fluorophors for 30 minutes. Flow cytometry was performed on FACSort equipped with CellQuest (BD Biosciences). Epidermal cells were gated for single events and viability, and then sorted according to their expression of CD34 and α6-integrin. Primary antibodies used for FACS: anti-mouse CD34 conjugated to biotin at 1μg per 4×106 cells (eBioscience); Streptavidin-Peridinin Chlorophyll-a Protein (SAv-PerCP) conjugate (1:200, BD Pharmingen), α6-integrin coupled to FITC at 1μg per 4×106 cells (Serotec).
BrdU Long-term Label Retaining Experiment
5’-Bromo-2’-deoxyuridine (BrdU, Sigma-Aldrich) pulse chase experiments were performed as described (Braun et al., 2003). Intra-peritoneal injections (50μg/g BrdU) were carried out twice a day for 3 days at post-natal day 4 and analyzed 10 weeks later for label retention. BrdU incorporation was detected by anti-mouse BrdU antibody (1:200, Roche). Six wild type and ten DN-Clim transgenic mice were used for long-term BrdU label retaining experiment.
Results
Clim co-factors are expressed in corneal epithelium, epidermis and hair follicles
Previous studies showed that transcripts for Clim2 and its interaction partner LMO4 are prominently expressed in epidermis and hair follicles (Bach et al., 1997; Sugihara et al., 1998). Similar to hair follicles and epidermis, the cornea is an actively renewing and stratified epithelial tissue that contains a population of slow-cycling stem cells required for homeostasis. Therefore, we investigated whether Clims are also expressed within the cornea. By means of immunostaining with an antibody that recognizes Clim1 and Clim2, we found that Clims are prominently expressed in the corneal epithelium, including the limbal epithelium, which contains the corneal stem cells (Fig. 1A-B). Both Clim2 and LMO4 transcripts are expressed throughout the initial hair follicle morphogenesis and during hair cycling (Fig. 1C); the temporal pattern of expression for both genes is similar with the first peak of expression during mid morphogenesis, on postnatal (PN) day 5, followed by decreased expression as the hair shaft differentiates on PN days 8 to 15. There is another peak of expression for both genes during catagen. Similar expression patterns were observed in a microarray expression study of the hair cycle (data not shown). To determine the spatial distribution of Clims within mouse back skin, we studied the expression of Clims by immunostaining (Becker et al., 2002). As early as embryonic (E) day 13.5, Clims are expressed in embryonic ectoderm and in the underlying mesenchyma (data not shown). In early stage hair follicles, Clim expression is prominent in cells of the hair germ and in the underlying mesenchymal aggregate (Fig. 1D). As the hair follicle forms, Clim expression becomes located to the basal cell layer of the epidermis and the outer root sheath (ORS) of the follicles (Fig. 1E-F). In addition, expression is prominent in cells of the hair follicle matrix and in a subset of dermal papilla cells (Fig. 1F). During catagen, high expression is found in cells of the matrix and ORS of the regressing follicle (Fig. 1G-I). In telogen follicles, Clim is highly expressed in the upper part of the ORS and epidermis, the bulge cells, and the dermal papilla (Fig. 1J). There is Clim expression in a subset of dermal fibroblasts (Fig. 1 E and G) but no expression within the subcutaneous fat (Fig. 1J). In hair follicles, the expression pattern of Clims is similar to that of LMO4 (Sugihara et al., 1998), and Clim2 and LMO4 co-immunoprecipitate in skin extracts, indicating that these proteins interact in vivo (Fig. 1K). In summary, the expression of Clims in three distinct epithelial tissues, cornea, epidermis and hair follicles, is similar in that it is primarily localized to proliferating epithelial cells and overlaps with the stem cell compartments. These data suggest that Clims may be involved in the development and/or homeostasis of cornea, epidermis and hair follicles.
Figure 1. The temporal and spatial expression pattern of Clims.
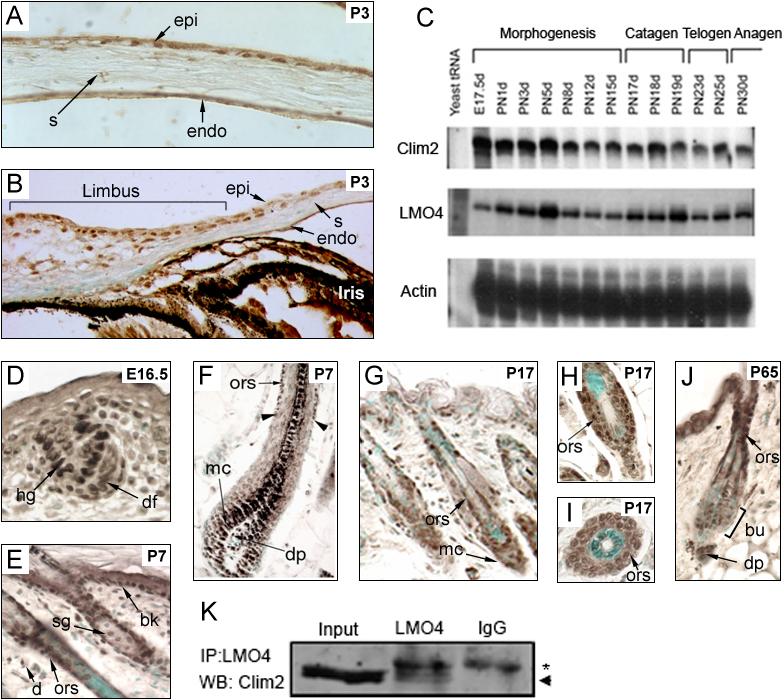
(A-B) Immunostaining of Clims in neonatal (P3) corneal epithelium. (C) Skin RNA was isolated from mice at the indicated developmental time points, and Clim2 and LMO4 transcript levels were determined with Clim2 and LMO4 cRNA probes in RNase protection assays. β-actin was used as a control. (D-J) Immunostaining of Clim expression during epidermis and hair follicle morphogenesis and the beginning of hair cycling. The dark pigment in the centrally located medulla of the hair follicle shown in panel F is endogenous melanin. The arrowheads in panel F point to the deepest point of Clim expression within the ORS. The counter stain in skin sections is methyl green. (K) Co-immunoprecipitation of endogenous Clim2 and LMO4. Skin extract was immunoprecipitated with either an LMO4 antibody or IgG as a control. Western blot of immunoprecipitate was probed with a Clim2 antibody. The arrowhead indicates the location of Clim2 whereas the asterisk indicates the IgG background band. bk, basal keratinocytes; bu, bulge; d, dermis; df, dermal fibroblast; dp, dermal papilla; E, embryonic day; endo, endothelium; epi, epithelium; hg, hair germ; mc, matrix cells; ors, outer root sheet; P, postnatal day; s, stroma; sg, sebaceous gland. Original magnification: A-C and L-M, 400X; E-K and N, 200X.
Mice with keratin 14-driven expression of a dominant-negative (DN) Clim develop hair loss and corneal opacity
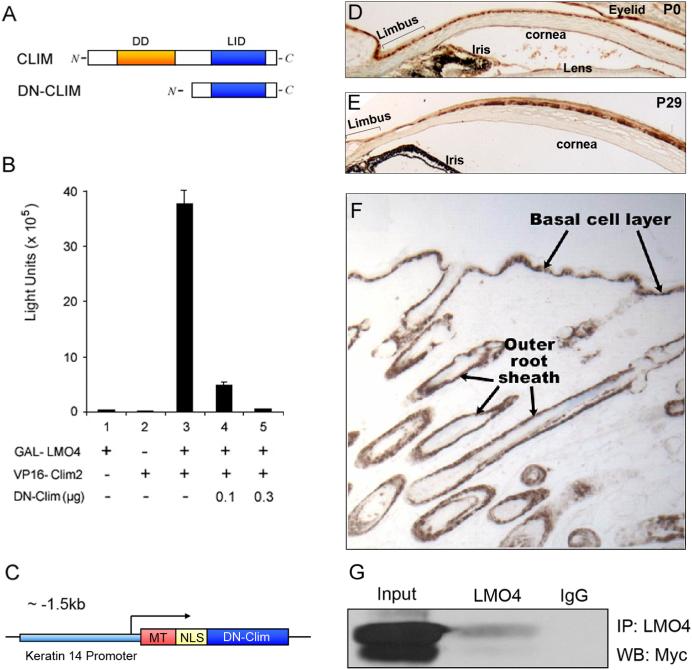
Clim proteins contain at least two domains that are essential for their normal function: a C-terminally located LIM-interaction domain (LID), which mediates high-affinity interactions with LIM domain proteins and other transcription factors, and an N-terminally located dimerization domain (DD), which mediates homodimerization of Clims (Fig. 2A). Experiments in diverse species have demonstrated that the singular expression of either of these two domains creates effective dominant negative Clims (Bach et al., 1999; Becker et al., 2002; Gimnopoulos et al., 2002). In our experiments, we took advantage of a Myc-tagged truncated version of Clim containing the highly conserved LID fused to a nuclear localization sequence (Becker et al., 2002). To demonstrate the dominant-negative (DN) activity of this protein, we tested its ability to disrupt interactions between Clim and LMO4 in a mammalian two-hybrid assay. LMO4 was fused to the DNA-binding domain of GAL4 which binds to GAL sites upstream of the TK promoter and luciferase. Clim2 was fused to the herpes simplex virus activator protein VP16. The hybrid protein (GAL4-VP16) activates transcription efficiently in mammalian cells, allowing the interaction between LMO4 and Clim2 to be detected and measured (Fig. 2B). As expected, transfection of LMO4-GAL4 or Clim2-VP16 alone had no effect on the transcription, whereas co-transfection of these expression plasmids strongly stimulated transcription. Transfection with increasing amounts of DN-Clim caused dose-dependent decrease of transcription, indicating that DN-Clim disrupts interactions between Clims and LIM domain proteins (Fig. 2B).
