Abstract
Lactobacillus reuteri is a commensal-derived anaerobic probiotic that resides in the human gastrointestinal tract. L. reuteri converts glycerol into a potent broad-spectrum antimicrobial compound, reuterin, which inhibits the growth of gram-positive and gram-negative bacteria. In this study, we compared four human-derived L. reuteri isolates (ATCC 55730, ATCC PTA 6475, ATCC PTA 4659, and ATCC PTA 5289) in their ability to produce reuterin and to inhibit the growth of different enteric pathogens in vitro. Reuterin was produced by each of the four L. reuteri strains and assessed for biological activity. The minimum inhibitory concentration (MIC) of reuterin derived from each strain was determined for the following enteric pathogens: enterohemorrhagic Escherichia coli, enterotoxigenic E. coli, Salmonella enterica, Shigella sonnei and Vibrio cholerae. We also analyzed the relative abilities of L. reuteri to inhibit enteric pathogens in a pathogen overlay assay. The magnitude of reuterin production did not directly correlate with the relative ability of L. reuteri to suppress the proliferation of enteric pathogens. Additional antimicrobial factors may be produced by L. reuteri, and multiple factors may act synergistically with reuterin to inhibit enteric pathogens.
Keywords: antibiotics, gastroenteritis, lactobacilli, lactic acid bacteria, reuterin
1. Introduction
Therapeutic microbiology is expanding and beneficial bacteria are being implemented as treatment and prevention strategies for immune disorders and infectious diseases. Important characteristics of probiotic bacteria include their abilities to suppress the proliferation and virulence of pathogenic organisms. Lactic acid bacteria (LAB) such as Lactobacillus spp. produce antimicrobial factors and bacteriocins including lantibiotics, small heat-stable, non-lanthionine containing membrane-active peptides, larger heat-labile proteins and complex bacteriocins containing one or more chemical moieties [1–3]. Due to the production of diverse antimicrobial agents, probiotics may be considered for the treatment and prevention of a variety of infectious diseases caused by oral, enteric and urogenital pathogens [4–8].
Lactobacillus reuteri is an established probiotic agent, the most widely distributed Lactobacillus species among animals and considered to be one of a limited number of indigenous Lactobacillus species in the human intestine [4]. A primary antimicrobial compound produced by L. reuteri, reuterin, lacks a peptide or protein component and is produced during glycerol fermentation. Reuterin, a β-hydroxypropionaldehyde (3-HPA) derivative of glycerol, is produced under anaerobic conditions and exhibits broad spectrum effects against gram-positive and gram-negative bacteria [9] as well as fungi, yeasts and protozoa [10]. The broad-spectrum antimicrobial activity of L. reuteri has been considered an important feature conferring therapeutic potential for the prevention or treatment of infections [6, 8, 11–14].
In this study, we analyzed four different human-derived L. reuteri strains for their relative abilities to produce reuterin and inhibit a variety of bacterial species including commensal organisms and diverse enteric pathogens. We also developed high-throughput versions of existing functional assays for measuring probiotic-mediated antimicrobial activities. High-throughput strategies may be important for functional screening of many candidate probiotic strains in the future. We have demonstrated strain-dependent variation in reuterin production among candidate probiotics and revealed the potential for multifactorial pathogen inhibition by L. reuteri. These studies provide an improved understanding of probiotic-mediated pathogen inhibition, and support future applications of beneficial bacteria for the prevention of diarrheal diseases.
2. Materials and methods
2.1. Bacterial strains and media
The strains used in this study are described in Table 1. L. reuteri strains ATCC 55730, ATCC PTA 6475, ATCC PTA 4659 or ATCC PTA 5289 will be referred to as strains 55730, 6475, 4659 and 5289, respectively, throughout this manuscript. Lactobacillus strains were cultured in DeMan-Rogosa-Sharpe (MRS) media (Becton Dickinson, Sparks, Md.) at 37°C for 24 to 48 hr in an anaerobic chamber (Model 1025 S/N, Forma Scientific, Inc., Marietta, Ohio). Enteric pathogen strains were cultured aerobically in Brain Heart Infusion (BHI) media (Becton Dickinson) for 24 hr at 37°C. Modifications for specific assays are detailed accordingly.
Table 1.
Listing of bacterial strains included in this study.
| Strains | Description | Source |
|---|---|---|
| Probiotic Lactobacillus strains: | ||
| L. reuteri ATCC 55730 | Isolate from Peruvian mother’s milk | Biogaia AB |
| L. reuteri ATCC PTA 6475 | Isoalte from Finnish mother’s milk | Biogaia AB |
| L. reuteri ATCC PTA 4659 | Isoalte from Finnish mother’s milk | Biogaia AB |
| L. reuteri ATCC PTA 5289 | Oral isolate from Japanese female | Biogaia AB |
| L. acidophilus ATCC 4356 | Probiotic strain, human isolate | ATCCa |
| L casei ATCC 334 | Probiotic strain isolated from Emmental cheese | ATCC |
| L. gasseri ATCC 33323 | DSM 20243, Probiotic strain, human isolate | ATCC |
| L. johnsonii ATCC 33200 | VPI 7960, Human blood isolate | ATCC |
| L. plantarum 42/6 | Fecal isolate from healthy Thai woman | S. Tumwasorn |
| Enteric pathogen strains: | ||
| EHEC | Clinical isolate of enterohemorrhagic E. coli | TCHb |
| ETEC | Clinical isolate of enterotoxigenic E. coli | TCH |
| Salmonella enterica | Clinical isolate | TCH |
| Shigella sonnei | Clinical isolate | TCH |
| Vibrio cholerae | Clinical isolate | TCH |
America Type Culture Collection (ATCC)
Microbiology Laboratories, Department of Pathology, Texas Children’s Hospital (TCH).
