SUMMARY
The recently sequenced genomes of several Aspergillus species have revealed that these organisms have the potential to produce a surprisingly large range of natural products, many of which are currently unknown. We have found that A. nidulans produces emericellamide A, an antibiotic compound of mixed origins with polyketide and amino acid building blocks. Additionally, we describe the discovery of four previously unidentified, related compounds that we designate emericellamide C-F. Using recently developed gene targeting techniques, we have identified the genes involved in emericellamide biosynthesis. The emericellamide gene cluster contains one polyketide synthase and one nonribosomal peptide synthetase. From the sequences of the genes, we are able to deduce a biosynthetic pathway for the emericellamides. The identification of this biosynthetic pathway opens the door to engineering novel analogs of this structurally complex metabolite.
INTRODUCTION
Secondary metabolites are a rich source of medically useful compounds. The recent sequencing of a number of fungal genomes reveals, surprisingly, that common fungi may be a rich and under-exploited source of secondary metabolites. The sequencing of the genomes of four species of the genus Aspergillus, for example, (Galagan et al., 2005; Machida et al., 2005; Nierman et al., 2005; Pel et al., 2007), revealed that they have many more secondary metabolite pathways than was suspected from decades of natural product chemistry. A. nidulans, for example, has 27 polyketide synthases (PKSs) and 14 nonribosomal peptide synthetases (NRPSs), but the products of fewer than ten biosynthesis gene clusters had been identified at the time the genome was completed (Bergmann et al., 2007; Bok et al., 2006; Haas, 2003; Hoffmeister and Keller, 2007; Marquez-Fernandez et al., 2007; Scherlach and Hertweck, 2006). Moreover, the genes of individual secondary metabolite pathways are almost always clustered together in the genome, which potentially facilitates study and molecular genetic manipulation (Brown et al., 1996).
The realization that common laboratory fungi are a likely source of many more natural products has, happily, been accompanied by important advances in the molecular genetic manipulation of some of these fungi. One was the development of fusion PCR techniques that have greatly facilitated the production of linear molecules for gene replacements, promoter replacements and gene tagging (e.g. Kuwayama et al., 2002; Nayak et al., 2006; Szewczyk et al., 2006; Yang et al., 2004; Zarrin et al., 2005). A second was the discovery that deletion of KU genes, the products of which are required for non-homologous end joining, greatly improves gene targeting frequencies (da Silva Ferreira et al., 2006; Krappmann et al., 2006; Nayak et al., 2006; Ninomiya et al., 2004; Takahashi and Smithies, 1999). We have applied these approaches to natural product discovery in A. nidulans and, in particular, have used them to identify and characterize the emericellamide biosynthesis pathway.
RESULTS AND DISCUSSION
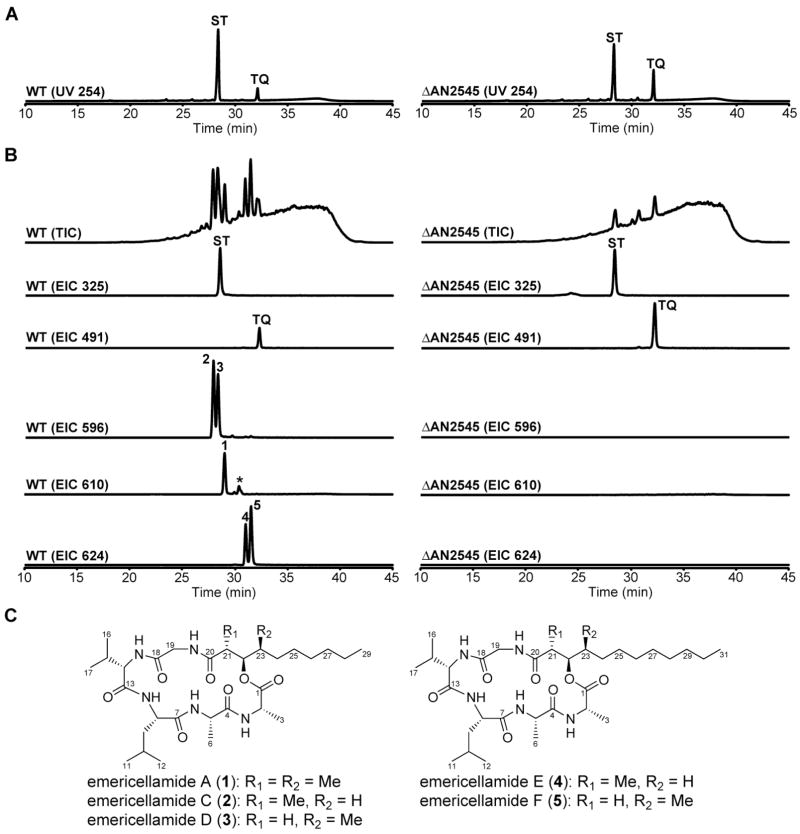