Figure 2. Construction of K14-DN-Clim mice.
(A) The upper panel is a schematic of the domain structure of Clims, showing the dimerization domain (DD) and the LIM-interacting domain (LID). The lower panel shows the dominant negative form of Clim (DN-Clim) containing the LID. (B) A GAL-luciferase plasmid was transiently cotransfected into HEK293T cells with the indicated combinations of expression plasmids encoding GAL-LMO4 fusion protein, VP16-Clim2 fusion protein, and DN-Clim. We determined relative luciferase activity 40h after the transfection. The results represent the mean and SEM from at least three different experiments. (C) A schematic of the transgene showing Myc-tagged (MT) DN-Clim containing a nuclear localization signal (NLS) under control of the K14 promoter. (D-E) Immunostaining for DN-Clim expression in corneal epithelium, including limbal epithelium, at P0 and P29. (F) Immunostaining for DN-Clim expression in transgenic skin. (G) Co-immunoprecipitation of DN-Clim and LMO4. The panel shows a Western blot with Myc-tag antibody after immunoprecipitaition with an LMO4 antibody. Original magnification: E and G, 100X; F, 40X.
To investigate the roles of Clims in epithelial tissues, we created transgenic mice expressing Myc-tagged DN-Clim under control of the well-characterized keratin 14 (K14) promoter (Fig. 2C). Immunostaining using anti-Myc antibody showed strong transgene expression in the basal layer of the corneal epithelium, both centrally and in the limbal region (Fig. 2D-E). In addition, the transgene was expressed in the basal layer of epidermis and outer root sheath (ORS) of hair follicles (Fig. 2F). An LMO4 antibody readily co-immunoprecipitated the DN-Clim protein in extracts from neonatal epidermis, indicating that the transgene interacts with LIM domains in vivo (Fig. 2G). In addition to several lines of mice that were analyzed as neonates, two independent lines were followed to adult age. Both lines of transgenic mice showed hair loss and corneal abnormalities; one of these lines was selected for further characterization.
Blisters, wounds and disrupted homeostasis of the corneal epithelium in K14-DN-Clim mice
The K14-DN-Clim transgenic mice developed striking corneal abnormalities. By one month of age, the ocular surface of approximately half of the transgenic mice became opacified. As the mice aged, corneal abnormalities became more severe and included neovascularization; by 6 months nearly all mice were affected (Fig. 3A). Histologically, the abnormal corneal epithelium in older mice was hyperplastic, cornified and the stroma was vascularized (Fig. 3B); sebaceous-like cells were observed in the corneal epithelium of one transgenic mouse (Fig. 3C). In the highly hyperplastic transgenic corneal epithelium, expression of the corneal-specific K12 was almost extinguished. In contrast, the epidermal markers K10 and filaggrin were expressed in the transgenic corneal epithelium (Fig. 3D). In addition, the corneal epithelium developed a basal layer which was morphologically similar to epidermal basal layer. This basal layer was highly proliferative as seen by BrdU staining and expressed the Myc-tagged transgene (Fig. 3D). These data show that the corneal epithelium in K14-DN-Clim transgenic mice ultimately converted to a keratinized epidermis-like epithelium, implying an important regulatory role for Clims in epithelial homeostasis in the cornea.
Figure 3. Epidermis-like differentiation of cornea in K14-DN-Clim mice.
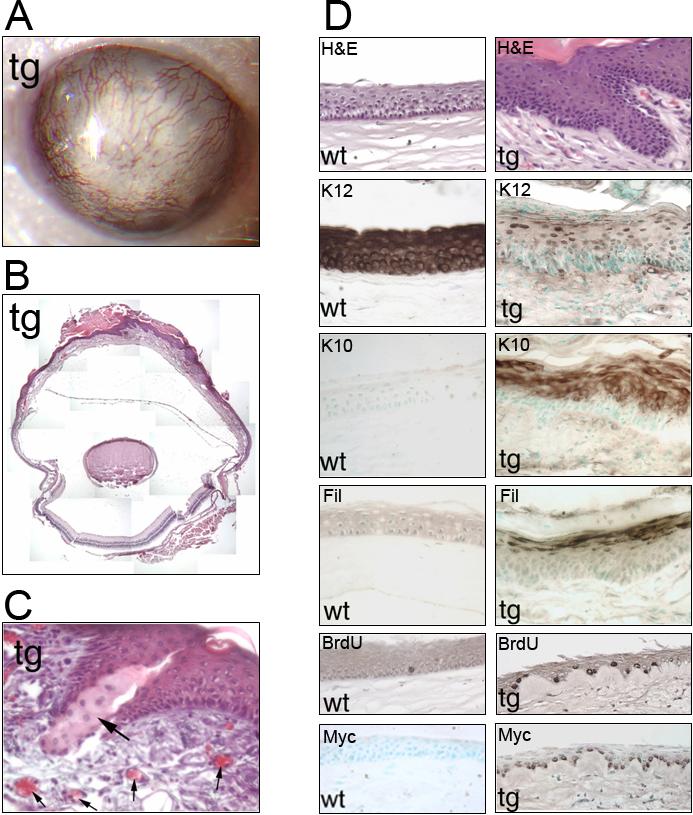
(A) Corneal opacity and neovascularization in a 1-year-old K14-DN-Clim mouse. (B) Overview of the histology of an eye from a 1-year-old K14-DN-Clim mouse. Iris and part of lens were removed prior to embedding. (C) Histological analysis of the cornea from a K14-DN-Clim transgenic mouse. The large arrow points to sebaceous-like cells. Small arrows point to blood vessels. (D) Histology and immunostaining of corneal epithelium from wild type and K14-DN-Clim mice. Antibodies to the following keratinocyte differentiation markers were used: K12, K10 and Filaggrin. The second to the bottom panel shows BrdU staining of corneal epithelium from wild type and K14-DN-Clim mice. The bottom panel shows expression of the DN-Clim transgene in wild type and K14-DN-Clim cornea as detected with anti-Myc antibody. The two lowest panels are from a stage when corneal thinning is prominent while the other panels are from corneas of one year old mice. tg, K14-DN-Clim transgenic; wt, wild type; Fil, Filaggrin. Original magnification: A, 100X; C, 400X; D, 200X.
To understand the pathogenesis of corneal abnormalities in DN-Clim mice, we examined the progressive development of the phenotype. As early as postnatal day (P) 0, approximately 50% of transgenic mice exhibited subepithelial edema of the stroma (Fig. 4A, B and Supplementary Table 1); during the first few postnatal days, the number of mice exhibiting subepithelial edema increased rapidly. Also, during this period, the corneal epithelium of the transgenic mice was much thicker than that of wild type controls (Fig. 4A, B). In addition, we observed occasional blisters, apparently caused by detachment of the corneal epithelial from the underlying stroma in the presence of edema (Fig. 4C), and wounds with intense inflammatory infiltrate, possibly caused by ruptured blisters (Fig. 4D). Between days 11 and 16, when the wild type epithelium had thickened to 3−5 cell layers, the transgenic corneal epithelium became as thin as one cell layer (Fig. 4E, F). The corneal thinning was common up to age 5 months, when corneas started to show features of epidermis-like differentiation. The penetrance of the corneal phenotype is incomplete in newborn mice but reaches 100% by P 10 (Supplementary Table 1). Some mice opened their eyes prematurely (data not shown) but corneal pathology was independent of this abnormality. We observed no CD45+ cells in neonatal corneas of transgenic mice exhibiting hyperplasia and blisters (Fig. 4G, H). In contrast, CD45 immunostaining illustrated abundant migration of leukocytes across the stroma and into the epithelium after wounds had developed (Fig. 4I). These data indicate that the inflammatory feature in the transgenic cornea is secondary to epithelial disruption and not the primary cause of the corneal abnormality.