EHEC, enterohemorrhagic E. coli; ETEC, enterotoxigenic E. coli.
2.2. Reuterin production by bacterial cultures
Reuterin was produced in order to assess minimum inhibitory concentrations (MICs) for various bacterial strains, as done previously [10, 15, 16]. Briefly, MRS was inoculated with L. reuteri and incubated anaerobically for 24 hr at 37°C. The bacteria were collected by centrifugation and washed with 50 mM sodium phosphate buffer (pH 7.4). The bacteria were resuspended to a concentration of ~1.5 × 1010 cells/mL in 250 mM glycerol and incubated anaerobically at 37°C for 2 hr. Simultaneously, samples were taken immediately after resuspension in glycerol to obtain viable cell counts. After the 2 hr incubation, the bacteria were pelleted and reuterin-containing supernatants were collected and filter-sterilized using a 0.22 μm polyvinylidine difluoride filter (Millipore, Billerica, MA). Reuterin solution was stored at 4°C as described previously [17, 18].
2.3. Quantification of reuterin by HPLC
The chromatographic separation of reuterin was performed using modified methods of Catassi, et al. [19] and Talarico, et al. [20]. In brief, the filter-sterilized reuterin solution was diluted 1:10 in sterile double-distilled water (ddH2O), and a 20 μL aliquot was injected into an Aminex HPX 87C 300 7.8 mm cation-exchange column protected with a “Deashing Guard Cartridge” (Cat # 1250118, BioRad Laboratories, Richmond, CA) and eluted with degassed pure water at a flow rate of 0.6 mL/min at 85°C. The column effluent was monitored with a differential refractometer (Dionex Corperation, Sunnyvale, CA) having an internal temperature of 50°C. This method was calibrated with 1 mg/mL (10.87 mM) glycerol, and the assay’s limit of detection was 1 μM of glycerol. The conversion of glycerol to reuterin occurs at a 1:1 ratio [20], and the concentration of reuterin (aldehyde form, 74 g/mol) in each sample was determined by subtracting the remaining amount of glycerol from the HPLC-determined concentration of the starting material.
2.4. Reuterin Quantification by Absorbance Spectrophotometry
Reuterin samples were analyzed colorimetrically as done previously [16, 21], with modifications to a 96-well plate format as follows. Reuterin samples were serially diluted in sterile ddH2O, mixed with 10 mM tryptophan (Fluka) solution (0.01 M in 0.05 M HCl, stabilized with a few drops of toluene) followed by addition of 12 M HCl (Fisher) and incubation at 37°C for 30 min. The optical density of each reaction was measured at 560 nm using a Spectramax 340PC (Molecular Devices Corporation, Sunnyvale, CA). A standard curve was generated using HPLC-quantified reuterin. The accuracy of this assay was confirmed by HPLC analyses of multiple reuterin samples from four different L. reuteri strains.
2.5. Minimum Inhibitory Concentration (MIC) Assay for Reuterin
The effects of reuterin on various bacterial strains were tested in a MIC assay. These experiments were performed three times in duplicate. This assay was described previously by Chung, et al. [10], but it has been modified to accommodate specific quantities of reuterin in a 96-well format. Enteric pathogen strains were grown aerobically for 16–18 hr in BHI broth at 37°C and subsequently diluted to a concentration of ~1 × 106 cells/mL. Aliquots (1 mL) were deposited into individual wells of 96-well, 2.5 mL deep-well plates (Cat # 37001-520, VWR International, West Chester, PA). Reuterin solution was serially diluted, and 1 mL of each dilution was added per well. The deep-well plates were incubated aerobically at 37°C for 18–24 hr, and aliquots from each well were transferred to individual wells of a 96-well microtiter plate (VWR, West Chester, PA). Optical densities were measured at 600 nm in a Spectramax 340PC (Molecular Devices Corporation). Positive growth controls were assayed using 1 mL BHI broth without reuterin.
2.6. Agar Spot Method of Pathogen Inhibition
An agar spot test was used to detect antimicrobial activities of L. reuteri against various bacterial strains. These assays were performed three times in triplicate. This assay is a modification of methods previously described by Schillinger and Lücke [22] and Jacobsen, et al. [23]. Assay optimization was based on previous studies indicating the importance of glucose in reuterin production [24]. Overnight cultures (18–24 hr) of test strain (e.g. L. reuteri) were spotted (2 μL) onto the surfaces of BHI agar supplemented with 20 mM glucose, and the spots were developed by anaerobic incubation at 37°C for 24 hr. Enteric pathogen strains were inoculated at a concentration of ~1 × 106 cells per mL in 7 mL of soft agar (BHI, 2% glycerol, 0.7% agar), overlain atop the L. reuteri spots and incubated anaerobically at 37°C for 1 hr in order to facilitate reuterin production by spots containing L. reuteri. The anaerobic incubation was followed by aerobic culture at 37°C for 18–24 hr. In contrast to enteric pathogens, Lactobacillus strains were inoculated at ~1 × 107 in MRS, 2% glycerol, 0.7% agar and incubated anaerobically at 37°C for 18–24 hr. Clear zones of inhibition measuring ≥ 1 mm around each spot were scored.
