Abstract
OBJECTIVES— To identify, map, clone, and functionally validate a novel mouse model for impaired glucose tolerance and insulin secretion.
RESEARCH DESIGN AND METHODS— Haploinsufficiency of the insulin receptor and associated mild insulin resistance has been used to sensitize an N-ethyl-N-nitrosourea (ENU) screen to identify novel mutations resulting in impaired glucose tolerance and diabetes. The new impaired glucose tolerance 4 (IGT4) model was selected using an intraperitoneal glucose tolerance test and inheritance of the phenotype confirmed by generation of backcross progeny. Segregation of the phenotype was correlated with genotype information to map the location of the gene and candidates sequenced for mutations. The function of the SRY-related high mobility group (HMG)-box 4 (Sox4) gene in insulin secretion was tested using another ENU allele and by small interfering RNA silencing in insulinoma cells.
RESULTS— We describe two allelic autosomal dominant mutations in the highly conserved HMG box of the transcription factor Sox4. Previously associated with pancreas development, Sox4 mutations in the adult mouse result in an insulin secretory defect, which exhibits impaired glucose tolerance in association with insulin receptor+/−–induced insulin resistance. Elimination of the Sox4 transcript in INS1 and Min6 cells resulted in the abolition of glucose-stimulated insulin release similar to that observed for silencing of the key metabolic enzyme glucokinase. Intracellular calcium measurements in treated cells indicate that this defect lies downstream of the ATP-sensitive K+ channel (KATP channel) and calcium influx.
CONCLUSIONS— IGT4 represents a novel digenic model of insulin resistance coupled with an insulin secretory defect. The Sox4 gene has a role in insulin secretion in the adult β-cell downstream of the KATP channel.
Type 2 diabetes is a heterogeneous metabolic disorder characterized by hyperglycemia resulting from defects in insulin action and/or insulin secretion with underlying genetic and environmental causes. There has been some success in identifying genetic factors underlying type 2 diabetes in some human populations (for example, calpain 10, PPARG, KCNJ11, TCF7L2, and ENPP1), and new whole-genome association studies are identifying further new candidates; however, many genes remain to be identified (1–8). One approach to identifying new genes is to use the mouse as a model. For example, using a combination of quantitative trait loci mapping and mutagenesis techniques, we recently identified a novel gene involved in insulin secretion (9–11).
Phenotype-driven N-ethyl-N-nitrosourea (ENU) mutagenesis screens have proven to be a powerful and efficient tool for the identification of novel models of human disease (12,13). Mutations are introduced at a rate of ∼1/1,000 (one functional mutation at any one locus in 1,000 mice), and each founder mutagenized mouse carries ∼30 such mutations (14–16). ENU is an alkylating agent that preferentially induces A/T to T/A transversion mutations at random across the genome; these point mutations can create a variety of alleles, including hypomorphs, hypermorphs, and loss or gain of function (17). Thus, ENU could be used to introduce mutations into potentially any gene in the genome that can be mutated to give rise to diabetes or diabetes-related traits such as glucose intolerance. As an example, new ENU-induced dominant mutations in the key metabolic enzyme glucokinase (maturity onset diabetes of the young 2 [MODY2]) have recently been described (18,19) from large-scale dominant ENU mutagenesis screens. Brüning et al. (20) showed that double heterozygotes of null alleles of the insulin receptor and insulin receptor substrate-1 led to the development of overt diabetes in ∼40% of animals in contrast to single heterozygotes. Diabetes development was dependent on genetic background with C57BL/6J showing ∼85% transition to diabetes (21). We therefore decided to sensitize our ENU mutagenesis screen by using mice heterozygous for an insulin receptor knockout (22) maintained on a C57BL/6J background, which exhibits mild insulin resistance but not diabetes. Here, we describe two ENU-induced mutations in the transcription factor Sox4 that result in impaired glucose-stimulated insulin release leading to a mild impairment in glucose tolerance, and we also show that one of these in combination with haploinsufficiency of the insulin receptor and associated insulin resistance provide a novel digenic model that may have relevance to the study of polygenic type 2 diabetes.
The SRY-related high mobility group (HMG)-box (SOX) gene family is a group of important transcriptional regulators of a number of developmental processes, including the development of the endocrine pancreas (23). Mice homozygous for a null mutation of Sox4 showed abnormal pancreatic bud formation and endocrine cell differentiation, and cultured pancreatic explants failed to form normal islets. In wild-type animals, levels of Sox4 expression peaked at embryonic day 13 and persisted at high levels in adult islets. No role has yet been elucidated for Sox4 in the adult pancreas.
RESEARCH DESIGN AND METHODS
Animal husbandry.
Mice were kept in accordance with U.K. Home Office welfare guidelines and project license restrictions. Insulin receptor knockout mice (22) were obtained from The Jackson Laboratories (Bar Harbor, ME) and maintained as heterozygotes on a C57BL/6J background. MUD91 mice (a line carrying an ENU-induced Sox4 allele from a developmental abnormality screen) maintained on a mixed genetic background have been previously reported (24).
ENU mutagenesis.
Male mice heterozygous for the insulin receptor knockout mutation were treated with 2 × 100 mg/kg doses of ENU at 7-day intervals (12). Lines were maintained by back-crossing to C3H/HeH mice.
Genotyping.
Genomic DNA was extracted from mouse tail tissue using a DNeasy tissue kit (Qiagen, West Sussex, U.K.) according to the manufacturer's instructions. A genome scan was performed with a panel of 140 microsatellite markers spanning the mouse genome. Insulin receptor knockout mice were genotyped using a generic neo PCR (The Jackson Laboratories). Genotypes for both Sox4 alleles were confirmed by sequencing, and gene fragments were amplified using the following primers listed in a 5′-3′ orientation and sequenced: Y123C, GCAAGATCATGGAGCAGTC (SOX4F2) and GAGAACGATGCAGCCG (SOX4Rbio); and S70P, GAGCACTTCAGCGGTGA (SOX4F1) and GGAGATCTCGGCCGTTGT (SOX4R1).
Micro–magnetic resonance imaging.
Embryos were dissected at 14.5 days post coitum (dpc) into 37°C Hanks solution plus 5.0 mmol/l EDTA and allowed to exsanguinate. Embryos were then fixed in 4% paraformaldehyde in PBS plus 2 mmol/l dimeglumine gadopentetate (Gd-DPTA; Magnevist) for several days at 4°C. Before magnetic resonance imaging (MRI), the fixed embryos were embedded in 1% agarose in water with 2 mmol/l Gd-DPTA in 13-mm (high-resolution single-embryo imaging) glass tubes. MRI was performed at an experimental resolution of 9.7 × 9.7 × 12.7 m/voxel. MRI images were archived as TIFF files on DVD and analyzed using AMIRA 4.1 software.