Figure 4. Subepithelial edema, blisters and wounds in K14-DN-Clim corneas.
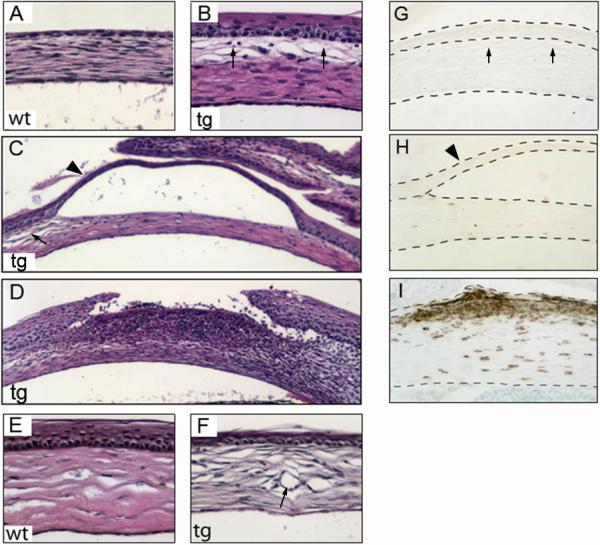
(A-F) Histological analysis of corneas from wild type and K14-DN-Clim mice during postnatal development (A-B, P0; C-D, P11; E-F, P29). (G-I) Immunostaining of cornea from K14-DN-Clim mice with pan-leukocyte marker CD45 (G, P0; H, P11; I P29). The dashed lines show the outlines of corneal epithelium and stroma. The arrows in B, C and G indicate the subepithelial edema; arrowheads in C and H indicate a blister; the arrow in F points to a blood vessel. wt, wild type; tg, K14-DN-Clim transgenic. Original magnification: A-B, E-F and G-I, 200X; C-D, 100X.
Defective hemidesmosomes and epithelial adhesion in corneas of K14-DN-Clim mice
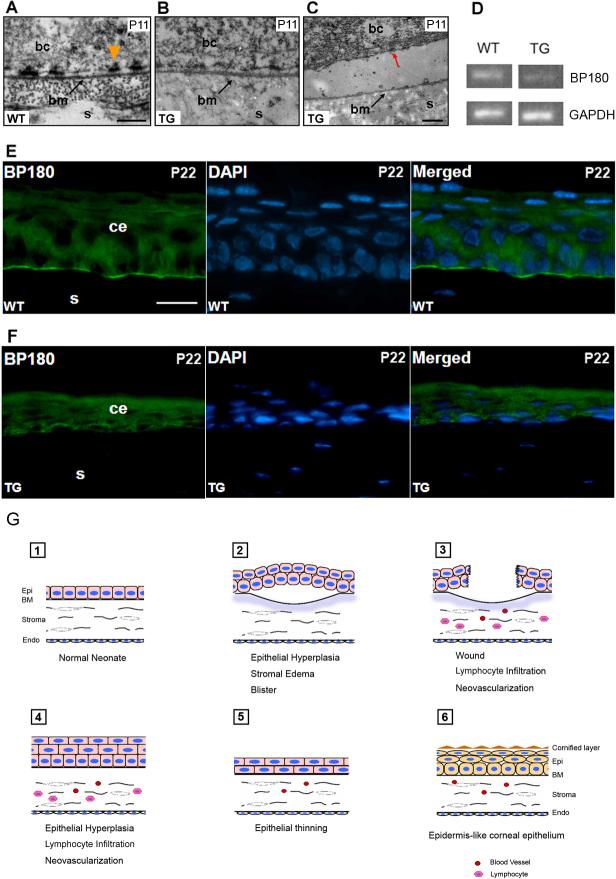
To understand the cause of corneal blister formation, we studied the epithelial-stromal junction of neonatal corneas by transmission electron microscopy. Hemidesmosomes in corneas of transgenic mice were smaller and thinner than that in corneas of wild type mice (Fig. 5A, B). This abnormality was found in short discontinuous stretches throughout the cornea, affecting approximately a third of each cornea. At the location of a blister, we observed separation of basal epithelial cells from the underlying basement membrane (Fig. 5C), suggesting that the hemidesmosome abnormality underlies epithelial-basement membrane separation and blister formation in the cornea. Transcripts encoding BP180, a transmembrane component of the hemidesmosome, were downregulated approximately 2-fold (Fig. 5D). Immunofluorescence study with anti-BP180 antibody showed that BP180 expression was downregulated in discontinuous stretches throughout the corneas of transgenic mice (not shown), and in some parts of the transgenic cornea, BP180 expression was nearly undetectable (Fig. 5E, F). Together, these data suggest that decreased BP180 protein expression and hemidesmosome dysfunction causes blister formation in transgenic corneas. We believe that this is the primary abnormality leading to repeated wounding, inflammatory infiltrate and hyperplasia. Later, corneal epithelial thinning is observed, similar to that found in human stem cell deficient corneas (Coster et al., 1995; Dua et al., 2000). Ultimately, differentiation of corneal epithelial cells is subverted to an epidermis-like pathway (Fig. 5G).
Figure 5. Defective hemidesmosomes and decreased BP180 expression in K14-DN-Clim mice.
(A-C) Electron microscopy ultrastructure of the epithelial-basement region, including hemidesmosomes, in corneas from mice of the indicated genotypes. Red arrow indicates a hemidesmosome on the basal surface of a basal epithelial cell that has separated from the underlying basal lamina. (D) Semiquantitative RT-PCR analysis of BP180 mRNA levels in cornea from wild type (WT) and K14-DN-Clim (TG) mice. GAPDH is included as a control. (E, F) Immunofluorescence analysis of BP180 expression in wild type and K14-DN-Clim corneal epithelium. DAPI staining shows nuclei in the same section. (G) Model for the progression of corneal abnormalities in K14-DN-Clim mice. bc, basal epithelial cell; bm (or BM), basement membrane; ce, corneal epithelium; Endo, endothelium; Epi, epithelium; s, stroma; Scale bar: A, 200nm; C, 500nm; E, 20μm.
Hair loss and aberrant hair follicle homeostasis in K14-DN-Clim mice
The K14-DN-Clim mice developed progressive age-dependent hair loss. To investigate the cause of hair loss, we examined hair follicles in relationship to the hair growth cycle (Morris et al., 2004; Muller-Rover et al., 2001). In mice, initial hair follicle morphogenesis is completed around P14, at which time the lower two-thirds of the follicle undergoes programmed cell death in a process referred to as catagen. The follicle then enters a resting phase (telogen), until a new cycle is initiated around P23; the growth phase of the follicle is referred to as anagen. The hair growth cycle is repeated continuously throughout the life of a mouse, with the first two cycles being synchronized. After completion of hair follicle morphogenesis and the first catagen, approximately at P20, the majority of K14-DN-Clim mice had normal coat, but occasional mice showed very mild hair loss in the head-neck and peri-tail regions. At the end of second hair cycle (P49), the coat was scraggly and, areas of local baldness were observed on the back. On the ventral surface, sparse hair was uniformly observed (data not shown). As the mice aged, the areas of baldness enlarged, both on the dorsal and ventral sides (Fig. 6A, B). Eventually, some of the transgenic mice became completely bald (Fig. 6C, D); the adult phenotype varies from mild hair loss with scraggly coat to complete baldness over the entire body. Some of the older transgenic mice also developed wounds on the back (Fig. 6D). Consistent with the postnatal onset of hair loss, hair follicle density was normal in K14-DN-Clim mice at all time points (Fig. 6E, F); even mice with nearly complete baldness had normal number of hair follicles. Therefore, the progressive hair-loss phenotype appears to be caused by hair follicle dysfunction rather than decreased number of follicles.
Figure 6. Hair loss in K14-DN-Clim mice.