3. Results
3.1. Production and quantification of reuterin from different strains of Lactobacillus reuteri
In this study, four human-derived probiotic L. reuteri strains (55730, 6475, 4659 and 5289) were assessed and compared for their ability to produce reuterin using the two-step fermentation process outlined previously by Lüthi-Peng, et al. [17]. Viable cell counts by colony plating were determined for each reuterin preparation.
Reuterin concentrations for each strain were determined in a modified Trp-HCl assay described by Doleyres, et al. [16]. Previous studies used the toxin, unstable chemical, acrolein, as a standard. To eliminate this hazardous chemical, we adapted this method to a 96-well format for high-throughput analyses and implemented HPLC-quantified reuterin as a standard. In order to validate the accuracy of this modified Trp-HCl assay, concentrations of multiple reuterin samples were determined by parallel HPLC analyses. The parallel concentrations determined by this colorimetric method and HPLC analyses were consistent with each other. Quantitative comparisons of concentrations obtained by colorimetric and HPLC studies yielded errors of ≤ 3% (Table 2). The data confirm that this high-throughput version of the Trp-HCl assay is an accurate and reliable method for determining reuterin concentrations.
Table 2.
Comparison of reuterin quantification methods.
| Reuterin Samplea | HPLC (mM) | Trp-HClb (mM) |
|---|---|---|
| 55730-1 | 258.53 | 266.36 |
| 55730-2 | 298.35 | 293.27 |
| 6475-1 | 103.66 | 99.81 |
| 6475-2 | 109.97 | 104.04 |
| 5289-1 | 119.19 | 122.82 |
| 5289-2 | 115.74 | 113.93 |
| 4659-1 | 97.92 | 101.13 |
| 4659-2 | 115.82 | 110.07 |
Reuterin was produced from each L. reuteri strain and analyzed by both quantitative methods. Duplicate samples from each L. reuteri strain are shown here and are representative of a larger data set. L. reuteri strains include ATCC 55730 (55730), ATCC PTA 6475 (6475), ATCC PTA 5289 (5289) and ATCC PTA 4659 (4659).
Each concentration determined by the colorimetric method was carried out in triplicate and is represented here as the mean.
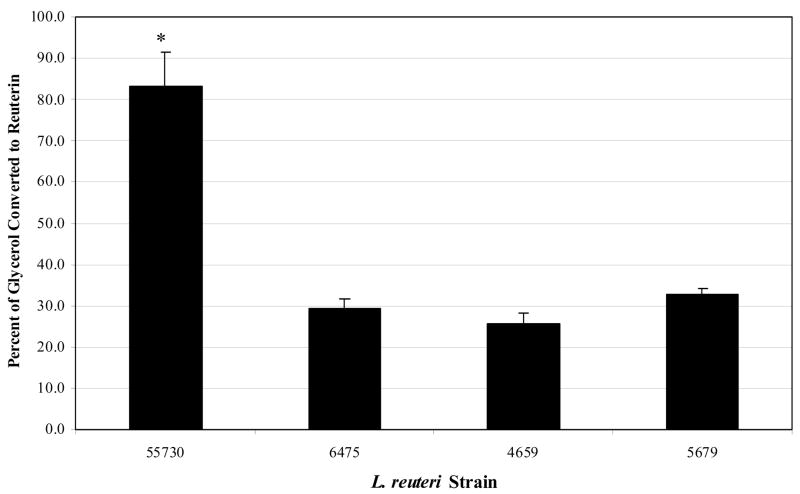
Reuterin was produced by each L. reuteri strain and the relative quantities were compared. Reuterin concentration was compared to the concentration of glycerol remaining in solution, and the differences in each L. reuteri strain’s ability to convert glycerol to reuterin are depicted in Figure 1. L. reuteri 55730 converts approximately 3 fold more glycerol to reuterin than L. reuteri strains 6475, 4659 and 5289. The amounts of reuterin produced by L. reuteri strains 6475, 4659 and 5289 were comparable to each other. Statistical analyses verified that differences in reuterin production between L. reuteri 55730 and each of the other three L. reuteri strains were significant (P < 0.05) (Fig. 1).
Figure 1. Comparison of reuterin production among probiotic L. reuteri strains.
Reuterin was produced from L. reuteri strains 55730, 6475, 4659 and 5289 in glycerol solution three times in triplicate using a two-step fermentation process. Reuterin concentrations were determined using a modified Trp-HCl assay and HPLC-quantified reuterin as the standard. Reuterin concentrations were compared against concentrations of glycerol remaining in solution to yield a percentage of glycerol converted to reuterin. An asterisk (*) indicates that the amount of reuterin produced by 55730 is significantly different (P < 0.05) than the amounts produced by the other strains.