Intraperitoneal glucose tolerance tests.
Mice were tested using the EMPReSS simplified intraperitoneal glucose tolerance test (IPGTT) (http://empress.har.mrc.ac.uk). Briefly, mice were fasted overnight, and a fasted blood sample was taken before intraperitoneal administration of a 2 g/kg glucose load. Subsequent blood samples were taken over time (60 and 120 min for glucose measurements or 10, 20, and 30 min for insulin measurements). Plasma glucose was measured using an Analox Glucose Analyser GM9. Plasma insulin was measured using a Mercodia Ultra-Sensitive Mouse ELISA kit (Mercodia, Uppsala, Sweden) according to the manufacturer's instructions.
INS1 and Min6 cell culture.
INS-1 cells (starting passage 30) were maintained in RPMI 1640 containing 5.5 mmol/l glucose (Gibco, Paisley, U.K.) and supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were incubated in humidified 5% CO2, 95% air at 37°C.
MIN6 cells (starting passage 30) were maintained in Dulbecco's modified Eagle's medium containing 25 mmol/l glucose and supplemented with 15% heat-inactivated FBS in humidified 5% CO2, 95% air at 37°C.
Islet isolation.
Mice were killed by cervical dislocation, the pancreases were removed, and islets were isolated by liberase digestion and handpicking (25). Cells were maintained in this medium at 37°C in a humidified atmosphere at 5% CO2 in air and used 1–2 days after the isolation.
Small interfering RNA.
Synthetic small interfering RNAs (siRNAs) were designed by Ambion and synthesized by both Ambion and Eurogentec. A total of four siRNAs were used: nonsense, glucokinase (Gck), and two Sox4 siRNAs. Each siRNA had a Cy3 tag on the 5′ end of the sense strand to enable visualization of transfected cells. siRNA duplexes (500 nmol/l) were transfected into INS1 and MIN6 cells using Lipofectamine 2000 (Invitrogen, Paisley, U.K.) at 100 μmol/l. Cells were cultured in RPMI 1640 (FBS free) for a further 24 h before experiment. siRNA sequences used listed in a 5′ to 3′ orientation as follows: Sox4(1), ′GGACAGCGACAAGAUUCCGtt′ (forward) and ′CGGAAUCUUGUCGCUGUCCtt′ (reverse); Sox4(2), ′GGAGAGGAGAGAGGGAAAAtt′ (forward) and ′UUUUCCCUCUCUCCUCUCCtg′ (reverse); Gck, ′ACGUAGGUGGGCAACAUCUtt′ (forward) and ′AGAUGUUGCCCACCUACGUtt′ (reverse); and Nonsense, ′GGUCUAGUCUUCGGUCUUGtt′ (forward) and ′CAAGACCGAAGACUAGACCtt′ (reverse).
Fura-2 calcium imaging.
INS1 cells were cultured on 35-mm Fluorodishes (World Precision Instruments) and incubated with 3 μmol/l Fura-2-AM (Molecular Probes) for 40 min at 37°C. They were imaged at room temperature (20–24°C) using an IonOptix fluorescence system (IonOptix, Boston, MA), with 340- and 380-nm dual excitation. The 510-nm emission ratio was collected at 1 Hz. Background subtraction was performed by measuring fluorescence from a cell-free region in the field of view. Cells were perfused continuously with extracellular solution containing 137 mmol/l NaCl, 5.6 mmol/l KCl, 2.6 mmol/l CaCl2, 1.2 mmol/l MgCl2, and 10 mmol/l HEPES (pH 7.4 with NaOH) plus glucose or tolbutamide as indicated. Only data from cells that responded to 200 μmol/l tolbutamide were analyzed.
Insulin secretion.
INS1 or Min6 cells (1 × 105) were re-plated, transfected with various siRNAs, and cultured for 24 h (as above) before being used in secretion assays. Islets were isolated from wild-type and mutant mice and cultured overnight (as described above) before insulin secretion was assessed. Insulin secretion per group of five islets was measured during 1-h static incubations in Krebs-Ringer buffer (118.5 mmol/l NaCl, 2.54 mmol/l CaCl2, 1.19 mmol/l KH2PO4, 4.74 mmol/l KCl, 25 mmol/l NaHCO3, 1.19 mmol/l MgSO4, and 10 mmol/l HEPES, pH 7.4) containing 0, 2, or 20 mmol/l glucose or 0.2 mmol/l tolbutamide in the presence of 2 mmol/l glucose. Each glucose concentration was replicated three times per individual. Samples of the supernatant were assayed for insulin. To determine total insulin content, insulin was extracted using 95:5 ethanol:acetic acid. Insulin was measured using an Ultra-Sensitive Mouse Insulin ELISA kit (Mecodia).
Quantitative RT-PCR.
Total RNA from isolated islets and siRNA-transfected INS1 and MIN6 cells was extracted using a RNeasy mini-kit (Qiagen). cDNA generated by Superscript II enzyme (Invitrogen) was analyzed by quantitative RT-PCR using the TaqMan system based on real-time detection of accumulated fluorescence (ABI Prism 7700; Perkin-Elmer). Gene expression was normalized relative to the expression of glyceraldehyde-3-phosphate dehydrogenase (Gapdh). Inventoried Taqman Probes with FAM tags were purchased from Applied Biosystems.
Samples were tested in triplicate, and results are expressed relative to Gapdh. The Student's t test was used to measure the significance between wild-type and Sox4 heterozygous islet cDNA (three biological replicates for each group) when amplified for Camk2b, Cdc42, Glut2, Glut4, Gpx-1, Ins1, Ins2, Kir6.2, NeuroD1, Nkx6.1, Pax6, PDX1, Prkci, Rab27, Sox4, Sur1, and Syt9 gene expression. Islet cDNA amplified for Glut4, Rab27, and Syt9 was first preamplified using a Taqman PreAmp Master Mix (Applied Biosystems). The Student's t test was also used to measure the significance between nonsense siRNA-transfected INS1 cells, Gck siRNA-transfected INS1 cells, and Sox4 siRNA-transfected INS1 cells when amplifying for either Gck or Sox4 gene expression.
Complementation test and embryonic phenotype analysis.
Males heterozygous for the Y123C Sox4 mutation were crossed to females heterozygous for the same mutation, the S70P mutation, and the Del(13)Svea36H deletion. Embryo dissections were carried out at 9.5, 13.5, and 14.5 dpc, and the embryonic phenotype was visually analyzed. Tissue was collected from individual embryos and the samples genotyped by sequencing as described above.
Electron microscopy.