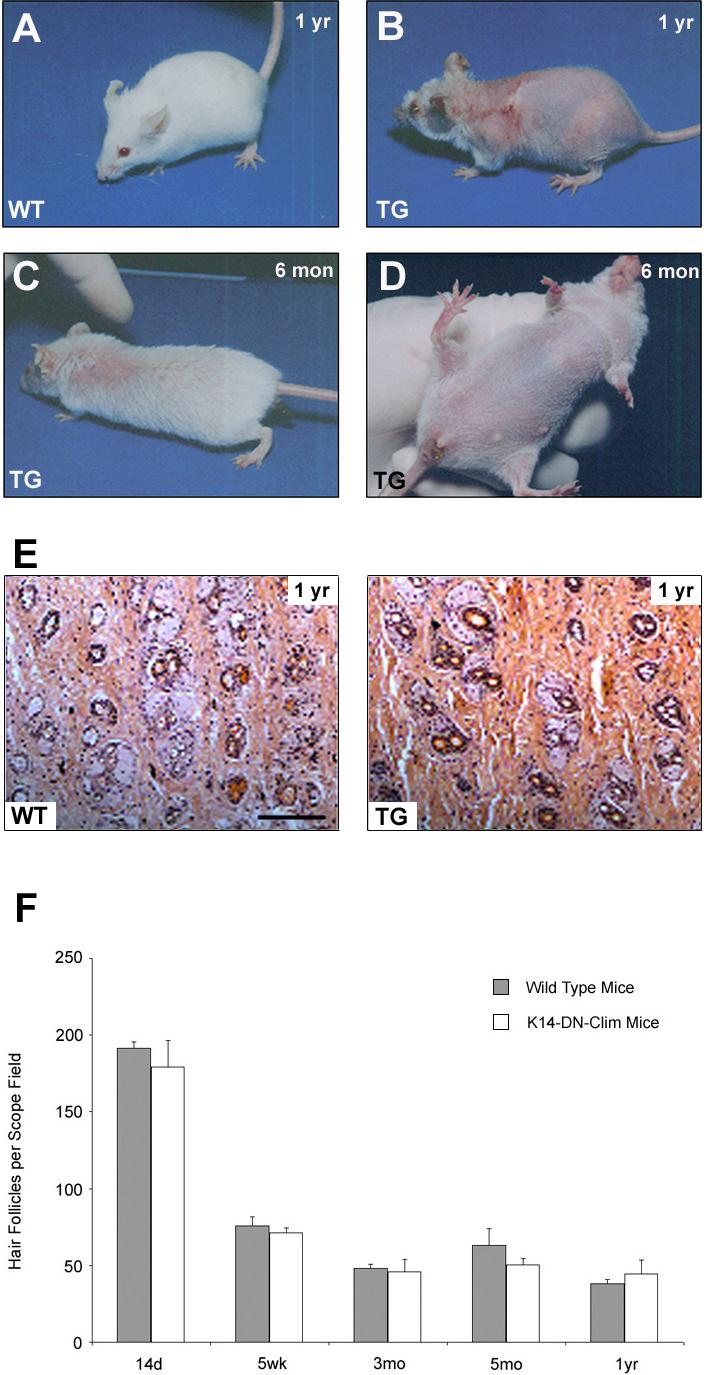
(A-D) Appearance of K14-DN-Clim (TG) mice and their littermate controls (WT) at indicated time points. (E) Histological analysis of hair follicle densities in back skin from 1-year-old wild-type and K14-DN-Clim mice. (F) Hair follicle number per scope field was counted at the indicated time points for wild type and K14-DN-Clim mice. Shown are means and standard errors from at least three different skin samples for each time point.
Hair follicle morphogenesis was normal in K14-DN-Clim mice and except for the slightly slower progression through catagen, the first two hair cycles progressed normally (Supplementary Fig. 1 and Supplementary Table 2). However, consistent with hair follicle dysfunction, hair follicles became clearly abnormal in adult transgenic mice. Most prominent were single large cysts or multiple small cysts within follicles (Fig. 7A-D). In addition, enlarged sebaceous glands became evident during the second hair cycle and in adult mice, sebaceous glands became prominent part of the follicle (Fig. 7E, F), easily detected with OilRedO staining (Fig. 7G, H). Instead of being close to the epidermis as in wild type mice, the sebocytes in K14-DN-Clim mice extended deep into the hair follicles (Fig. 7E, F). No expression of epidermal markers, such as K10, filaggrin and loricrin, was detected in cysts or other parts of the abnormal transgenic follicles (data not shown), indicating that aberrant differentiation along the sebocyte lineage was the most prominent differentiation abnormality.
Figure 7. Abnormal hair follicle structures in K14-DN-Clim mice.
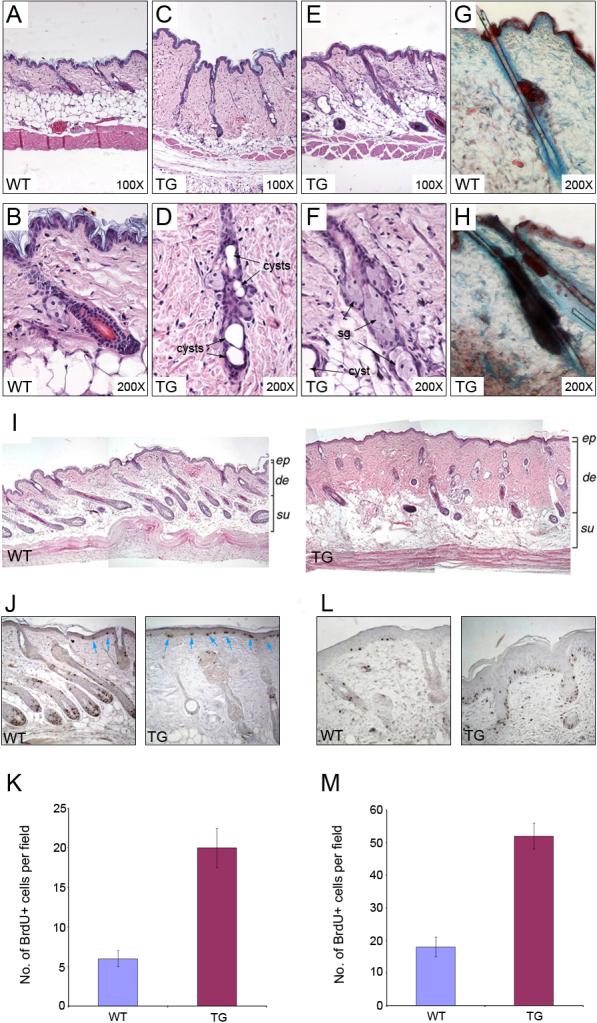
(A-F) Histological analysis of hair follicles in 7-month-old wild type (WT) and K14-DN-Clim (TG) mice. B, D and F are higher magnifications of A, C and E, respectively. (G, H) Oil Red O staining for sebaceous glands in 49d wild type and K14-DN-Clim skin. (I) Histology of skin 5 days after hair depilation in wild type (upper panel) and K14-DN-Clim (lower panel) mice. (J) BrdU staining in wild type and K14-DN-Clim mice after hair plucking. Blue arrows point to the BrdU positive cells in epidermis. (K) Quantification of BrdU staining in wild type and K14-DN-Clim mice after hair plucking. (L) BrdU staining in wild type and K14-DN-Clim mice 4 days after wounding. (M) Quantification of BrdU staining in wild type and K14-DN-Clim mice after wounding. de, dermis; ep, epidermis; sg, sebaceous gland; su, subcutis. Original magnification: A, C, E and I-K, 100X; B, D, F, G and H, 200X.
Unlike wild type mice that underwent re-growth of hair follicles quickly after hair plucking, transgenic hair follicles only occasionally entered anagen (Fig. 7I). Hair plucking also led to an exaggerated dermal reaction in K14-DN-Clim mice (Fig. 7I). Consistent with an enhanced wounding response, both hair plucking (Fig. 7J, K) and incisional wounding (Fig. 7L, M) caused increased epidermal proliferation compared to wild type mice. Interestingly the abnormal hair follicles in transgenic mice clearly showed decreased proliferation after plucking, perhaps due to the structural abnormalities resulting in a lack of normal matrix compartment where proliferation is most prominent in wild type follicles (Fig. 7J). In unperturbed skin there was no difference in epidermal proliferation between the K14-DN-Clim and wild type mice (data not shown). Prior to morphological abnormalities of hair follicles, the expression of AE 13 (anti-acidic hair keratin) (Lynch et al., 1986) and AE15 (anti-trychohyalin) (O'Guin et al., 1992) was normal in transgenic hair follicles (Supplementary Fig. 2), suggesting that a differentiation defect is not the primary reason for hair follicle dysfunction. Together, these data indicate that Clims play pleiotropic roles in hair follicle regulation, affecting hair cycling control, keratinocyte differentiation and structure of the follicle. We conclude that aberrant hair follicle differentiation and disrupted hair follicle structure contribute to the hair loss phenotype.