With the expectation that deleting each biosynthetic gene will result in the inability to synthesize one or more secondary metabolites in the same biosynthetic pathway, we randomly selected six NRPS genes, using the Broad Institute designation (http://www.broad.mit.edu/annotation/genome/Aspergillus_group/MultiHome.html), i.e. AN0607.3, AN1242.3, AN2545.3, AN2621.3, AN8412.3 and AN9244.3 (Table S1 and S2), and deleted each of them. One of the NRPS genes AN8412.3 has recently been shown to be involved in asypridone biosynthesis (Bergmann et al., 2007). The selective disruption of each gene was verified by diagnostic PCR (data not shown). The results of HPLC profiles show that only the AN2545.3 deletant exhibited a different profile from the control while the other five NRPS deletants were the same as the wild type (data not shown). These results highlight the fact that most biosynthetic gene clusters are cryptic, at least in common laboratory culture conditions. While both wild type and the AN2545.3 deletant still produce sterigmatocystin (ST) and terrequinone (TQ) (Figure 1A), five compounds with m/z = 609 (m/z = 610 [M+H]+, compound 1), m/z = 595 (m/z = 596 [M+H]+, compound 2 and 3), and m/z = 623 (m/z = 624 [M+H]+, compound 4 and 5) were missing in the AN2545.3 deletant (Figure 1B). A search of known A. nidulans secondary metabolites from CrossFire Beilstein database revealed that these five compounds had not been previously described in A. nidulans.
Figure 1. LC-DAD-MS analysis of wild-type A. nidulans and ΔAN2545 mutant metabolites.
(A) UV trace at 254 nm
(B) Positive ion mode: Total ion current (TIC); extracted ion current (EIC) at m/z 325, sterigmatocystin [M+H]+; EIC at m/z 491, terrequinone [M+H]+; EIC at m/z 596, emericellamides C & D [M+H]+; EIC at m/z 610, emericellamide A [M+H]+; EIC at m/z 624, emericellamides E & F [M+H]+
(C) Chemical structures of emericellamides
(*) A minor compound which has same MS/MS fragment with emericellamide A
All five compounds were isolated from large-scale cultivation of wild-type A. nidulans by purification, initially by flash chromatography and then by preparative HPLC. Full one- and two-dimensional NMR analysis of compound 1 identified the product as emericellamide A (Figure 1C, Tables S3 and S4), a mixed cyclic polyketide nonribosomal peptide (active against methicillin-resistant Staphylococcus aureus) produced by a marine Emericella species (Oh et al., 2007). (Emericella is the genus designation generally used for species of Aspergillus with a demonstrated sexual cycle.) The four other compounds were completely new emericellamides (Figure 1C), and we solved their structures by a combination of one- and two-dimensional NMR analyses and by comparison to emericellamide A (1) (For detailed structural elucidation, see Supplemental Data). The MS fragmentation also supports the assigned structures (Figures S2 and S3). Interestingly, the intensities of dehydration fragments in compounds 3 and 5 are stronger than compounds 2 and 4 (m/z = 295, 323, 408, 436, and 507 for 2 and 3; m/z = 323, 351, 436, 464, and 535 for 4 and 5). These fragmentation data also support the assigned structures of the aliphatic side chains, since compounds 3 and 5 have two carbonyl α-protons that increase the probability of H2O elimination to generate αβ-unsaturated carbonyl fragments. Similarity in structures and the single biosynthetic origin leads us to name compounds 2–5 emericellamides C-F (Figure 1C). In addition to emericellamide A (1), a minor peak having the same MS/MS fragments (data not shown) with emericellamide A (1) was also observed in the EIC of 610 (Figure 1B). Although not present in sufficient quantity to allow full NMR characterization, this compound could be another long-chained emericellamide which did not undergo methyltransfer.