After preparation (see above), isolated islets (30–40 per animal) were left to recover in pancreatic islet medium (hCell Technology) for 1 h at 37°C. Islets were then washed in PBS and fixed in cold 2.5% glutaraldehyde in PBS for 1 h at 4°C, after which they were washed twice in PBS. Osmium tetroxide at 1% in PBS was then added to the islets for 1 h at 4°C, followed by washing twice with PBS. The islets were then left in PBS at 4°C overnight. Finally, islets were dehydrated in increasing concentrations of ethanol (50, 70, 90, and 100%) for 5–10 min each and then embedded in increasing concentrations of resin (50% ethanol/50% Agar 100 resin mix [48% Agar 100 resin, 26% dodecenyl succinic anhydride, 26% methyl nadic anhydride; Agar Scientific], 25% ethanol/75% resin, and 100% resin) for 5–10 min each. Islets were left in 100% resin (resin plus 2% benzyl dimethylamine) at least overnight at room temperature before ultra-thin sections were cut for electron microscopy.
Islet immunohistochemistry.
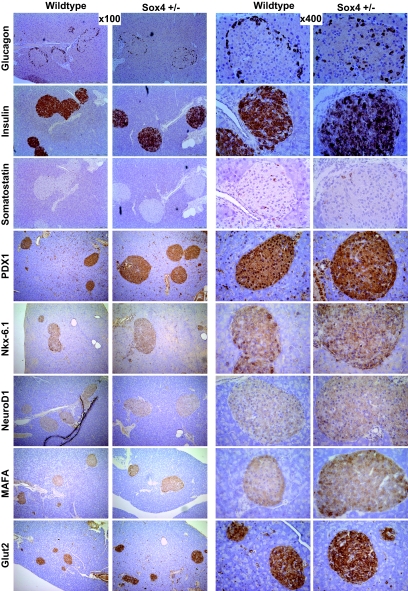
Immunohistochemistry was carried out on paraffin sections of 17-week-old pancreas. A rabbit or goat (depending on primary antibody) ABC staining system (Santa Cruz Biotechnology) was used according to manufacturer's instructions. The following primary antibodies were used: rabbit anti-insulin (1:50; Santa Cruz Biotechnology), rabbit anti-human glucagon (1:40; AbD Serotec), rabbit anti-somatostatin (2 μg/ml; Chemicon International), rabbit anti-PDX1 (1:500; Abcam), goat anti–Nkx-6.1 (1:50; Santa Cruz Biotechnology), goat anti-NeuroD (1:50; Santa Cruz Biotechnology), rabbit anti-MafA (1:500; Bethyl Laboratory), and rabbit anti-Glut2 (1:2,000; Chemicon International). Sections were counterstained with Gill's formulation no. 2 hematoxylin.
Islet area analysis.
The pancreas from each mouse was fixed in neutral buffered formaldehyde (Surgipath Europe) and mounted in wax longitudinally using the spleen for orientation. Serial sections were cut from each pancreas, and every 20th section was stained with hematoxylin-eosin (H-E) (mounting, cutting, and staining was carried out by Medical Solutions, Nottingham, U.K.). Ten H-E–stained sections from each mouse were photographed completely, and islet area calculated using Adobe Photoshop to measure islet area and total pancreas section area in each image.
RESULTS
Sensitized ENU mutagenesis screen.
Haploinsufficiency of the insulin receptor leading to insulin resistance but not diabetes was used for a sensitized ENU phenotype-driven screen. Male INSR+/− mice heterozygous for the insulin receptor knockout (insulin receptor+/−) on a C57BL/6J genetic background were treated with two doses of 100 mg/kg ENU separated by 7 days. Upon regaining fertility, they were mated with C3H/HeH females, and up to 50 F1 offspring per male were generated. A total of 789 male F1 offspring heterozygous for the insulin receptor knockout mutation were assayed for glucose tolerance at 12 and 24 weeks of age by IPGTT. Fourteen male mice were identified with impaired glucose tolerance and were tested for inheritance of the phenotype by backcrossing to C3H/HeH females (impaired glucose tolerance [IGT] lines 1–14). Five phenotypes were confirmed as inherited in the first backcross generation (data not shown).
Mapping line IGT4 and identification of a mutation.
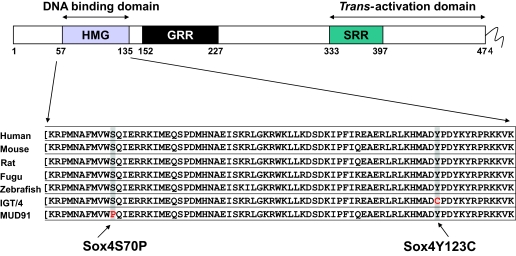
One of the lines clearly associated with an impaired glucose tolerance phenotype was IGT4. The causative mutation was initially mapped to chromosome 13 using 16 backcross animals and a panel of 140 microsatellite markers. An additional 67 second-backcross animals refined the candidate region to 5.4 Mb (25.9–31.3 Mbp). Analysis of the candidate region using the Ensembl database (http://www.ensembl.org/Mus_musculus/index.html Build 27) identified 39 genes, including a large cluster of prolactin-like genes that were excluded from analysis because they are unlikely candidates for a glucose tolerance phenotype. The remaining eight genes were sequenced in the F1 founder mouse to identify the ENU-induced causative mutation. A single A to G mutation was found at nucleotide position 1,025 (A1025G), resulting in a tyrosine to cysteine substitution at amino acid 123 (Y123C), which lies in the highly conserved HMG-binding domain of the transcription factor Sox4 (Fig. 1).
FIG. 1.
Schematic of Sox4 gene structure and position of Sox4 mutations within the highly conserved HMG DNA binding domain.
Sox4 homozygous mutant mice have an embryonic-lethal phenotype.
Embryos homozygous for the IGT4 Sox4 Y123C mutation die in utero at 14.5 dpc. An additional mutant allele of Sox4 (MUD91) carrying a T to C transition at nucleotide 869 (T869C), resulting in a serine to proline substitution at amino acid 70 (S70P), was available from a developmental phenotype screen at Harwell (24). Both mutations alter highly conserved amino acids within the HMG binding domain of Sox4 and are predicted to abolish the DNA binding ability of the mutant proteins (Fig. 1). Embryos homozygous for Mud91 Sox4 S70P die in utero at 14.5 dpc because of circulatory failure (24).
Analysis of the embryonic phenotype shows no difference between embryos homozygous for either of the two alleles. We therefore decided to test whether the two mutations complement each other by breeding compound heterozygotes. We found that Y123C/S70P mice exhibit the same embryonic phenotype as the homozygotes for either mutation (Table 1). We also tested the IGT4 Sox4 Y123C mutation by breeding it in trans with a deletion that spans the Sox4 gene (Del(13)Svea36H) and again observed embryonic lethality indistinguishable from that of the two homozygous Sox4 mutants. The Mud91 Sox4 S70P allele in trans with a deletion that spans the Sox4 gene (Del(13)Svea36H) was previously shown to be embryonic lethal (26).