Transcripts normally enriched in hair follicle stem cells are downregulated in telogen skin of K14-DN-Clim mice
To gain insights into the molecular mechanisms of hair loss in K14-DN-Clim mice, we used Affymetrix DNA microarrays to profile mRNA expression in mouse back skin from 3 time points, representing the initial hair follicle morphogenesis (P6 and P14) and the first telogen (P23); hair growth is synchronized during these time points. We used the Cyber-T program to identify statistically significant differentially regulated genes in the K14-DN-Clim mice (Fig. 8A). At P6, P14 and P23, we found, respectively, 55, 182 and 248 statistically significant differentially expressed genes.
Figure 8. Abnormal gene expression profiles in K14-DN-Clim skin.
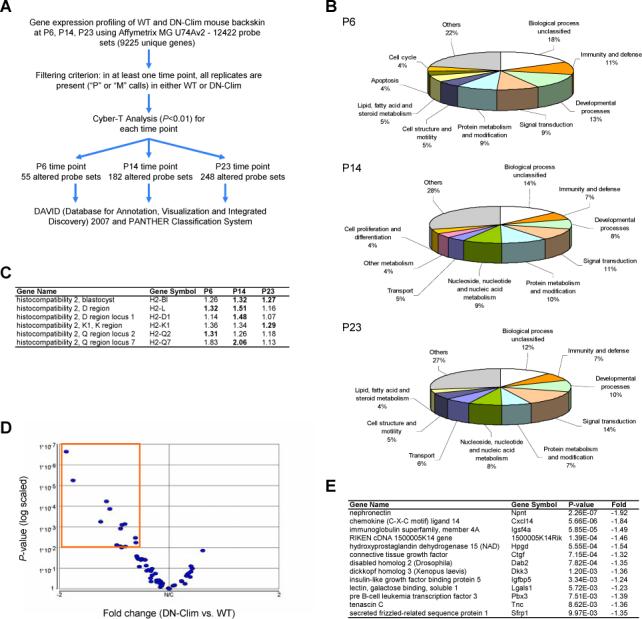
(A) Overview of microarray data processing and statistical tests to identify significantly differentially expressed genes in K14-DN-Clim transgenic mouse back skin. (B) Breakdown of biological process categories (PANTHER/X ontology) for the significantly differentially expressed genes for each of the three profiled time points: pP6, P14, and P23. (C) Members of the major histocompatibility complex genes are significantly differentially expressed at the three profiled time points. Shown are the fold changes between DN-Clim and wild-type mouse back skin and the significant differential expression is highlighted in bold. (D) Volcano plot of the significance (p-value) and degree of differential expression (fold change) between DN-Clim and wild-type P23 mouse backskin for genes known to be enriched in hair follicle stem cells. Dots within the orange outlined box represent significantly differentially expressed genes (p<0.01). These genes are listed in (E).
Analysis of over-representation of Gene Ontology categories revealed that immunity and defense genes are overrepresented as early as day 6 and persist throughout all three time points (Fig. 8B). These include histocompatibility genes H2-L, H2-D1, H2-K1, H2-Q2 and H2-Q7, all of which are upregulated (Fig. 8C). Most of these genes encode MHC I antigens, indicating that the MHC class I-dependent pathway of antigen presentation is upregulated prior to observable histological abnormalities. Activation of this antigen presentation pathway suggests that subtle epithelial injury exist in K14-DN-Clim mouse skin prior to morphological abnormalities.
The progressive hair follicle dysfunction suggested the possibility that hair follicle stem cells might be affected in the K14-DN-Clim mice. We therefore tested whether genes enriched in hair follicle stem cells (Morris et al., 2004; Tumbar et al., 2004) were preferentially affected in the K14-DN-Clim mice. To minimize false positives, we used a strict criterion to define genes enriched in hair follicle stem cells: the genes must be identified by both previous hair follicle stem cell expression profiling studies in mice (Morris et al., 2004; Tumbar et al., 2004). For Morris et al. 2004, we used the list of 107 enriched probe sets, and for Tumbar et al. 2004, we used the list of 154 enriched probe sets. There are 62 probe sets in common between the two lists, and 58 of these are expressed in our DN-Clim microarray dataset. Intriguingly, of these 58 genes, thirteen were found to be significantly downregulated at P23 in K14-DN-Clim mice; none of the stem cell genes were upregulated (Fig. 8D, E). Given the number of hair follicle stem cell signature genes of the genome and the total number of differentially expressed genes in K14-DN-Clim mice at P23, this fraction of differentially expressed stem cell genes is significantly overrepresented (p=2.03E-10, Fisher's exact test). These data suggest that stem cells may be affected in the K14-DN-Clim mice.
Clims interact with Lhx2 and regulate the number of adult hair follicle stem cells
The specific decrease in the expression of stem cell-enriched genes prior to progressive hair follicle dysfunction suggested that stem cell depletion might cause hair follicle abnormalities in the K14-DN-Clim mice. To investigate hair follicle stem cells, we examined this cell population by fluorescence-activated cell sorting (FACS). Epithelial cells directly isolated from the skin of 8-week-old wild type and transgenic mice were subjected to FACS analyses with antibodies recognizing stem cell markers CD34 and integrin α6. FACS analysis showed that the CD34 and integrin α6 double positive population was dramatically decreased in the K14-DN-Clim transgenic mice (Fig. 9A); the number of follicle stem cells in transgenic mice was only 30% of that in wild type controls (Fig. 9B). We also examined hair follicle stem cells with long-term BrdU label retention. Ten weeks after the BrdU injection, mouse back skins were dissected and stained with anti-BrdU antibody. Label-retaining cells (Fig. 9C) in the hair follicles of K14-DN-Clim mice were only 20% of that in wild type controls (Fig. 9D). In combination with the microarray expression data, these experiments indicate that Clims are required for hair follicle stem cell maintenance. One potential interaction partner of Clims is LIM-homeodomain transcription factor Lhx2, which plays a role in the maintenance of hair follicle stem cells (Rhee et al., 2006). To examine whether there is an interaction between Clim2 and Lhx2 in skin, we performed immunoprecipitation with mouse skin tissues. In co-IP studies, Clim2 was precipitated with an Lhx2 antibody, indicating interaction between Clim2 and Lhx2 in skin (Fig. 9E; upper panel). In transgenic skin tissues, the myc-tagged dominant-negative Clim was precipitated with Lhx2 antibody (Fig. 9E; lower panel). These data show that Clim2 interacts with Lhx2 in vivo, and that DN-Clim is capable of competing for this binding. We conclude that the Clim/Lhx2 complex is required for maintenance of hair follicle stem cells and that disruption of either component of the complex leads to decreased number of hair follicle stem cells (Fig. 9F).
Figure 9. Decreased number of hair follicle stem cells in K14-DN-Clim mice.
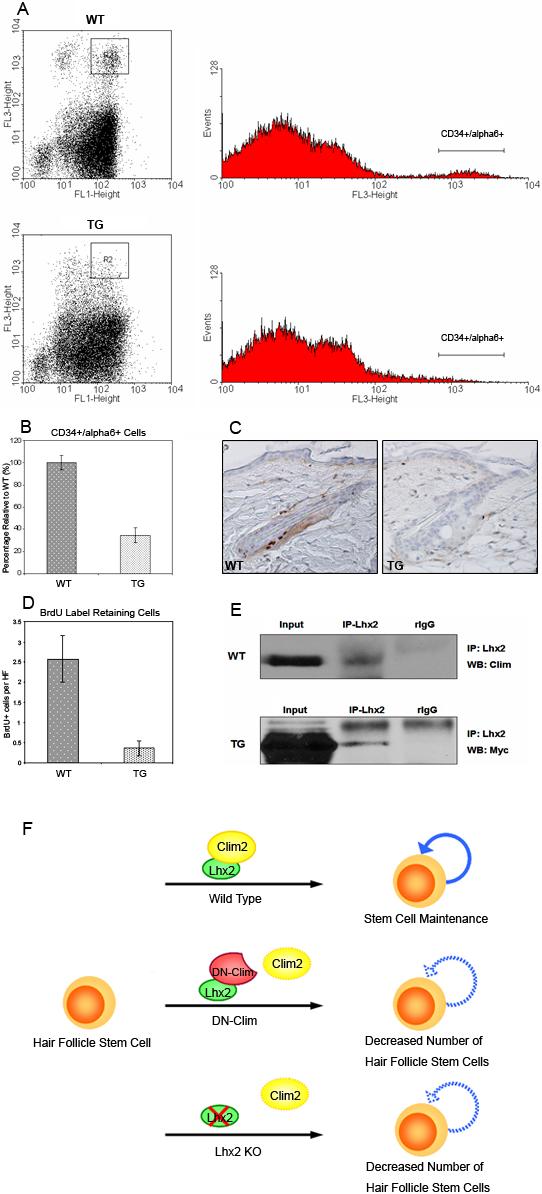
(A) FACS analysis of CD34 and integrin α6 double positive cells in 8-week-old wild type (WT) and K14-DN-Clim (TG) skin. Epithelial cells were directly isolated from wild-type and transgenic skin. FL1 indicates Integrin α6; FL3 indicates CD34. The double positive population is highlighted with boxes in the dotplot graphs. The histograms show the proportion of CD34/integrin α6 positive cells. (B) The number of double positive cells in transgenic skin is only 30% of that of wild type skin (P<0.01). 13 wild type and 9 transgenic mice were assayed. (C) BrdU long-term label retaining of hair follicle stem cells in wild type and K14-DN-Clim mice. BrdU was injected into 4-day-old pups twice a day for 3 days. After 10 week chasing period, back skin was dissected and stained with BrdU antibody. (D) BrdU positive cells per hair follicle were counted for both wild type and K14-DN-Clim mice. Shown are means and standard errors from skin samples of 6 WT and 10 TG mice (P<0.01). (E) Co-immunoprecipitation of endogenous Clim2 (upper panel) and DN-Clim (lower panel) with Lhx2 in epidermal cell extracts. (F) A model representing the role of a Clim/Lhx2 complex for hair follicle stem cell maintenance. Either interfering with binding of Clim to Lhx2, as in the experiments described in this paper, or deletion of Lhx2 as previously described (Rhee et al., 2006), leads to decreased number of stem cells. Original magnification: 200X .