In fungi, biosynthesis genes of secondary metabolites are generally clustered. To identify genes in the emericellamide biosynthesis pathway, additional genes surrounding the NRPS (from AN2542.3 to AN10325.3, Figure 2A) were deleted and the mutant strains analyzed (Figure 2B and data not shown). The data show that, using the BLAST NCBI protein database, AN2547.3 (a type I PKS homolog), AN2548.3 (an acyltransferase homolog), and AN2549.3 (an AMP dependent CoA-ligase homolog) are involved in the emericellamide biosynthetic pathway. While the structures of emericellamides clearly suggest the involvement of a PKS, the involvement of the two additional genes provides evidence that the transfer of the polyketide intermediates, i.e. the acyl side chains, from the PKS to the first thiolation domain of the NRPS is mediated by additional enzymes. Our data show that deletants of AN2544.3 (a sugar transporter homolog) and AN2546.3 (a hypothetical protein) continue to produce emericellamides.
Figure 2. Boundary of emericellamide synthesis (eas) cluster.
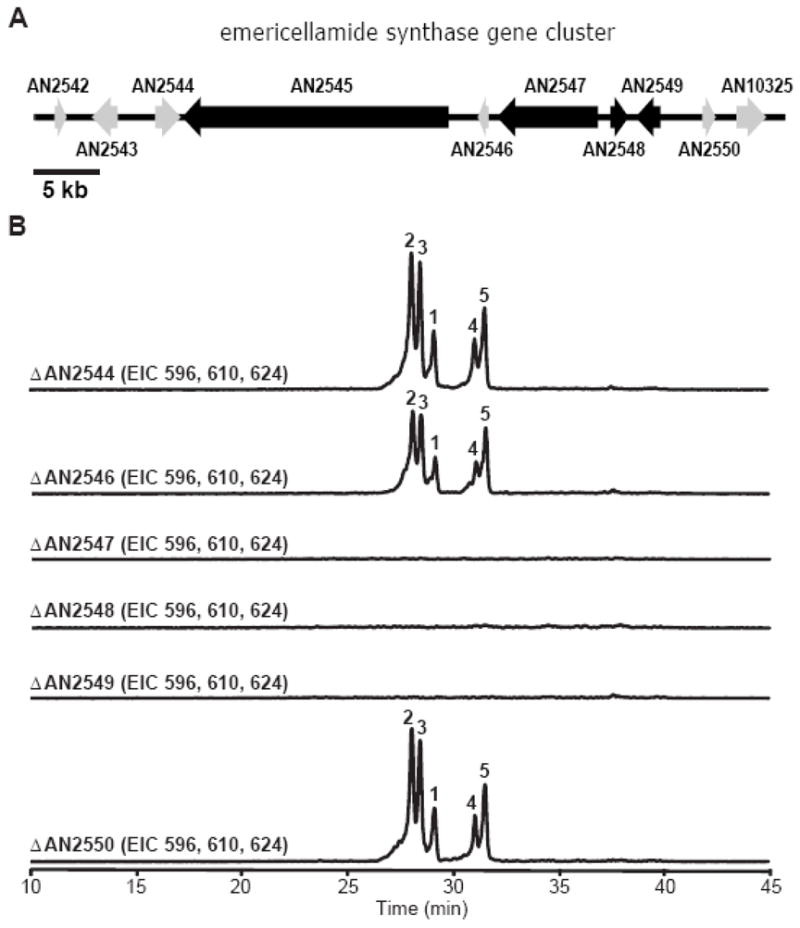
(A) Organization of the emericellamide synthase gene cluster in A. nidulans. Black open reading frame (ORF), genes involved in emericellamide biosynthesis; gray ORF, genes not involved in the pathway
(B) LC-MS analysis of A. nidulans mutant metabolites in positive ion mode, EIC at m/z 596, emericellamides C & D [M+H]+; EIC at m/z 610, emericellamide A [M+H]+; EIC at m/z 624, emericellamides E & F [M+H]+
The biosynthetic pathway of emericellamides can be deduced by analyzing the four genes in the gene cluster, which we designate the eas (emericellamide synthesis) cluster. The biosynthesis of emericellamide initiates from the highly reducing iterative type I polyketide synthase EasB (AN2547.3) (Figure 3). The enzyme contains the ketosynthase (KS), acyl transferase (AT), methyltransferase (MT), dehydrogenase (DH), enoylreductase (ER), ketoreductase (KR) and acyl carrier protein (ACP) domains. The single module fungal polyketide iteratively catalyzes the formation of the linear polyketide chain. Isolation and identification of emericellamide derivatives differing in the polyketide chain length and degree of methylation provide evidence that EasB produces several polyketides that can be further processed by the downstream enzymes. The polyketide intermediate is not transferred directly to the NRPS EasA (AN2545.3) but instead is mediated by two additional enzymes as supported by our genetic data. The polyketide is released from EasB as a linear polyketide carboxylic acid which is converted to a CoA thioester by the acyl-CoA ligase EasD (AN2549.3). The