TABLE 1.
Result of embryonic phenotype analysis and complementation tests
| Heterozygous cross | Stage (dpc) | Affected embryos (two mutant alleles) | Unaffected embryos (two mutant alleles) | Unaffected embryos (wild-type alleles) |
|---|---|---|---|---|
| S70P X Y123C | 9.5 | 1 | 1 | 9 |
| S70P X Y123C | 13.5 | 5 | 1 | 18 |
| Y123C X Y123C | 9.5 | 2 | 2 | 16 |
| Y123C X Y123C | 14.5 | 2 | 0 | 3 |
| Del X Y123C | 13.5 | 2 | 1 | 5 |
Data are n. Males heterozygous for the Y123C Sox4 mutation were crossed to females heterozygous for the same mutation, the S70P mutation, and the Del(13)Svea36H deletion. Embryo dissections were carried out at 9.5, 13.5, and 14.5 dpc, and the embryonic phenotype was visually analyzed.
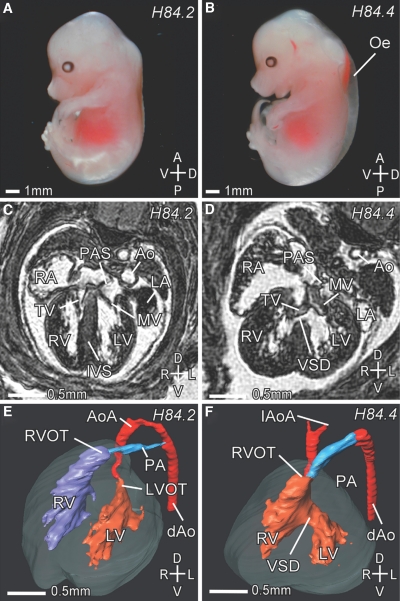
To further ascertain the nature of the IGT4 cardiac phenotype, we analyzed IGT4 Sox4 Y123C/Del(13)Svea36H embryos at 14.5 dpc by micro-MRI (μMRI). IGT4 Sox4 Y123C/Del(13)Svea36H embryos showed external edema (Fig. 2). Cardiac defects observed by μMRI included an outlet ventricular septal defect, dysplastic atrioventricular valves, a double-outlet right ventricle, and a hypoplastic or interrupted aortic arch (Fig. 2). Of the IGT4 Sox4 Y123C/Del(13)Svea36H embryos, 60% also showed a cleft palate, and on one occasion we found bilateral renal hypoplasia and bilateral hypoplasia of the thymus. Apart from a primum atrial septal defect, only found in the Mud91 Sox4 S70P, these MRI results are consistent with the findings in embryos homozygous for Mud91 Sox4 S70P (24). The results are a phenocopy of the range of cardiac defects previously detected in the targeted allele of Sox4, but additionally we detected dysplastic atrioventricular valves that have not been described in the Sox4 knockout embryos (27,28).
FIG. 2.
IGT4 embryos at 14.5 dpc. A: Wild-type embryo. B: Mutant IGT4 Sox4 Y123C/Del(13)Svea36H embryo showing severe edema. C and D: Transverse μMRI sections through a Sox4 wild-type (C) and mutant (D) embryo at the level of the AV valves at 14.5 dpc. C: In the wild-type embryo, the left (LV) and right (RV) ventricle are separated by the interventricular septum (IVS), and the left (LA) and right (RA) atrium are separated by the primary atrial septum (PAS). D: In a corresponding section of a mutant embryo, a ventricular septal defect (VSD) can be seen. The mitral valve (MV) and the tricuspidal valve (TV) are dysplastic. E and F: Three-dimensional reconstructions of a wild-type (E) and a mutant (F) heart at 14.5 dpc. In the wild-type heart (E), the aorta (Ao) originates from the left ventricle (LV), and the pulmonary artery (PA) originates from the right ventricle (RV). The left (LVOT) and right ventricular outflow tract (RVOT) are indicated. The PA connects to the descending aorta (dAo) via the ductus arteriosus. In the mutant (F), a ventricular septal defect (VSD) connects the LV and RV. The Ao and PA both originate from the RV (double-outlet right ventricle). The aortic arch (AoA) does not connect to the descending aorta (dAo), showing an interrupted aortic arch (IAoA). Note also the dilated PA. Axes: D, dorsal; V, ventral; A, anterior; P, posterior; L, left; R, right. (Please see http://dx.doi.org/10.2337/db07-0337 for a high-quality digital representation of this figure.)
The failure of these mutations to complement either each other or a deletion of the Sox4 gene demonstrates that IGT4 Sox4 Y123C mutation affects gene function. Moreover, the fact that the mutation gives the same embryonic phenotype when in trans with a deletion suggests that IGT4 Sox4 Y123C mutation is a null.
Sox4 heterozygous mutant mice show impaired glucose tolerance and reduced insulin secretion.
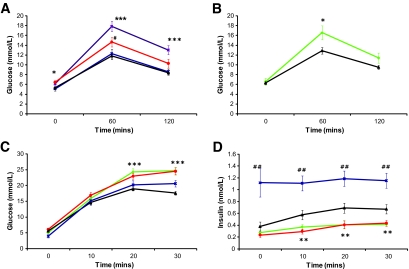
Glucose tolerance was assayed using an IPGTT with glucose measured at 0 min (T0), 60 min (T60), and 120 min (T120). Male mice heterozygous for both the insulin receptor knockout (insulin receptor+/−) and the IGT4 Sox4 Y123C mutant allele exhibited impaired glucose tolerance in an IPGTT test at 12 weeks of age (Fig. 3A). Glucose levels were significantly different from wild-type or insulin receptor+/− littermates at all three time points examined (T0, P < 0.05; T60, ≤0.001; and T120, ≤0.001). The existence of the Mud91 Sox4 S70P allele allowed us to test whether a different Sox4 mutation also affected glucose tolerance (Fig. 3B). Inheritance of either of the Sox4 mutant alleles (Y123C or S70P) alone and in the absence of the insulin receptor mutation increased glucose levels significantly compared with wild-type or insulin receptor+/− littermates 60 min after glucose administration (P < 0.05), with levels failing to return to normal levels in Y123C mice after 120 min (Fig. 3A and B). Glucose intolerance observed in two independently derived alleles of Sox4 confirms that the phenotype is caused by the identified mutations. In the case of the IGT4 Sox4 Y123C mutation, the presence of the insulin receptor+/− significantly increased blood glucose concentrations in an IPGTT, demonstrating a sensitizing effect of the insulin receptor+/− knockout (Fig. 3A). To investigate whether there was a defect in insulin secretion that might account for the glucose intolerance, we measured insulin secretion for both Sox4 alleles (Y123C or S70P). This was carried out in the absence of the insulin receptor+/− mutation using a 30-min glucose tolerance test with blood samples taken and glucose and insulin levels measured at T0, T10, T20, and T30 (Fig. 3C and D). Both lines showed significantly elevated glucose levels (Fig. 3C) at T20 and T30 (P < 0.001) and secreted significantly lower levels of insulin (Fig. 3D) in response to a glucose challenge compared with wild type at T10, T20, and T30 (P < 0.01).