3.2. Compatibility of L. reuteri with other Lactobacillus species
We assessed the potential inhibitory properties of L. reuteri on other Lactobacillus species using the agar spot method. This method allows for the direct assessment of secreted factors by L. reuteri on a test strain. L. reuteri strains (55730, 6475, 4659 and 5289) were spotted and grown on BHI agar supplemented with 20 mM glucose and subsequently overlain with test strains of choice. First, each L. reuteri strain was tested against itself and other L. reuteri strains. By defining a zone of inhibition with a minimum diameter ≥ 1 mm, no zones of inhibition were observed for L. reuteri strains 55730, 6475, 4659 or 5289 when overlain with the same or different L. reuteri strain (data not shown). These results demonstrate that L. reuteri strains are resistant to one another’s antimicrobial effects.
A variety of lactobacilli, including but not limited to L. acidophilus, L. casei, L. gasseri and L. johnsonii are available in commercial products [25]. Combinations of different probiotic strains may be useful for probiotic formulations, and investigations of compatibilities of L. reuteri strains with other Lactobacillus species may be useful. The following strains were included in this study: L. acidophilus ATCC 4356, L. casei ATCC 334, L. gasseri ATCC 33323 and L. johnsonii ATCC 33200 (Table 1). Each of these strains is a human-derived isolate, except for L. casei ATCC 334, which was isolated from Emmental cheese (www.atcc.org, Table 1). These strains were also assessed for inhibitory effects by L. reuteri using the agar spot method.
Mild inhibition by each L. reuteri strain was observed for L. acidophilus ATCC 4356 and L. gasseri ATCC 33323. Moderate inhibition by L. reuteri was observed for L. johnsonii ATCC 33200, while each L. reuteri strain exhibited potent inhibition versus L. casei ATCC 334 (Table 3). All four L. reuteri strains inhibited the same Lactobacillus test strain in a similar manner, lacking strain-dependent variation in growth inhibition of different lactobacilli. By contrast, L. reuteri was not inhibited by L. acidophilus ATCC 4356, L. casei ATCC 334, L. gasseri ATCC 33323 or L. johnsonii ATCC 33200 in the same assay (data not shown). These results show that the proliferation of specific Lactobacillus species including L. acidophilus and L. gasseri is not inhibited by L. reuteri. An improved understanding of inter-species and inter-strain combinations in the laboratory may be important as multi-strain probiotic formulations are currently being used and are being considered for future applications. More importantly, the data do not yield evidence of suppression of L. reuteri by other probiotic Lactobacillus species.
Table 3.
Effects of L. reuteri on the growth of Lactobacillus spp.
| Lactobacillus reuteria |
||||
|---|---|---|---|---|
| Lactobacillus spp | 55730 | 6475 | 4659 | 5289 |
| L. acidophilus | 6.72 ± .44 | 6.78 ± 0.54 | 6.33 ± 0.19 | 6.33 ± 0.44 |
| L. casei | 14.39 ± 0.67 | 14.28 ± 0.69 | 14.44 ± 0.72 | 14.22 ± 0.91 |
| L. gasseri | 7.44 ± 0.67 | 7.17 ± 0.73 | 7.22 ± 0.54 | 7.06 ± 0.69 |
| L. johnsonii | 7.22 ± 0.10 | 8.89 ± 0.89 | 7.78 ± 0.19 | 7.94 ± 0.69 |
The diameter of each zone of inhibition around the spotted L. reuteri was measured for each pathogen. The data are representative of three triplicate assays with standard deviations.
3.3. Differential growth inhibition of enteric pathogens by L. reuteri
The relative abilities of L. reuteri strains to inhibit enterohemorrhagic E. coli (EHEC), enterotoxigenic E. coli (ETEC), Salmonella enterica, Shigella sonnei and Vibrio cholerae were evaluated using the agar spot assay. All enteric pathogens tested were susceptible to L. reuteri, and L. reuteri strains demonstrated differential inhibitory activities when tested against enteric pathogens (Table 4). In all cases, L. reuteri strains 6475 and 4659 demonstrated the most potent growth inhibition of each enteric pathogen, while L. reuteri 55730 consistently yielded smaller zones of inhibition when compared to the other L. reuteri strains. L. reuteri 5289 maintained inhibitory zones either greater than or equal to strain 55730. Although L. reuteri 55730 produces abundant amounts of reuterin, this strain had the weakest inhibitory effect on each pathogen, which is clearly demonstrated by Salmonella enterica (Table 4). Variability was also noted with respect to susceptibilities of different enteric pathogens to L. reuteri. EHEC and ETEC were the least susceptible enteric pathogens, while Shigella sonnei showed the greatest susceptibility to L. reuteri by in vitro testing (Table 4). As a control, internal reference strain L. plantarum 42/6 was tested for pathogen inhibition in the same assays. Treatment with L. plantarum 42/6 resulted in minimal pathogen growth inhibition when compared to L. reuteri in the overlay assay (data not shown).
Table 4.