substrate is then loaded onto the acyltransferase EasC (AN2548.3) which shuttles the polyketide intermediate to the first thiolation (T) domain of the NRPS EasA (AN2545.3). Examples of transferase mediated transfer between carrier protein domains have recently been characterized in the coronamic acid biosynthesis pathway (Strieter et al., 2007). The NRPS EasA is a multimodular enzymatic assembly that contains 18 domains grouped into five modules corresponding to each of the five amino acid monomers incorporated. The second to the fifth modules contain the three core domains, the condensation (C) domain responsible for catalyzing peptide bond formation, adenylation (A) domain responsible for selecting the amino acid monomer substrate, and the thiolation (T) domain. The first module in the NRPS contains a unique T-E-C-A-T-E structure where (E) domains are epimerization domains. We propose that the first of the two thiolation domains is responsible for accepting the incoming polyketide from the acyltransferase EasC. The second thiolation domain is responsible for accepting the glycine amino acid adenylated by the A domain in the module. The condensation domain then catalyzes the C-N bond formation between the upstream acyl thioester and the downstream aminoacyl thioester. The roles of the two epimerization domains located in the first module of the NRPS are unclear since glycine is the amino acid activated by the module. Interestingly the NRPS does not contain a TE domain at the end of module 5, presumably suggesting that TE is not necessary for the cyclization of the emericellamides. It should be noted that of the NRPS in the A. nidulans genome only ACV NRPS (AN2621.3), terrequinone NRPS (AN8513.3) and the unannotated NRPS AN3396.3 contain a TE domain as part of the enzyme. The nine residues that define the signature sequence of A domain selectivity were identified and analyzed using two publicly available bioinformatic software programs (Table 1) (Rausch et al., 2005; Yadav et al., 2003). Both software tools, Search NRPS/PKS and NRPSpredictor, correctly predicted that the first module codes for glycine. However, the specificity of the A domain in the second module was only correctly predicted by NRPSpredictor to be valine. The A domain selectivity of the last three modules was predicted incorrectly by both programs. Many characterized NRPS used by these bioinformatic software tools are bacterial in origin and understanding of fungal NRPS is still in its infancy. Identification of the products of fungal NRPS from genome sequencing could help in the development of a better understanding of the fungal NRPS code.