FIG. 3.
Plasma glucose measured in IPGTT in male mice at 12 weeks of age. A: Animals heterozygous for the Sox4Y123C mutation show impaired glucose tolerance (#P < 0.05) compared with wild-type littermates. The phenotype is exacerbated in mice also carrying a heterozygous insulin receptor (IR) knockout. Insulin receptor and Sox4 Y123C, purple squares, n = 20; IR+/− only, blue stars, n = 21; Sox4 Y123C, red circles, n = 15; wild-type littermates, black triangles, n = 15. B: Animals heterozygous for the Sox4S70P mutation show impaired glucose tolerance compared with wild-type littermates. Sox4S70P, green diamonds, n = 15; compared with wild-type littermate, black triangles, n = 23. C: Both lines exhibit significantly higher plasma glucose concentrations measured at 20 and 30 min of an IPGTT. Sox4 Y123C, red circles, n = 12; Sox4S70P, green diamonds, n = 9; IR only, blue stars, n = 9; wild-type littermate, black triangle, n = 24. D: Insulin secretion measured during a 30-min IPGTT. Both lines show impaired glucose stimulated insulin release. Statistical significance of Sox4 Y123C mice versus wild-type littermates and Sox4S70P mice versus wild-type littermates is indicated by *P < 0.05, **P < 0.01, and ***P < 0.001 (Student's t test). Insulin secretion from IR+/− mice was significantly different at all time points (##P < 0.01). Wild-type littermates from S70P and Y123P lines were not statistically significantly different from each other and were pooled in C and D. All data are expressed as means ± SE.
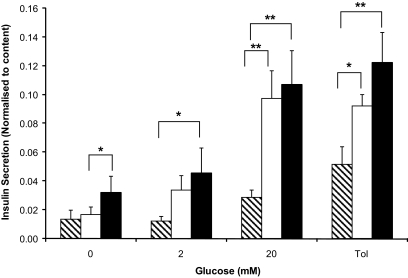
To further confirm a defect in insulin secretion, we measured glucose-stimulated insulin secretion in isolated islets from mice carrying the IGT4 Sox4 Y123C mutation (Fig. 4). Basal insulin secretion was the same as wild-type littermate control islets. However, incubation in 20 mmol/l glucose stimulated significantly less insulin secretion from mutant compared with wild-type littermate islets (P < 0.05). Furthermore, the response to the sulfonylurea tolbutamide was also significantly impaired, but not to the same extent as observed in 20 mmol/l glucose (Fig. 4). There was no significant difference in total insulin content between mutant and wild-type islets (Supplemental Fig. 1, available in the online appendix at http://dx.doi.org/10.2337/db07-0337). Because tolbutamide closes ATP-sensitive K+ channels (KATP channels) directly, this result suggests that the insulin secretory defect observed in Sox4 heterozygous mutant mice lies downstream of KATP channel closure. Insulin secretion in insulin receptor heterozygous controls was significantly higher at all points, consistent with increased in vivo insulin secretion (Fig. 3D). Additionally, insulin receptor islets showed reduced total insulin content (P < 0.001; Supplemental Fig. 1).
FIG. 4.
Insulin secretion from islets isolated from Sox4 Y123C mutant (black diagonal stripe bars), insulin receptor heterozygotes (black bars), and wild-type littermates (white bars) in response to glucose (0, 2, or 20 mmol/l) or 200 μmol/l tolbutamide (tol) normalized to total insulin content. Islets were isolated from five mice of each genotype. The data represent the mean of five animals (with three technical replicates per animal and each replicate comprising five islets ± SE). Statistical analysis by Student's t test, where *P < 0.05 and **P < 0.01.
Sox4 heterozygous animals form normal islets.
Pancreatic sections from Sox4-heterozygous animals and wild-type littermates were examined for differences in both islet mass and architecture. There was no significant difference between wild-type and heterozygous IGT4 Sox4 Y123C mice in their percentage of islet areas (the percentage of a histological section identified as islet; Table 2). Histological staining showed no qualitative difference in islet architecture in terms of arrangement or relative numbers of α-, β-, or δ-cells (Fig. 5). Transmission electron microscopy of islets from Sox4-heterozygous animals and wild-type littermates at 12 weeks of age showed no obvious differences (Supplemental Fig. 2). Quantitative RT-PCR analysis of islet RNA showed an increase in Sox4 expression that just reaches significance (P = 0.045) and no difference in expression levels of a number of key β-cell target genes, including insulin (Supplemental Fig. 3).
TABLE 2.
Pancreatic islet area in 17-week-old male mice
| Genotype | Total sectional area* | Total islet area* | Islets (n) | Islet area (%) |
|---|---|---|---|---|
| Sox4 Y123C | 6,707,744 | 76,493 | 173 | 1.14 |
| Sox4 Y123C | 4,582,464 | 56,548 | 124 | 1.23 |
| Sox4 Y123C | 7,077,281 | 141,769 | 199 | 2.00 |
| Total | 18,367,489 | 274,810 | 496 | 1.50 |
| Wild-type littermate | 6,960,257 | 138,588 | 245 | 1.99 |
| Wild-type littermate | 6,163,259 | 58,314 | 170 | 0.95 |
| Wild-type littermate | 5,752,898 | 97,481 | 151 | 1.69 |
| Total | 18,876,414 | 294,383 | 566 | 1.56 |
Data are n and percent. The pancreas was mounted longitudinally and serially sectioned with every 20th section stained with H-E, resulting in 10 sections from each of 3 mutant mice (Sox4Y123C) and 3 wild-type mice (60 whole sections).
Arbitrary units based on pixel values.
FIG. 5.
Immunohistochemistry on paraffin sections of 17-week-old pancreas. Images show wild-type and Sox4 heterozygous animals stained with insulin, glucagon, somatostatin, PDX1, Nkx-6.1, NeuroD1, MafA, and Glut-2 at ×100 and ×400 magnification. Sections were counterstained with Gill's formulation no. 2 hematoxylin. (Please see http://dx.doi.org/10.2337/db07-0337 for a high-quality digital representation of this figure.)
siRNA knockdown of Sox4 in INS1 cells.