Effects of L. reuteri on the growth of enteric pathogens.
| Lactobacillus reuteria |
||||
|---|---|---|---|---|
| Enteric Pathogens | 55730 | 6475 | 4659 | 5289 |
| EHEC | 7.28 ± 0.92 | 11.56 ± 1.21 | 12.67 ± 2.34 | 9.78 ± 1.58 |
| ETEC | 7.94 ± 0.63 | 11.89 ± 1.35 | 11.56 ± 0.19 | 10.28 ± 1.60 |
| Salmonella enterica | 9.66 ± 2.64 | 14.17 ± 0.88 | 14.28 ± 1.06 | 13.17 ± 1.72 |
| Shigella sonnei | 16.33 ± 2.03 | 18.11 ± 0.38 | 18.11 ± 0.38 | 18.00 ± 0.96 |
| Vibrio cholerae | 13.56 ± 1.84 | 15.00 ± 1.21 | 14.56 ± 1.84 | 13.22 ± 1.03 |
The diameter of each zone of inhibition around the spotted L. reuteri was measured for each pathogen. The data are representative of three triplicate assays with standard deviations.
EHEC, enterohemorrhagic E. coli; ETEC, enterotoxigenic E. coli.
3.4. Reuterin demonstrates growth inhibitory effects, but not differential inhibition, of enteric pathogens
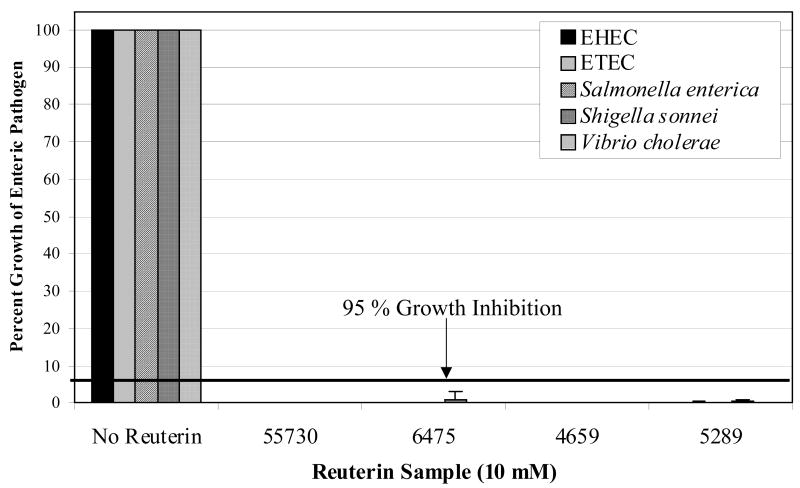
L. reuteri strain-dependent inhibition of enteric pathogens prompted a closer examination of reuterin as a central pathogen-inhibitory factor produced by L. reuteri. Pathogen-inhibitory effects of reuterin secreted by the human-derived L. reuteri strains were evaluated with enteric pathogens in order to determine if strain-dependent variation in pathogen inhibition could be detected. We modified a previously described MIC assay [10] for analyses of quantified reuterin in a high-throughput manner. Test strains were incubated in the presence and absence of serially-diluted reuterin samples obtained from L. reuteri strains 55730, 6475, 4659 and 5289, and their growth was analyzed by measuring optical densities at 600 nm (A600). The MIC assay showed that reuterin derived from each L. reuteri strain inhibited the enteric pathogens in the same manner, with concentrations as low as 10 mM capable of exhibiting ≥ 95% growth inhibition (Fig. 2). As a control, L. plantarum strain 42/6 was tested for reuterin production and pathogen inhibition in the same assays. L. plantarum strain 42/6 produced no reuterin and did not inhibit pathogen growth (data not shown). The data showed that reuterin had similar effects on pathogen growth regardless of the source strain, and that each pathogen demonstrates comparable susceptibilities to reuterin. These results also demonstrate that reuterin only accounts for part of the pathogen-inhibitory activities of probiotic L. reuteri, and other antimicrobial factors may be important for differential inhibition of bacterial pathogens by L. reuteri.
Figure 2. Reuterin inhibits proliferation of enteric pathogens.
Reuterin was produced by L. reuteri strains 55730, 6475, 4659 and 5289 and tested for inhibitory effects on various enteric pathogens in an MIC assay. The growth of each pathogen was determined by measuring the A600 after 18–24 hr incubation in the presence or absence of reuterin. Pathogen growth at 18–24 hr in the absence of reuterin was normalized to 100% for each replicate. The concentration of reuterin that consistently resulted in > 95% growth inhibition for each of the pathogens tested was 10 mM for each L. reuteri strain. EHEC, enterohemorrhagic E. coli; ETEC, enterotoxigenic E. coli.
Lactobacillus species produce lactic acid, resulting in reduced pH that may suppress bacterial growth. L. reuteri strains may differ in the amount of lactic acid produced, which would cause variations in their environmental pH and suppression of pathogenic bacteria. Therefore, the pH values of overnight cultures of each L. reuteri strain were measured. All L. reuteri strains reduced the pH of the media to ~5.06 ± 0.02 with no strain-dependent variation, suggesting that the quantity of lactic acid produced was similar for each strain.