Discussion
While Clim co-factors are known to be highly expressed in epithelial tissues, including epidermis and hair follicles, their potential roles within epithelia remain unknown. The findings in this paper indicate important roles for Clims in the regulation of corneal and hair follicle homeostasis.
Pathogenesis of corneal abnormalities in K14-DN-Clim mice
A time course study reveals a potential model for the pathogenesis of corneal abnormalities in the K14-DN-Clim mice (Fig. 5G). Detachment of corneal epithelium from the underlying basal lamina, resulting in blister formation, is an early abnormality, apparently caused by defective hemidesmosomes. The hemidesmosome abnormalities may be caused, at least in part, by decreased BP180 expression. Because its transcripts are downregulated in both cornea and skin in K14-DN-Clim mice, BP180 may be a target gene of Clims. In the skin, BP180 transcript expression is consistently downregulated by about 50% in all three time points in our gene expression profiling study. However, we believe that the moderate two-fold transcriptional downregulation of BP180 is insufficient to account for the dramatic discontinuous loss of BP180 protein in the transgenic cornea. While we never observed blisters in the transgenic skin, we did observe wounding in older animals, which could be due to weak tissue adhesion leading to tissue breakdown.
Sub-epithelial edema is another corneal abnormality found as early as P0 and at all time points, suggesting that edema is an early event in the transgenic cornea. The subepithelial location of the stromal edema suggests that it is caused by a defect in the corneal epithelium, possibly by the adhesion defect. Also, stromal edema may contribute to adhesion defects and blisters (Friedman, 1974; Green, 1969; Levenson, 1975), thus creating a vicious cycle. Corneal wounds are most likely due to the adhesion defect and ruptured blisters. These wounds then lead to recruitment of inflammatory cells, epithelial hyperplasia and stromal neovascularization. As postnatal development progresses and the wild type corneal epithelium becomes 3−5 cell layers thick, the transgenic corneal epithelium become as thin as one cell layer, suggesting the possibility of stem cell deficiency. In the terminal phenotype, which develops after the age of several months, the corneal epithelium can undergo a change to an epidermis-like epithelium. While it is tempting to speculate that this differentiation change might originate in corneal stem cells, it is known that the more differentiated basal cells of the adult corneal epithelium possess the capacity to follow an alternative differentiation pathway; in response to embryonic dermal stimuli, the basal cells of adult corneal epithelium are able to activate epidermal, pilosebaceous and sweat gland genetic programs (Pearton et al., 2004; Pearton et al., 2005; Powell et al., 2005).
K14-DN-Clim mice as a model for corneal diseases
The corneal abnormalities of K14-DN-Clim mice are quite similar to the clinical features of limbal stem cell deficiency, a difficult clinical problem, which is associated with thinning of the epithelium, stromal vascularization, chronic inflammation and keratinisation (Dua et al., 2003). Similar to the K14-DN-Clim mice, the condition of human corneal stem cell deficiency is often related to recurrent epithelial defects and ulcerations in the cornea (Ang and Tan, 2004; Araujo and Flowers, 1984). Therefore, our K14-DN-Clim mouse model may help elucidate the pathogenesis of corneal stem cell deficiency and the development of possible treatments. The K14-DN-Clim mice may also be a useful model for studying blistering diseases in the cornea. While corneal blistering does not appear to be a prominent feature in patients with BP180 mutations, it has been reported that the expression of BP180 is reduced in the epithelial basement membrane zone in keratoconus corneas, a disease characterized by thinning of the central and paracentral cornea and scarring in advanced cases (Cheng et al., 2001; Kenney and Chwa, 1990). Corneal blisters are also found in patients with recessive dystrophic epidermolysis bullosa (DEB) (Matsumoto et al., 2005), bullous keratopathy (Kenney and Chwa, 1990; Kenyon, 1969) and junctional epidermolysis bullosa (Herlitz-Pearson) (Hammerton et al., 1984). In addition, corneal cells of K14-DN-Clim mice differentiate into a hyperplastic, keratinized, skin-like epithelium. Vitamin A deficiency in humans is known to induce severe xerophthalmia, which is a similar corneal defect in humans (Diniz Ada and Santos, 2000). Interestingly, Notch1 signaling affects vitamin A metabolism by regulating the expression of cellular retinol binding protein 1 (CRBP1); Notch1 deficient mice develop a corneal phenotype that is similar to that of older K14-DN-Clim mice (Nicolas et al., 2003; Vauclair et al., 2007).
Decreased number of stem cells in the hair follicle bulge of K14-DN-Clim mice
In view of the expression of Clims during the first stages of hair follicle development, and the important developmental roles of LIM domain transcription factors, we expected to find abnormalities in hair follicle morphogenesis. However, initial morphogenesis of hair is normal in the K14-DN-Clim mice, and the hair loss phenotype appeared progressively in subsequent hair cycles, suggesting the possibility that the hair loss might relate to stem cell dysfunction. This hypothesis is supported by the microarray gene expression data showing decreased expression of stem cell-enriched genes prior to macroscopic or microscopic phenotypes. Further support for stem cell abnormalities come from experiments showing a decrease in CD34/α6-integrin positive and BrdU label retaining cells. These data indicate that Clims may play an important role in maintaining the “stemness” of hair follicle stem cells. In addition, we show that in the skin, Clim2 can interact with Lhx2, a LIM homeodomain protein that has been recently demonstrated to maintain stem cell character in hair follicles (Rhee et al., 2006). We, therefore, propose a model in which the Clim/Lhx2 transcriptional complex is required for stem cell maintenance (Fig. 9F).
In the K14-DN-Clim mice, we also find abnormal hair follicles with cysts and enlarged sebaceous glands, which invade the central space where hair shafts originally occupy. These morphological abnormalities bear similarity to those of several other transgenic mouse models that are also thought to affect stem cells. For instance, it has been shown that bulge stem cells of the mutant skin contribute to the follicle-derived cysts in β-catenin mutant mice (Huelsken et al., 2001). Deregulation of c-Myc expression targeted to epidermal stem cells leads to increased sebaceous gland and epidermal differentiation at the expense of hair follicles (Arnold and Watt, 2001; Waikel et al., 2001). K14-△NLef1 transgenic mice show large cysts and dislocated sebaceous glands which appear at the tips of hair follicles (Merrill et al., 2001). In addition, RXR-alpha mutant mice and Notch1 deficient mice develop hair loss, histologically characterized by the destruction of hair follicle architecture and the formation of cysts in adult mice (Li et al., 2001; Vauclair et al., 2005). Hair follicle cyst formation and preferential sebaceous-like differentiation may be a common response to hair follicle stem cell depletion.
A defect in hair differentiation is an alternative explanation for the lack of capacity of hair follicles to generate hair shafts. However, we observed normal expression of AE 13 and AE15 prior to morphological abnormalities of hair follicles (Supplementary Fig. 2). Also, our microarray results show essentially normal expression of hair shaft markers as late as p23, indicating normal hair differentiation during hair follicle morphogenesis. Therefore, while abnormal hair follicles in K14-DN-Clim mice show clear abnormalities in hair follicle differentiation, it is unlikely that a terminal differentiation defect is the primary abnormality in K14-DN-Clim mice.