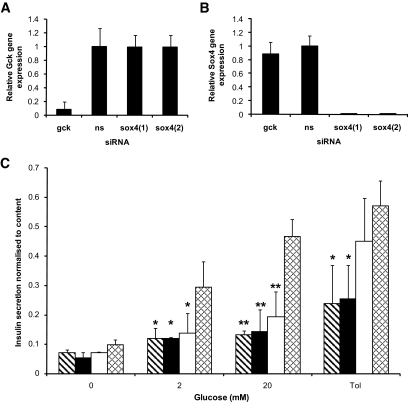
To further validate the in vivo and isolated islet data, we investigated the effect of loss of Sox4 gene expression in a rat insulin-secreting cell line INS1 using RNA interference (siRNA) to knock down the expression of the endogenous Sox4 gene. INS1 cells were transfected with one of four siRNA duplexes: nonsense (negative control), Gck (positive control), or two different Sox4-specific siRNAs [Sox4(1) and Sox4(2)]). All siRNAs were tagged with Cy3 fluorescent dye to enable transfected cells to be easily detected. Gene-specific knockdown of the corresponding RNAs was confirmed by quantitative real-time PCR. Expression of Gck and Sox4 mRNAs was abolished by siRNAs targeted against these genes, but neither was affected in INS1 cells transfected with nonsense siRNA (Fig. 6A and B). Basal insulin secretion was the same for all transfected cells (Fig. 6C), and siRNA knockdown had no effect on total insulin content compared with nonsense transfected cells (Supplemental Fig. 4). Nonsense siRNA-transfected INS1 cells responded to incubation in both 2 and 20 mmol/l glucose by secreting increasing amounts of insulin. In comparison, glucose-stimulated insulin secretion at 2 and 20 mmol/l glucose was significantly reduced for both nonoverlapping Sox4 siRNA duplexes and for the Gck-positive control compared with wild type (Fig. 6C). There was a significant reduction in insulin secretion from Sox4 siRNA-transfected INS1 cells in response to tolbutamide compared with nonsense siRNA-transfected cells. This was not observed with Gck-transfected cells (Fig. 6C). These results resemble those found for isolated Sox4 mutant islets (Fig. 4). These experiments have been repeated in the mouse insulinoma cell line Min6 with the same results (Supplemental Figs. 4–6).
FIG. 6.
A and B: Quantitative PCR of corresponding RNAs from gene-specific knockdowns in Ins1 cells. A: Gck gene expression is markedly reduced in cells transfected with Gck siRNA compared with nonsense (ns) or Sox4 siRNA–transfected cells. B: Sox4 gene expression is markedly reduced in cells transfected with Sox4 siRNA compared with nonsense or Gck siRNA–transfected cells. Data represent the mean of three experimental (i.e., biological) replicates with three technical replicates of each. C: Insulin secretion in response to glucose (0, 2, or 20 mmol/l) or 200 μmol/l tolbutamide (Tol) from INS1 cells transfected with Sox4 siRNA (black diagonal stripe bars, n = 6; and black bars, n = 6), Gck siRNA (white bars, n = 6), or nonsense siRNA (cross hatch bars, n = 6). The data are the mean of three separate experiments each of six replicates of 1 × 105 cells. Statistical analysis by Student's t test compared with nonsense siRNA control, where *P < 0.05 and **P < 0.01.
Intracellular calcium measurements.
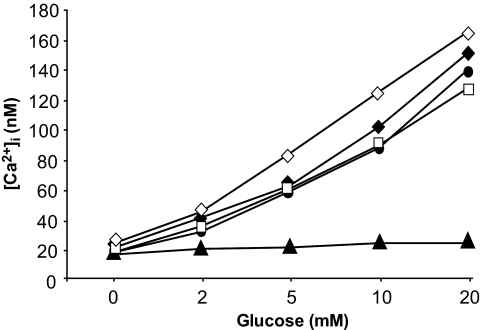
To further characterize the glucose-stimulated insulin secretory defect in Sox4 siRNA-transfected INS1 cells, we measured changes in intracellular calcium concentration [Ca2+]i in response to incubation in increasing glucose concentrations. In Sox4 (duplexes 1 and 2), nonsense, and mock-transfected cells, glucose concentrations >2 mmol/l elicited a rise in [Ca2+]i, and this was abolished, as expected, in cells transfected with Gck siRNA (Fig. 7). These data suggest that the secretory defect lies downstream of the KATP channels, calcium influx that follows channel closure, and membrane depolarization.
FIG. 7.
Mean change in [Ca2+]i in response to 2, 5, 10, and 20 mmol/l glucose for mock-transfected (unfilled diamond) INS1 cells or INS1 cells transfected with nonsense (unfilled square), Sox4(1) (filled diamond), Sox4(2) (filled circle), or Gck (filled triangle) siRNA. Each data point is the mean of 14–24 cells. The data are representative of two separate experiments.
DISCUSSION
Sox4 is a transcription factor that is expressed in the pancreas (29,23). Embryos null for Sox4 show normal pancreatic bud formation and endocrine cell differentiation up to 12.5 dpc (23). However, after this stage, they failed to form normal islets, and there was a lack of proper expansion of the endocrine cell population and particularly of β-cells (23). Breeding of compound heterozygotes (IGT4 Sox4 Y123C and Mud91 Sox4 S70P) and crossing both alleles with the chromosome deletion del36H confirms that both Sox4 mutations, as predicted, affect the function of the protein. The observation of glucose intolerance in the two independent heterozygous ENU alleles also confirms that the ENU mutation accounts for the phenotype, and the likelihood of other ENU mutations being present within the mapped region is very low (16). The role of Sox4 in the adult islet is unknown.
Random ENU mutagenesis has proven to be an effective tool in the identification of novel mouse models of human disease (12,13). In our sensitized approach, which used a mouse knockout exhibiting insulin resistance but not diabetes and a sensitive phenotypic assay (IPGTT), we have successfully identified a novel mouse model for impaired glucose tolerance and cloned the underlying gene. Mice heterozygous for either ENU mutated allele of Sox4 (Y123C or S70P) exhibited a statistically significant impairment in glucose tolerance at 12 weeks of age.
IPGTT testing of mice heterozygous for a null mutation of Sox4 has been reported to have nonsignificant differences in glucose tolerance at the 30-min time point of the test; however, there was a clear trend of increased blood glucose concentrations in heterozygotes at all of the IPGTT time points after fasting shown (23). It should be noted that the genetic background of these mice is different. Furthermore, Wilson et al. (23) carried out IPGTT tests using a 1 g/kg glucose challenge, lower than the 2 g/kg that we used, and this may have also contributed to the differences we observed.