Figure 3. Model for emericellamide A (1) biosynthesis.
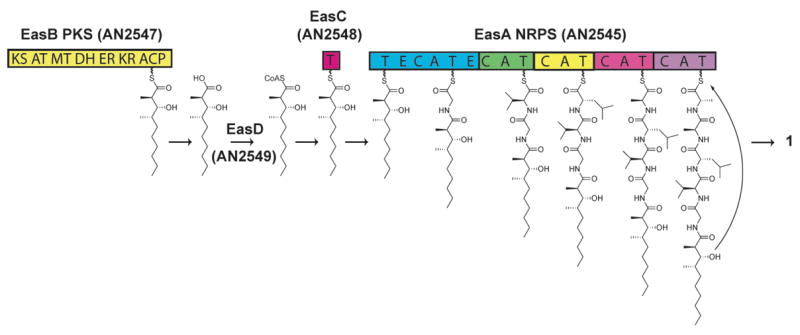
Table 1. Signature sequences of the adenylation (A) domains.
| Amino Acid | Module | Signature Sequence Position
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 235 | 236 | 239 | 278 | 299 | 301 | 322 | 330 | 331 | ||
| Gly | 1 | D | I | Q | G | V | L | A | M | Q |
| Val | 2 | D | A | S | Q | I | G | G | I | Y |
| Leu | 3 | D | I | H | F | V | G | A | I | A |
| Ala | 4 | D | L | L | V | V | A | G | I | L |
| Ala | 5 | D | I | A | I | L | V | A | I | L |
SIGNIFICANCE
The rapid completion of fungal genome sequencing provides an unprecedented opportunity to study fungal secondary metabolism pathways. In this communication, we report the application of gene targeting approaches to functionally characterize a pathway containing both a PKS and an NRPS working to produce the cyclic polyketide peptide emericellamides. Previously emericellamide A was only discovered in a marine Emericella and produced in very small amounts when the fungus was cultured alone. When the marine fungus was stimulated by co-culturing with a bacterium, production of emericellamide A was increased 100 fold (Oh et al., 2007). By comparison, A. nidulans is a well studied, established laboratory model fungus and the emericellamides are produced by the A. nidulans cultured alone in levels comparable to the bacterium stimulated marine fungus. Our efficient gene deletion approach allows rapid annotation of the genes in A. nidulans and can be applied to other secondary metabolite clusters. Replacement of the native promoter with either inducible or strong constitutive promoters is feasible and can provide a rational route to increase the production of emericellamides. The identification of the emericellamide pathway opens the door to the engineering of the PKS and NRPS for the production of novel analogs. Finally, additional functional annotation in A. nidulans and other fungi will improve both our understanding of fungal secondary metabolite biosynthesis and bioinformatic analysis of fungal NRPS and PKS.
EXPERIMENTAL PROCEDURES
Molecular Genetic Procedures
Construction of fusion PCR products, protoplast production and transformation were carried out as described (Szewczyk et al., 2006). For construction of the fusion PCR fragments, two ~1000 base pair fragments upstream and downstream of the targeted gene were amplified from genomic A. nidulans DNA by PCR. Fusion PCR was set up with the two amplified flanking sequences and the A. fumigatus pyrG selectable marker cassette. The three fragments were fused into a single molecule and amplified with two nested primers (Figure S1 and Table S2). The ΔnkuA strain TN02A3 (Nayak et al., 2006) was used as a recipient strain. Diagnostic PCR of deletant strains was performed using the external primers used in the first round of the PCR. The difference in the size between the gene replaced by the selective marker and the native gene allowed the determination of correct gene replacement. In cases when the sizes of both the wild-type and deletant products were similar, diagnostic PCR was performed using one of the external primers and a primer located inside the marker gene. In those cases, the deletant gave the PCR product of the expected size versus no product in the wild-type strain.