The target genes of the Lhx2/Clim2 complex in hair follicle stem cells remain to be discovered. One interesting finding of our study is upregulation of genes in the MHC class I-dependent pathway of antigen presentation as early as P6, long before hair loss is observed, suggesting cellular injury/stress prior to morphological abnormalities. One possibility is that downregulation of cell adhesion molecules, such as BP180, causes subtle cellular stress. Among the stem cell signature genes that are downregulated in K14-DN-Clim mice are several genes involved in the Wnt pathway, but also cell adhesion molecules such as tenascin C. Since hair follicle stem cells reside in a specific niche, it is tempting to speculate that dysfunctional stem cell-matrix adhesion may play a role in the phenotypes observed in K14-DN-Clim mice. In this respect it is interesting to note that hair loss is found in some patients with BP180 mutations (Jonkman et al., 1995; McGrath et al., 1995; Woo et al., 2000). While we do not believe that immune mechanisms are the primary cause of hair and skin abnormalities in K14-DN-Clim mice, the upregulation of MHC class I genes suggests the possibility that immunological mechanisms may contribute to progression of the phenotype.
In summary, we have shown that interfering with binding of Clim co-factors to transcription factors leads to dramatic phenotypes in cornea and hair follicles, indicating important roles for Clims in homeostasis of these two organs. In both cornea and epidermis, Clims are important for BP180 expression. In the cornea, hemidesmosome function and adhesion of the corneal epithelium to the underlying basal lamina is disrupted in K14-DN-Clim mice. In the hair follicles, Clims are required for maintenance of bulge stem cells, most likely due to their interactions with the LIM homeodomain factor Lhx2.
Acknowledgements
This work was supported by NIH grants AR44882 and EY016425 to BA. We thank Zhi Liu for BP180 antibody, Ingolf Bach for Clim antibody, Charles Ribak for advice on EM, and Chris Kenney, Liz Rugg, Xing Dai and Mike Demetriou for helpful advice and review of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
References
- Agulnick AD, Taira M, Breen JJ, Tanaka T, Dawid IB, Westphal H. Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature. 1996;384:270–2. doi: 10.1038/384270a0. [DOI] [PubMed] [Google Scholar]
- Ang LP, Tan DT. Ocular surface stem cells and disease: current concepts and clinical applications. Ann Acad Med Singapore. 2004;33:576–80. [PubMed] [Google Scholar]
- Araujo OE, Flowers FP. Stevens-Johnson syndrome. J Emerg Med. 1984;2:129–35. doi: 10.1016/0736-4679(84)90332-9. [DOI] [PubMed] [Google Scholar]
- Arnold I, Watt FM. c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr Biol. 2001;11:558–68. doi: 10.1016/s0960-9822(01)00154-3. [DOI] [PubMed] [Google Scholar]
- Bach I. The LIM domain: regulation by association. Mech Dev. 2000;91:5–17. doi: 10.1016/s0925-4773(99)00314-7. [DOI] [PubMed] [Google Scholar]
- Bach I, Carriere C, Ostendorff HP, Andersen B, Rosenfeld MG. A family of LIM domain-associated cofactors confer transcriptional synergism between LIM and Otx homeodomain proteins. Genes Dev. 1997;11:1370–80. doi: 10.1101/gad.11.11.1370. [DOI] [PubMed] [Google Scholar]
- Bach I, Rodriguez-Esteban C, Carriere C, Bhushan A, Krones A, Rose DW, Glass CK, Andersen B, Izpisua Belmonte JC, Rosenfeld MG. RLIM inhibits functional activity of LIM homeodomain transcription factors via recruitment of the histone deacetylase complex. Nat Genet. 1999;22:394–9. doi: 10.1038/11970. [DOI] [PubMed] [Google Scholar]
- Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–19. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- Becker T, Ostendorff HP, Bossenz M, Schluter A, Becker CG, Peirano RI, Bach I. Multiple functions of LIM domain-binding CLIM/NLI/Ldb cofactors during zebrafish development. Mech Dev. 2002;117:75–85. doi: 10.1016/s0925-4773(02)00178-8. [DOI] [PubMed] [Google Scholar]
- Braun KM, Niemann C, Jensen UB, Sundberg JP, Silva-Vargas V, Watt FM. Manipulation of stem cell proliferation and lineage commitment: visualisation of label-retaining cells in wholemounts of mouse epidermis. Development. 2003;130:5241–55. doi: 10.1242/dev.00703. [DOI] [PubMed] [Google Scholar]
- Bulchand S, Subramanian L, Tole S. Dynamic spatiotemporal expression of LIM genes and cofactors in the embryonic and postnatal cerebral cortex. Dev Dyn. 2003;226:460–9. doi: 10.1002/dvdy.10235. [DOI] [PubMed] [Google Scholar]
- Cheng EL, Maruyama I, SundarRaj N, Sugar J, Feder RS, Yue BY. Expression of type XII collagen and hemidesmosome-associated proteins in keratoconus corneas. Curr Eye Res. 2001;22:333–40. doi: 10.1076/ceyr.22.5.333.5491. [DOI] [PubMed] [Google Scholar]
- Coster DJ, Aggarwal RK, Williams KA. Surgical management of ocular surface disorders using conjunctival and stem cell allografts. Br J Ophthalmol. 1995;79:977–82. doi: 10.1136/bjo.79.11.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotsarelis G. Epithelial stem cells: a folliculocentric view. J Invest Dermatol. 2006;126:1459–68. doi: 10.1038/sj.jid.5700376. [DOI] [PubMed] [Google Scholar]
- Dawid IB, Breen JJ, Toyama R. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 1998;14:156–62. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- Dennis G,, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Diniz Ada S, Santos LM. [Vitamin A deficiency and xerophtalmia]. J Pediatr (Rio J) 2000;76(Suppl 3):S311–22. doi: 10.2223/jped.169. [DOI] [PubMed] [Google Scholar]
- Dua HS, Joseph A, Shanmuganathan VA, Jones RE. Stem cell differentiation and the effects of deficiency. Eye. 2003;17:877–85. doi: 10.1038/sj.eye.6700573. [DOI] [PubMed] [Google Scholar]
- Dua HS, Saini JS, Azuara-Blanco A, Gupta P. Limbal stem cell deficiency: concept, aetiology, clinical presentation, diagnosis and management. Indian J Ophthalmol. 2000;48:83–92. [PubMed] [Google Scholar]
- Friedman MH. A physical description of the pathogenesis, histopathology and treatment of corneal epithelial edema. J Theor Biol. 1974;45:153–69. doi: 10.1016/0022-5193(74)90048-4. [DOI] [PubMed] [Google Scholar]
- Gimnopoulos D, Becker CG, Ostendorff HP, Bach I, Schachner M, Becker T. Expression of the zebrafish recognition molecule F3/F11/contactin in a subset of differentiating neurons is regulated by cofactors associated with LIM domains. Mech Dev. 2002;119(Suppl 1):S135–41. doi: 10.1016/s0925-4773(03)00106-0. [DOI] [PubMed] [Google Scholar]
- Green K. Anatomic study of water movement through rabbit corneal epithelium. Am J Ophthalmol. 1969;67:110–6. doi: 10.1016/0002-9394(69)90015-4. [DOI] [PubMed] [Google Scholar]
- Hammerton ME, Turner TW, Pyne RJ. A case of junctional epidermolysis bullosa (Herlitz-Pearson) with corneal bullae. Aust J Ophthalmol. 1984;12:45–8. doi: 10.1111/j.1442-9071.1984.tb01123.x. [DOI] [PubMed] [Google Scholar]
- Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105:533–45. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- Jonkman MF, de Jong MC, Heeres K, Pas HH, van der Meer JB, Owaribe K, Martinez de Velasco AM, Niessen CM, Sonnenberg A. 180-kD bullous pemphigoid antigen (BP180) is deficient in generalized atrophic benign epidermolysis bullosa. J Clin Invest. 1995;95:1345–52. doi: 10.1172/JCI117785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata LW, Gill GN. Functional analysis of the nuclear LIM domain interactor NLI. Mol Cell Biol. 1997;17:5688–98. doi: 10.1128/mcb.17.10.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata LW, Kenny DA, Gill GN. Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proc Natl Acad Sci U S A. 1996;93:11693–8. doi: 10.1073/pnas.93.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–31. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- Kaya G, Rodriguez I, Jorcano JL, Vassalli P, Stamenkovic I. Selective suppression of CD44 in keratinocytes of mice bearing an antisense CD44 transgene driven by a tissue-specific promoter disrupts hyaluronate metabolism in the skin and impairs keratinocyte proliferation. Genes Dev. 1997;11:996–1007. doi: 10.1101/gad.11.8.996. [DOI] [PubMed] [Google Scholar]