Both ENU-induced mutations in Sox4 (S70P and Y123C) were significantly glucose intolerant because of impaired glucose-stimulated insulin secretion. This reduction in glucose-stimulated insulin release was also observed in isolated islets from IGT4 Sox4 Y123C mutant mice when compared with wild-type littermates. Knockdown of Sox4 in insulinoma cells with two different siRNAs targeted to separate regions of the gene abolished glucose-stimulated insulin release, as did siRNA knockdown of the key metabolic enzyme Gck. Changes in [Ca2+]i evoked by glucose, however, were normal in Sox4 mutant cells, indicating that KATP channels close normally. This suggests the insulin secretory defect lies downstream of calcium signal and may represent, for example, a defect in triggering exocytosis. Electron microscopy analysis, however, did not reveal any gross abnormalities in vesicles or their distribution.
Tolbutamide stimulates insulin secretion in pancreatic β-cells by closing KATP channels in the plasma membrane directly, leading to membrane depolarization and activation of voltage-gated calcium channels. This results in calcium influx and a rise in intracellular calcium ([Ca2+]i) that triggers insulin release. We observed a significant reduction in tolbutamide-stimulated insulin secretion both in isolated mutant islets and in Sox4 siRNA–treated INS1 or Min6 cells compared with wild-type cells, consistent with a defect downstream of the KATP channel. However, insulin secretion by mutant islets or siRNA-treated insulinoma cells was significantly more stimulated by tolbutamide than by 20 mmol/l glucose treatment (Figs. 4 and 6; Supplemental Fig. 5). It is known that at higher tolbutamide concentrations, there is stimulation of insulin exocytosis by direct interaction with the secretory machinery independently of closure of the plasma membrane KATP channel. This mechanism may explain why the inhibition of tolbutamide-stimulated insulin secretion by the Sox4 mutation is not as strong as that observed for glucose alone (30).
The glucose intolerance caused by the IGT4 Sox4 Y123C mutation is made worse by the presence of the heterozygous insulin receptor null mutation. This may in part be a contribution of insulin receptor haploinsufficiency to insulin resistance in key tissues, such as liver, adipose tissue, and muscle, that reduces glucose tolerance, further compounding lower glucose-stimulated insulin secretion. However, the insulin receptor is important in β-cell function, as shown by tissue-specific knockout of this gene that results in decreased acute insulin secretion and in impairment of glucose tolerance (31). Martinez et al. (32) have shown that the transcription factor Foxo1 is regulated in the β-cell by glucose but that this requires the insulin receptor. The Sox4 mutations outlined in this work have an effect on glucose-stimulated insulin secretion in the absence of the insulin receptor heterozygous null mutation, so although we cannot rule out a sensitizing effect in the β-cell when the insulin receptor+/− mutation is present, it is not a requirement for impairment. The IGT4 model in combination with insulin receptor+/− shows the effect of multiple mutations on glucose intolerance. This is important because it is thought that multiple variants in susceptibility genes contribute to impairment of glucose homeostasis in diabetes through the sum of multiple negative variants (33).
Interestingly, insulin receptor+/− heterozygotes secrete more insulin and have lower isolated islet insulin content. This may reflect the higher levels of secretion as a result of insulin resistance.
Sox proteins are dependent on other transcription factors as partner proteins for efficient gene activation (34,35). Specificity of Sox proteins are therefore reliant on different binding partners, which may explain the diverse roles in different tissues and developmental stages (36). Elucidation of the binding partner of Sox4 in adult islets may lead to the identification of the genes it regulates.
Sinner et al. (37) have recently reported that in colon carcinoma cells, there is an interaction between Sox4 and Sox17 with β-catenin and TCF4. They propose a role for these Sox genes in regulating Wnt signaling. Sox4 was shown to interact with either β-catenin or TCF4. In contrast, Sox17 bound cooperatively with both β-catenin and TCF4. It will be very interesting to determine whether these interactions are found in the β-cell, given the implication of TCF7L2 and Wnt signaling in insulin secretion (rev. in 38).
Type 2 diabetes is a multifactorial disease resulting from the interaction of genetic and environmental factors and develops as a result of both insulin resistance and defects in insulin secretion. We have clearly demonstrated a role for Sox4 in glucose-stimulated insulin release in adult pancreatic β-cells and in causing mild but significant glucose intolerance. It will be important to assess whether Sox4 has a role in contributing to type 2 diabetes in humans or to monogenic MODY type diabetes. Further characterization of the role of Sox4 in the adult β-cell will aid us in understanding the complex picture of transcriptional regulation in this important cell.
Supplementary Material
Acknowledgments
H.F. has received a studentship from the Medical Research Council. This work has received Diabetes U.K. Grant BDA:RD00/002072 and European Union FP6 Grant LHSM-CT-2003-503041.
We thank the staff of the Mary Lyon Centre for their excellent technical support. We thank Stuart Townsend (Medical Research Council, Harwell) and Judith Sheldon (Oxford Brookes University) for assistance with electron microscopy.
Published ahead of print at http://diabetes.diabetesjournals.org on 13 May 2008.