4. Discussion
New functional assays were developed in order to test the relative abilities of probiotics to generate specific antimicrobial factors and suppress the proliferation of bacterial pathogens. Probiotic L. reuteri strains produced different amounts of a key antimicrobial compound, reuterin, and are highly resistant to antimicrobial factors secreted by different strains belonging to the same species. Probiotic L. reuteri strains lacked robust inhibitory effects when combined with other probiotic Lactobacillus species such as L. acidophilus, L. gasseri and L. johnsonii. By contrast, L. reuteri potently inhibited L. casei and highlights the importance of species compatibility considerations when creating mixed species probiotic formulations (Table 3). While L. reuteri strains varied with respect to reuterin production, the magnitude of reuterin production did not correlate with the differential abilities of L. reuteri strains to inhibit enteric pathogens. Reuterin produced by each L. reuteri strain was sufficiently and equally potent at similar concentrations. Differential antimicrobial effects were evident with enteric pathogens, but not with Lactobacillus spp. Genetic diversity among strains of EHEC, ETEC, Salmonella enterica and Vibrio cholerae may account for relative differences in susceptibilities of pathogenic isolates to probiotics. The data show that L. reuteri strains vary significantly in the production of reuterin and in their ability to inhibit the growth of enteric pathogens. Variability in pathogen susceptibility on agar may suggest that each pathogen is affected differently by the coordinated expression of reuterin and other antimicrobial factors. Future studies implementing L. reuteri strains deficient in reuterin production will provide more clarity with regards to the differential antimicrobial properties of these strains.
Our data imply that L. reuteri strains vary in their ability to convert glycerol to reuterin (Fig. 1), yielding differences in reuterin synthesis between strains at the molecular level. Microarray data has indicated that differences in gene expression seem to correlate with differences in reuterin production between L. reuteri strains (Saulnier, D., personal communication). While the reuterin MIC values differ from previous studies, it is quite clear that there is no inherent qualitative difference in the reuterin produced from each L. reuteri strain. Previous studies have shown that much lower concentrations of reuterin can inhibit the growth of pathogens. However, these studies were either performed with different pathogens in co-culture systems using viable cells [9] or by adding filter-sterilized spent media [10, 17], each of which are quite different from the reuterin:glycerol suspension used here. Reuterin is known to exist in three forms, with the aldehyde form being biologically relevant. The dimeric, form (biologically inactive) of purified reuterin is predominant at high concentrations (4.9 M), while the hydrated molecule is prevalent from 1.4 M to 0.030 M [18]. Vollenweider, et al. [18] also suggests that the biologically active monomeric form of reuterin is dominant at concentrations in the micromolar range. Reuterin at a concentration of 10 mM may include various reuterin forms. The Trp-HCl assay quantifies all three reuterin forms, but the structure of reuterin in a glycerol solution has not been determined.
Probiotics may suppress the proliferation and virulence of bacterial pathogens by different mechanisms. Commensal-derived probiotics may produce diverse antimicrobial factors including bacteriocins and non-peptide compounds, and these strains may offer therapeutic alternatives in the era of multidrug-resistant pathogens. While lactic acid and reuterin are established antimicrobial effectors in lactobacilli [4], some strains of L. reuteri are known to secrete bacteriocins. L. reuteri LA 6 secretes a high-molecular weight bacteriocin, reutericin 6, that exhibits both bactericidal and bacteriolytic activity against other LAB species [26]. A second low-molecular weight compound, reutericyclin isolated from L. reuteri LTH2584, has bactericidal and bacteriostatic activity against many gram-positive species, but does not affect gram-negative bacteria [27] and has not been identified in reuterin-producing L. reuteri strains [28]. While no significant homology to reutericin 6 exists in our L. reuteri genomes (genetic information for reutericyclin is currently unknown), the narrow and restricted ranges of antimicrobial activities displayed by reutericin 6 and reutericyclin make them unlikely candidates for the synergistic antimicrobial compound(s) proposed to exist in our strains. Preliminary genomic comparisons for antimicrobial genes within L. reuteri 55730 and 6475 have identified plantaracin within 6475 but not in 55730 (Storm, M., personal communication). Future studies will determine if plantaracin is functioning as a fundamental antimicrobial compound in 6475.
Recent in vitro studies have enhanced our confidence in the ability of L. reuteri and reuterin to reshape the intestinal milieu. Cleusix, et al. have demonstrated that the addition of L. reuteri ATCC 55730 and glycerol decrease E. coli populations in an in vitro colonic fermentation model [29]. Human clinical trials are being pursued in an effort to identify candidate probiotics useful in treating a variety of ailments. The most recent Cochrane report on Probiotics for treating infectious diarrhoea [30] reviewed 23 studies that included 1917 participants, concluding that probiotics are useful supplements to rehydration therapy and can reduce infectious diarrhea by three days. A recent infant colic study further substantiated the health benefits of L. reuteri 55730, whereby 95% of the probiotic-treated infants responded to treatment for colic as compared to 7% in the simethicone-treated control group [31]. Clinical trials involving Nigerian women diagnosed with bacterial vaginosis (BV) showed that L. reuteri RC-14 augmented metronidazole treatment of bacterial vaginosis (BV) [32] and resulted in an effective cure of BV without added antibiotic treatment [6].
The development of probiotics-based strategies for human medicine increased the need for high-throughput screening methods that allow for the identification of new bacterial strains possessing specific probiotic features. In this study, we modified two phenotypic assays and one quantitative assay for high-throughput analyses of anti-infective properties of L. reuteri. Elucidation of the molecular basis of probiotic properties important for treating specific diseases will empower us to develop systematic screening methods that allow for the identification of candidate probiotic strains inherently tailored for suppression of different spectra of infectious agents. Future studies will include functional genomics approaches to elucidate the molecular mechanisms responsible for the variation in reuterin production between L. reuteri strains. Experiments elucidating the phenotypic events described here will facilitate an improved understanding of molecular mechanisms of probiotic functions of L. reuteri.