Fermentation and purification
Spores of A. nidulans wild type R153 and deletion strains were inoculated (1 X 106/ml) into 100 ml glucose minimal medium (6 g/l NaNO3, 0.52 g/l KCl, 0.52 g/l MgSO4• 7H2O, 1.52 g/l KH2PO4, 10 g/l D-glucose, 1 ml/l trace element solution (Cove, 1966) supplemented with pyridoxine (0.5 μg/ml) and cultivated at 37°C with shaking at 250 rpm. After 4 days, culture medium was collected by filtration and extracted with the same volume of EtOAc two times. The combined EtOAc layer was evaporated in vacuo and re-dissolved in MeOH (1 mg/ml) for HPLC-DAD-MS analysis. For structure elucidation, a 10 L fermentation volume was extracted with EtOAc to yield a gray powder (350 mg), which was applied to a Si gel column (Merck 230–400 mesh, ASTM, 10 × 100 mm) and eluted with CHCl3-MeOH mixtures of increasing polarity (frac. A, 1:0, 300 ml; frac. B, 19:1, 300 ml; frac. C, 9:1, 300 ml; frac. D, 7:3, 300 ml). The solvent of frac. B, containing emericellamides, was evaporated in vacuo and resuspended in CHCl3. The emericellamides, which do not dissolve in CHCl3, were collected by filtration. Enriched emericellamides were separated by preparative HPLC [Phenomenex Luna 5 μm C18 (2), 250 × 21.2 mm] with a flow rate of 10.0 ml/min and measured by a UV detector at 200 nm. The gradient system was MeCN (solvent B) in 5% MeCN/H2O (solvent A): 60 to 100% B from 0 to 15 min, 100% B from 15 to 20 min, and re-equilibration with 60% B from 20 to 22 min. A mixture of emericellamides C and D (6.6 mg), emericellamide A (1.2 mg), and a mixture of emericellamides E and F (5.6 mg) were eluted at 11.4, 12.5 and 14.8 min, respectively. Emericellamides C (2.8 mg, TR = 36.2 min) and D (2.5 mg, TR = 37.8 min) were further purified by the same HPLC using isocratic 50% B solvent. Emericellamides E (1.4 mg, TR = 47.0 min) and F (2.6 mg, TR = 49.1 min) were further purified by the same HPLC using isocratic 55% B solvent.
Accession codes
A. nidulans Broad Institute database easA (AN2545.3), easB (AN2547.3), easC(AN2548.3), and easD (AN2549.3). Accession codes were deposited as part of previous studies.
SUPPLEMENTAL DATA
Supplemental Data including detailed structural characterization of emericellamides, A. nidulans strains and primers used in this study, fusion PCR for gene disruption, 1H and 13C NMR spectral data, and MS/MS fragmentation of emericellamides, can be found with this article free of charge via the internet.
Acknowledgments
This work was financially supported by NIH grants GM 075857 (C.C.C.W.), GM 031837 (B.R.O.) and GM 084077 (C.C.C.W. and B.R.O.). H.S. is a recipient of the USC School of Pharmacy Undergraduate Summer Fellowship. E.K. is a recipient of the Rose Hill Undergraduate Fellowship. K.W. is a recipient of fellowships from the Japan Antibiotics Research Association, Pfizer Infectious Disease Foundation, and from the Agricultural Chemical Research Foundation. High resolution mass spectrometry analyses were performed in the Mass Spectrometry Laboratory of the Division of Chemistry and Chemical Engineering at the California Institute of Technology, supported in part by the National Science Foundation Materials and Research Science and Engineering Program. We also thank Brian Stoltz for access to a digital polarimeter and IR spectrophotometer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nature chemical biology. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- Bok JW, Hoffmeister D, Maggio-Hall LA, Murillo R, Glasner JD, Keller NP. Genomic mining for Aspergillus natural products. Chem Biol. 2006;13:31–37. doi: 10.1016/j.chembiol.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Brown DW, Yu JH, Kelkar HS, Fernandes M, Nesbitt TC, Keller NP, Adams TH, Leonard TJ. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc Natl Acad Sci USA. 1996;93:1418–1422. doi: 10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove DJ. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim Biophys Acta. 1966;113(1):51–56. doi: 10.1016/s0926-6593(66)80120-0. [DOI] [PubMed] [Google Scholar]