- Kenney MC, Chwa M. Abnormal extracellular matrix in corneas with pseudophakic bullous keratopathy. Cornea. 1990;9:115–21. [PubMed] [Google Scholar]
- Kenyon KR. The synthesis of basement membrane by the corneal epithelium in bullous keratopathy. Invest Ophthalmol. 1969;8:156–68. [PubMed] [Google Scholar]
- Leask A, Rosenberg M, Vassar R, Fuchs E. Regulation of a human epidermal keratin gene: sequences and nuclear factors involved in keratinocyte-specific transcription. Genes Dev. 1990;4:1985–98. doi: 10.1101/gad.4.11.1985. [DOI] [PubMed] [Google Scholar]
- Levenson JE. Corneal edema: cause and treatment. Surv Ophthalmol. 1975;20:190–204. doi: 10.1016/0039-6257(75)90002-8. [DOI] [PubMed] [Google Scholar]
- Li M, Chiba H, Warot X, Messaddeq N, Gerard C, Chambon P, Metzger D. RXR-alpha ablation in skin keratinocytes results in alopecia and epidermal alterations. Development. 2001;128:675–88. doi: 10.1242/dev.128.5.675. [DOI] [PubMed] [Google Scholar]
- Lin KK, Chudova D, Hatfield GW, Smyth P, Andersen B. Identification of hair cycle-associated genes from time-course gene expression profile data by using replicate variance. Proc Natl Acad Sci U S A. 2004;101:15955–60. doi: 10.1073/pnas.0407114101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MH, O'Guin WM, Hardy C, Mak L, Sun TT. Acidic and basic hair/nail (“hard”) keratins: their colocalization in upper cortical and cuticle cells of the human hair follicle and their relationship to “soft” keratins. J Cell Biol. 1986;103:2593–606. doi: 10.1083/jcb.103.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Dogru M, Tsubota K. Ocular surface findings in Hallopeau-Siemens subtype of dystrophic epidermolysis bullosa: report of a case and literature review. Cornea. 2005;24:474–9. doi: 10.1097/01.ico.0000151722.84634.bf. [DOI] [PubMed] [Google Scholar]
- Matthews JM, Visvader JE. LIM-domain-binding protein 1: a multifunctional cofactor that interacts with diverse proteins. EMBO Rep. 2003;4:1132–7. doi: 10.1038/sj.embor.7400030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JA, Gatalica B, Christiano AM, Li K, Owaribe K, McMillan JR, Eady RA, Uitto J. Mutations in the 180-kD bullous pemphigoid antigen (BPAG2), a hemidesmosomal transmembrane collagen (COL17A1), in generalized atrophic benign epidermolysis bullosa. Nat Genet. 1995;11:83–6. doi: 10.1038/ng0995-83. [DOI] [PubMed] [Google Scholar]
- Merrill BJ, Gat U, DasGupta R, Fuchs E. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev. 2001;15:1688–705. doi: 10.1101/gad.891401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, Lin JS, Sawicki JA, Cotsarelis G. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol. 2004;22:411–7. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay M, Teufel A, Yamashita T, Agulnick AD, Chen L, Downs KM, Schindler A, Grinberg A, Huang SP, Dorward D, Westphal H. Functional ablation of the mouse Ldb1 gene results in severe patterning defects during gastrulation. Development. 2003;130:495–505. doi: 10.1242/dev.00225. [DOI] [PubMed] [Google Scholar]
- Muller-Rover S, Handjiski B, van der Veen C, Eichmuller S, Foitzik K, McKay IA, Stenn KS, Paus R. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J Invest Dermatol. 2001;117:3–15. doi: 10.1046/j.0022-202x.2001.01377.x. [DOI] [PubMed] [Google Scholar]
- Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, Hui CC, Clevers H, Dotto GP, Radtke F. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33:416–21. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- O'Guin WM, Sun TT, Manabe M. Interaction of trichohyalin with intermediate filaments: three immunologically defined stages of trichohyalin maturation. J Invest Dermatol. 1992;98:24–32. doi: 10.1111/1523-1747.ep12494172. [DOI] [PubMed] [Google Scholar]
- Ostendorff HP, Tursun B, Cornils K, Schluter A, Drung A, Gungor C, Bach I. Dynamic expression of LIM cofactors in the developing mouse neural tube. Dev Dyn. 2006;235:786–91. doi: 10.1002/dvdy.20669. [DOI] [PubMed] [Google Scholar]
- Pearton DJ, Ferraris C, Dhouailly D. Transdifferentiation of corneal epithelium: evidence for a linkage between the segregation of epidermal stem cells and the induction of hair follicles during embryogenesis. Int J Dev Biol. 2004;48:197–201. [PubMed] [Google Scholar]
- Pearton DJ, Yang Y, Dhouailly D. Transdifferentiation of corneal epithelium into epidermis occurs by means of a multistep process triggered by dermal developmental signals. Proc Natl Acad Sci U S A. 2005;102:3714–9. doi: 10.1073/pnas.0500344102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AM, Sakuma-Oyama Y, Oyama N, Black MM. Collagen XVII/BP180: a collagenous transmembrane protein and component of the dermoepidermal anchoring complex. Clin Exp Dermatol. 2005;30:682–7. doi: 10.1111/j.1365-2230.2005.01937.x. [DOI] [PubMed] [Google Scholar]
- Rhee H, Polak L, Fuchs E. Lhx2 maintains stem cell character in hair follicles. Science. 2006;312:1946–9. doi: 10.1126/science.1128004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Degenstein L, Copenhaver C, Fuchs E. Defining the regulatory factors required for epidermal gene expression. Mol Cell Biol. 2000;20:2543–55. doi: 10.1128/mcb.20.7.2543-2555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara TM, Bach I, Kioussi C, Rosenfeld MG, Andersen B. Mouse deformed epidermal autoregulatory factor 1 recruits a LIM domain factor, LMO-4, and CLIM coregulators. Proc Natl Acad Sci U S A. 1998;95:15418–23. doi: 10.1073/pnas.95.26.15418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–41. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–63. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Rosenberg M, Ross S, Tyner A, Fuchs E. Tissue-specific and differentiation-specific expression of a human K14 keratin gene in transgenic mice. Proc Natl Acad Sci U S A. 1989;86:1563–7. doi: 10.1073/pnas.86.5.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vauclair S, Majo F, Durham AD, Ghyselinck NB, Barrandon Y, Radtke F. Corneal Epithelial Cell Fate Is Maintained during Repair by Notch1 Signaling via the Regulation of Vitamin A Metabolism. Dev Cell. 2007;13:242–53. doi: 10.1016/j.devcel.2007.06.012. [DOI] [PubMed] [Google Scholar]
- Vauclair S, Nicolas M, Barrandon Y, Radtke F. Notch1 is essential for postnatal hair follicle development and homeostasis. Dev Biol. 2005;284:184–93. doi: 10.1016/j.ydbio.2005.05.018. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Mao X, Fujiwara Y, Hahm K, Orkin SH. The LIM-domain binding protein Ldb1 and its partner LMO2 act as negative regulators of erythroid differentiation. Proc Natl Acad Sci U S A. 1997;94:13707–12. doi: 10.1073/pnas.94.25.13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waikel RL, Kawachi Y, Waikel PA, Wang XJ, Roop DR. Deregulated expression of c-Myc depletes epidermal stem cells. Nat Genet. 2001;28:165–8. doi: 10.1038/88889. [DOI] [PubMed] [Google Scholar]
- Wang N, Kudryavtseva E, Ch'en IL, McCormick J, Sugihara TM, Ruiz R, Andersen B. Expression of an engrailed-LMO4 fusion protein in mammary epithelial cells inhibits mammary gland development in mice. Oncogene. 2004;23:1507–13. doi: 10.1038/sj.onc.1207288. [DOI] [PubMed] [Google Scholar]
- Wang X, Zinkel S, Polonsky K, Fuchs E. Transgenic studies with a keratin promoter-driven growth hormone transgene: prospects for gene therapy. Proc Natl Acad Sci U S A. 1997;94:219–26. doi: 10.1073/pnas.94.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HJ, Lee JH, Kim SC, Kim CW, Kim TY. Generalized atrophic benign epidermolysis bullosa--poor prognosis associated with chronic renal failure. Clin Exp Dermatol. 2000;25:212–4. doi: 10.1046/j.1365-2230.2000.00617.x. [DOI] [PubMed] [Google Scholar]
- Xie W, Chow LT, Paterson AJ, Chin E, Kudlow JE. Conditional expression of the ErbB2 oncogene elicits reversible hyperplasia in stratified epithelia and up-regulation of TGFalpha expression in transgenic mice. Oncogene. 1999;18:3593–607. doi: 10.1038/sj.onc.1202673. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.