Acknowledgments
We thank Chandra Iyer and Angela Major for their technical efforts in method optimization, Martin Storm for his assistance with L. reuteri genomes and Sabine Vollenweider at the Institute of Food Science and Nutrition in Zurich, Switzerland for advice and detailed protocols regarding the colorimetric detection of reuterin. We also acknowledge Beverly Vispo at TCH for her efforts in HPLC analyses of reuterin samples, Tiffany Morgan and Sabeen Raza for administrative support, and Robert Britton and Delphine Saulnier for their thoughtful review. The authors acknowledge Eamonn Connolly (Biogaia AB) for provision of L. reuteri strains.
This work was supported by the National Institutes of Health (R01 DK065075), Senior Research Award from the Crohn’s & Colitis Foundation of America (CCFA), the Defense Advanced Research Projects Agency (DARPA) and the Texas Medical Center Digestive Diseases Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Klaenhammer TR. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev. 1993;12:39–85. doi: 10.1111/j.1574-6976.1993.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 2.Servin AL. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol Rev. 2004;28:405–440. doi: 10.1016/j.femsre.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Reid G, Burton J. Use of Lactobacillus to prevent infection by pathogenic bacteria. Microbes Infect. 2002;4:319–324. doi: 10.1016/s1286-4579(02)01544-7. [DOI] [PubMed] [Google Scholar]
- 4.Casas IA, Dobrogosz WJ. Validation of the probiotic concept: Lactobacillus reuteri confers broad-spectrum protection against disease in humans and animals. Microbial Ecology in Health and Disease. 2000;12:257–285. [Google Scholar]
- 5.Teitelbaum JE. Probiotics and the treatment of infectious diarrhea. Pediatr Infect Dis J. 2005;24:267–268. doi: 10.1097/01.inf.0000156408.17683.a6. [DOI] [PubMed] [Google Scholar]
- 6.Anukam KC, Osazuwa E, Osemene GI, Ehigiagbe F, Bruce AW, Reid G. Clinical study comparing probiotic Lactobacillus GR-1 and RC-14 with metronidazole vaginal gel to treat symptomatic bacterial vaginosis. Microbes Infect. 2006b doi: 10.1016/j.micinf.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 7.Caglar E, Kavaloglu Cildir S, Ergeneli S, Sandalli N, Twetman S. Salivary mutans streptococci and lactobacilli levels after ingestion of the probiotic bacterium Lactobacillus reuteri ATCC 55730 by straws or tablets. Acta Odontol Scand. 2006;64:314–318. doi: 10.1080/00016350600801709. [DOI] [PubMed] [Google Scholar]
- 8.Rosenfeldt V, Michaelsen KF, Jakobsen M, Larsen CN, Moller PL, Pedersen P, Tvede M, Weyrehter H, Valerius NH, Paerregaard A. Effect of probiotic Lactobacillus strains in young children hospitalized with acute diarrhea. Pediatr Infect Dis J. 2002;21:411–416. doi: 10.1097/00006454-200205000-00012. [DOI] [PubMed] [Google Scholar]
- 9.Axelsson LT, Chung TC, Dobrogosz WJ, Lindgren SE. Production of a broad spectrum antimicrobial substance by Lactobacillus reuteri. Microbial Ecology in Health and Disease. 1989;2:131–136. [Google Scholar]
- 10.Chung TC, Axelsson L, Lindgren E, Dobrogosz WJ. In vitro studies on reuterin synthesis by Lactobacillus reuteri. 1989:137–144. [Google Scholar]
- 11.Yunmbam MK, Roberts JF. In vivo evaluation of reuterin and its combinations with suramin, melarsoprol, DL-alpha-difluoromethylornithine and bleomycin in mice infected with Trypanosoma brucei brucei. Comp Biochem Physiol C. 1993;105:521–524. doi: 10.1016/0742-8413(93)90095-3. [DOI] [PubMed] [Google Scholar]
- 12.Waters WR, Harp JA, Wannemuehler MJ, Carbajal NY, Casas IA. Effects of Lactobacillus reuteri on Cryptosporidium parvum infection of gnotobiotic TCR-alpha-deficient mice. J Eukaryot Microbiol. 1999;46:60S–61S. [PubMed] [Google Scholar]
- 13.Alak JI, Wolf BW, Mdurvwa EG, Pimentel-Smith GE, Kolavala S, Abdelrahman H, Suppiramaniam V. Supplementation with Lactobacillus reuteri or L. acidophilus reduced intestinal shedding of cryptosporidium parvum oocysts in immunodeficient C57BL/6 mice. Cell Mol Biol (Noisy-le-grand) 1999;45:855–863. [PubMed] [Google Scholar]
- 14.Rosenfeldt V, Michaelsen KF, Jakobsen M, Larsen CN, Moller PL, Tvede M, Weyrehter H, Valerius NH, Paerregaard A. Effect of probiotic Lactobacillus strains on acute diarrhea in a cohort of nonhospitalized children attending day-care centers. Pediatr Infect Dis J. 2002;21:417–419. doi: 10.1097/00006454-200205000-00013. [DOI] [PubMed] [Google Scholar]