- da Silva Ferreira ME, Kress MR, Savoldi M, Goldman MH, Hartl A, Heinekamp T, Brakhage AA, Goldman GH. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryotic cell. 2006;5:207–211. doi: 10.1128/EC.5.1.207-211.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- Haas H. Molecular genetics of fungal siderophore biosynthesis and uptake: the role of siderophores in iron uptake and storage. Appl Microbiol Biotechnol. 2003;62:316–330. doi: 10.1007/s00253-003-1335-2. [DOI] [PubMed] [Google Scholar]
- Hoffmeister D, Keller NP. Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat Prod Rep. 2007;24:393–416. doi: 10.1039/b603084j. [DOI] [PubMed] [Google Scholar]
- Krappmann S, Sasse C, Braus GH. Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end-joining-deficient genetic background. Eukaryotic cell. 2006;5:212–215. doi: 10.1128/EC.5.1.212-215.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwayama H, Obara S, Morio T, Katoh M, Urushihara H, Tanaka Y. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 2002;30:E2. doi: 10.1093/nar/30.2.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- Marquez-Fernandez O, Trigos A, Ramos-Balderas JL, Viniegra-Gonzalez G, Deising HB, Aguirre J. Phosphopantetheinyl transferase CfwA/NpgA is required for Aspergillus nidulans secondary metabolism and asexual development. Eukaryotic cell. 2007;6:710–720. doi: 10.1128/EC.00362-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, Hynes MJ, Osmani SA, Oakley BR. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics. 2006;172:1557–1566. doi: 10.1534/genetics.105.052563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- Ninomiya Y, Suzuki K, Ishii C, Inoue H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc Natl Acad Sci USA. 2004;101:12248–12253. doi: 10.1073/pnas.0402780101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DC, Kauffman CA, Jensen PR, Fenical W. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J Nat Prod. 2007;70:515–520. doi: 10.1021/np060381f. [DOI] [PubMed] [Google Scholar]
- Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol. 2007;25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucleic Acids Res. 2005;33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherlach K, Hertweck C. Discovery of aspoquinolones A-D, prenylated quinoline–2-one alkaloids from Aspergillus nidulans, motivated by genome mining. Organic & biomolecular chemistry. 2006;4:3517–3520. doi: 10.1039/b607011f. [DOI] [PubMed] [Google Scholar]
- Strieter ER, Vaillancourt FH, Walsh CT. CmaE: a transferase shuttling aminoacyl groups between carrier protein domains in the coronamic acid biosynthetic pathway. Biochemistry. 2007;46:7549–7557. doi: 10.1021/bi700243h. [DOI] [PubMed] [Google Scholar]
- Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. Fusion PCR and gene targeting in Aspergillus nidulans. Nature protocols. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Smithies O. Gene targeting approaches to analyzing hypertension. J Am Soc Nephrol. 1999;10:1598–1605. doi: 10.1681/ASN.V1071598. [DOI] [PubMed] [Google Scholar]
- Yadav G, Gokhale RS, Mohanty D. SEARCHPKS: a program for detection and analysis of polyketide synthase domains. Nucleic Acids Research. 2003;31:3654–3658. doi: 10.1093/nar/gkg607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Ukil L, Osmani A, Nahm F, Davies J, De Souza CP, Dou X, Perez-Balaguer A, Osmani SA. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryotic cell. 2004;3:1359–1362. doi: 10.1128/EC.3.5.1359-1362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrin M, Leeder AC, Turner G. A rapid method for promoter exchange in Aspergillus nidulans using recombinant PCR. Fungal Genet Biol. 2005;42:1–8. doi: 10.1016/j.fgb.2004.10.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data including detailed structural characterization of emericellamides, A. nidulans strains and primers used in this study, fusion PCR for gene disruption, 1H and 13C NMR spectral data, and MS/MS fragmentation of emericellamides, can be found with this article free of charge via the internet.