- 15.Talarico TL, Dobrogosz WJ. Chemical characterization of an antimicrobial substance produced by Lactobacillus reuteri. Antimicrob Agents Chemother. 1989;33:674–679. doi: 10.1128/aac.33.5.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doleyres Y, Beck P, Vollenweider S, Lacroix C. Production of 3-hydroxypropionaldehyde using a two-step process with Lactobacillus reuteri. Appl Microbiol Biotechnol. 2005;68:467–474. doi: 10.1007/s00253-005-1895-4. [DOI] [PubMed] [Google Scholar]
- 17.Luthi-Peng Q, Scharer S, Puhan Z. Production and stability of 3-hydroxypropionaldehyde in Lactobacillus reuteri. Appl Microbiol Biotechnol. 2002;60:73–80. doi: 10.1007/s00253-002-1099-0. [DOI] [PubMed] [Google Scholar]
- 18.Vollenweider S, Grassi G, Konig I, Puhan Z. Purification and structural characterization of 3-hydroxypropionaldehyde and its derivatives. J Agric Food Chem. 2003;51:3287–3293. doi: 10.1021/jf021086d. [DOI] [PubMed] [Google Scholar]
- 19.Catassi C, Pierani P, Natalini G, Gabrielli O, Coppa GV, Giorgi PL. Clinical application of a simple HPLC method for the sugar intestinal permeability test. J Pediatr Gastroenterol Nutr. 1991;12:209–212. doi: 10.1097/00005176-199102000-00012. [DOI] [PubMed] [Google Scholar]
- 20.Talarico TL, Casas IA, Chung TC, Dobrogosz WJ. Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri. Antimicrob Agents Chemother. 1988;32:1854–1858. doi: 10.1128/aac.32.12.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Circle SJ, Stone L, Boruff CS. Acrolein determination by means of tryptophane. A colorimetric method. Industrial and engineering chemistry. 1945;17:259–262. [Google Scholar]
- 22.Schillinger U, Lucke FK. Antibacterial activity of Lactobacillus sake isolated from meat. Appl Environ Microbiol. 1989;55:1901–1906. doi: 10.1128/aem.55.8.1901-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobsen CN, Rosenfeldt Nielsen V, Hayford AE, Moller PL, Michaelsen KF, Paerregaard A, Sandstrom B, Tvede M, Jakobsen M. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl Environ Microbiol. 1999;65:4949–4956. doi: 10.1128/aem.65.11.4949-4956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luthi-Peng Q, Dileme FB, Puhan Z. Effect of glucose on glycerol bioconversion by Lactobacillus reuteri. Appl Microbiol Biotechnol. 2002;59:289–296. doi: 10.1007/s00253-002-1002-z. [DOI] [PubMed] [Google Scholar]
- 25.Holzapfel WH, Haberer P, Snel J, Schillinger U, Huis in’t Veld JH. Overview of gut flora and probiotics. Int J Food Microbiol. 1998;41:85–101. doi: 10.1016/s0168-1605(98)00044-0. [DOI] [PubMed] [Google Scholar]
- 26.Kabuki T, Saito T, Kawai Y, Uemura J, Itoh T. Production, purification and characterization of reutericin 6, a bacteriocin with lytic activity produced by Lactobacillus reuteri LA6. Int J Food Microbiol. 1997;34:145–156. doi: 10.1016/s0168-1605(96)01180-4. [DOI] [PubMed] [Google Scholar]
- 27.Ganzle MG, Holtzel A, Walter J, Jung G, Hammes WP. Characterization of reutericyclin produced by Lactobacillus reuteri LTH2584. Appl Environ Microbiol. 2000;66:4325–4333. doi: 10.1128/aem.66.10.4325-4333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ganzle MG. Reutericyclin: biological activity, mode of action, and potential applications. Appl Microbiol Biotechnol. 2004;64:326–332. doi: 10.1007/s00253-003-1536-8. [DOI] [PubMed] [Google Scholar]
- 29.Cleusix V, Lacroix C, Vollenweider S, Duboux M, Le Blay G. Inhibitory activity spectrum of reuterin produced by Lactobacillus reuteri against intestinal bacteria. BMC Microbiol. 2007;7:101. doi: 10.1186/1471-2180-7-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen SJ, Okoko B, Martinez E, Gregorio G, Dans LF. Probiotics for treating infectious diarrhoea. Cochrane Database Syst Rev. 2004:CD003048. doi: 10.1002/14651858.CD003048.pub2. [DOI] [PubMed] [Google Scholar]
- 31.Savino F, Pelle E, Palumeri E, Oggero R, Miniero R. Lactobacillus reuteri (American Type Culture Collection Strain 55730) versus simethicone in the treatment of infantile colic: a prospective randomized study. Pediatrics. 2007;119:e124–130. doi: 10.1542/peds.2006-1222. [DOI] [PubMed] [Google Scholar]
- 32.Anukam K, Osazuwa E, Ahonkhai I, Ngwu M, Osemene G, Bruce AW, Reid G. (a)) Augmentation of antimicrobial metronidazole therapy of bacterial vaginosis with oral probiotic Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14: randomized, double-blind, placebo controlled trial. Microbes Infect. 2006;8:1450–1454. doi: 10.1016/j.micinf.2006.01.003. [DOI] [PubMed] [Google Scholar]