A.H. and H.F. contributed equally to this work.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.McCarthy MI: Progress in defining the molecular basis of type 2 diabetes mellitus through susceptibility-gene identification. Hum Mol Genet 13: R33–R41, 2004 [DOI] [PubMed] [Google Scholar]
- 2.Barroso I: Genetics of type 2 diabetes. Diabet Med 22: 517–535, 2005 [DOI] [PubMed] [Google Scholar]
- 3.Florez JC, Hirschhorn J, Altshuler D: The inherited basis of diabetes mellitus: implications for the genetic analysis of complex traits. Annu Rev Genomics Hum Genet 4: 257–291, 2003 [DOI] [PubMed] [Google Scholar]
- 4.O'Rahilly S, Barroso I, Wareham NJ: Genetic factors in type 2 diabetes: the end of the beginning? Science 307: 370–373, 2005 [DOI] [PubMed] [Google Scholar]
- 5.Lyssenko V, Almgren P, Anevski D, Orho-Melander M, Sjogren M, Saloranta C, Tuomi T, Groop L, the Botnia Study Group: Genetic prediction of future type 2 diabetes. PloS Med 2: e345, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turner MD, Cassell PG, Hitman GA: Calpain-10: from genome search to function. Diabete Metab Res Rev 21: 505–514, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Freeman H, Cox RD: Type-2 diabetes: a cocktail of genetic discovery. Hum Mol Genet 15: R202–R209, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P: A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445: 881–885, 2007 [DOI] [PubMed] [Google Scholar]
- 9.Toye AA, Lippiat J, Proks P, Shimomura K, Bentley L, Hugill A, Mijat V, Goldsworthy M, Moir L, Haynes A, Quarterman J, Freeman HC, Ashcroft FM, Cox RD: A genetic and physiological study of impaired glucose homeostasis control in C57BL/6J mice. Diabetologia 48: 675–686, 2005 [DOI] [PubMed] [Google Scholar]
- 10.Freeman H, Shimomura K, Horner E, Cox RD, Ashcroft FM: Nicotinamide nucleotide transhydrogenase: a key role in insulin secretion. Cell Metab 3: 35–45, 2006 [DOI] [PubMed] [Google Scholar]
- 11.Freeman HC, Hugill A, Dear NT, Ashcroft FM, Cox RD: Deletion of nicotinamide nucleotide transhydrogenase: a new quantitative trait locus accounting for glucose intolerance in C57BL/6J mice. Diabetes 55: 2135–2136, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Nolan PM, Peters J, Strivens M, Rogers D, Hagan J, Spurr N, Gray IC, Vizor L, Brooker D, Whitehill E, Washbourne R, Hough T, Greenaway S, Hewitt M, Liu X, McCormack S, Pickford K, Selley R, Wells C, Tymowska-Lalanne Z, Roby P, Glenister P, Thornton C, Thaung C, Stevenson JA, Arkell R, Mburu P, Hardisty R, Kiernan A, Erven A, Steel KP, Voegeling S, Guenet JL, Nickols C, Sadri R, Nasse M, Isaacs A, Davies K, Browne M, Fisher EM, Martin J, Rastan S, Brown SD, Hunter J: A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nat Genet 25: 440–443, 2000 [DOI] [PubMed] [Google Scholar]
- 13.Hrabe de Angelis MH, Flaswinkel H, Fuchs H, Rathkolb B, Soewarto D, Marschall S, Heffner S, Pargent W, Wuensch K, Jung M, Reis A, Richter T, Alessandrini F, Jakob T, Fuchs E, Kolb H, Kremmer E, Schaeble K, Rollinski B, Roscher A, Peters C, Meitinger T, Strom T, Steckler T, Holsboer F, Klopstock T, Gekeler F, Schindewolf C, Jung T, Avraham K, Behrendt H, Ring J, Zimmer A, Schughart K, Pfeffer K, Wolf E, Balling R: Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat Genet 25: 444–447, 2000 [DOI] [PubMed] [Google Scholar]
- 14.Coghill EL, Hugill A, Parkinson N, Davison C, Glenister P, Clements S, Hunter J, Cox RD, Brown SD: A gene-driven approach to the identification of ENU mutants in the mouse. Nat Genet 30: 255–256, 2002 [DOI] [PubMed] [Google Scholar]
- 15.Quwailid MM, Hugill A, Dear N, Vizor L, Wells S, Horner E, Fuller S, Weedon J, McMath H, Woodman P, Edwards D, Campbell D, Rodger S, Carey J, Roberts A, Glenister P, Lalanne Z, Parkinson N, Coghill EL, McKeone R, Cox S, Willan J, Greenfield A, Keays D, Brady S, Spurr N, Gray I, Hunter J, Brown SD, Cox RD: A gene-driven ENU-based approach to generating an allelic series in any gene. Mamm Genome 15: 585–591,2004 [DOI] [PubMed]
- 16.Keays DA, Clark TG, Flint J: Estimating the number of coding mutations in genotypic- and phenotypic-driven N-ethyl-N-nitrosourea (ENU) screens. Mamm Genome 17: 230–238, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Justice MJ, Noveroske J, Weber JS, Zheng B, Bradley A: Mouse ENU mutagenesis. Hum Mol Genet 8: 1955–1963, 1999 [DOI] [PubMed] [Google Scholar]
- 18.Toye AA, Moir L, Hugill A, Bentley E, Quaterman J, Mijat V, Hough T, Goldsworthy M, Haynes A, Hunter JA, Browne M, Spurr N, Cox RD: A new mouse model of type 2 diabetes, produced by N-ethyl-nitrosourea mutagenesis, is the result of a missense mutation in the glucokinase gene. Diabetes 53: 1577–1583, 2004 [DOI] [PubMed] [Google Scholar]
- 19.Inoue M, Sakuraba Y, Motegi H, Kubota N, Toki H, Matsui J, Toyoda Y, Miwa I, Terauchi Y, Kadowaki T, Shigeyama Y, Kasuga M, Adachi T, Fujimoto N, Matsumoto R, Tsuchihashi K, Kagami T, Inoue A, Kaneda H, Ishijima J, Masuya H, Suzuki T, Wakana S, Gondo Y, Minowa O, Shiroishi T, Noda T: A series of maturity onset diabetes of the young, type 2 (MODY2) mouse models generated by a large-scale ENU mutagenesis program. Hum Mol Genet 13: 1147–1157, 2004 [DOI] [PubMed] [Google Scholar]
- 20.Brüning JC, Winnay J, Bonner-Weir S, Taylor SI, Accili D, Kahn CR: Development of a novel polygenic model of NIDDM in mice heterozygous for IR and IRS-1 null alleles. Cell 88: 561–572, 1997 [DOI] [PubMed] [Google Scholar]
- 21.Kulkarni RN, Almind K, Goren HJ, Winnay JN, Ueki K, Okada T, Kahn CR: Impact of genetic background on development of hyperinsulinemia and diabetes in insulin receptor/insulin receptor substrate-1 double heterozygous mice. Diabetes 52: 1528–1534, 2003 [DOI] [PubMed] [Google Scholar]
- 22.Accili D, Drago J, Lee EJ, Johnson MD, Cool MH, Salvatore P, Asico LD, Jose PA, Taylor SI, Westphal H: Early neonatal death in mice homozygous for a null allele of the insulin receptor gene. Nat Genet 12: 106–109, 1996 [DOI] [PubMed] [Google Scholar]
- 23.Wilson ME, Yang KY, Kalousova A, Lau J, Kosaka Y, Lynn FC, Wang J, Mrejen C, Episkopou V, Clevers HC, German MS: The HMG box transcription factor Sox4 contributes to the development of the endocrine pancreas. Diabetes 54: 3402–3409, 2005 [DOI] [PubMed] [Google Scholar]
- 24.Bogani D, Willoughby C, Davies J, Kaur K, Mirza G, Paudyal A, Haines H, McKeone R, Cadman M, Pieles G, Schneider JE, Bhattacharya S, Hardy A, Nolan PM, Tripodis N, Depew MJ, Chandrasekara R, Duncan G, Sharpe PT, Greenfield A, Denny P, Brown SD, Ragoussis J, Arkell RM: Dissecting the genetic complexity of human 6p deletion syndromes by using a region-specific, phenotype-driven mouse screen. Proc Natl Acad Sci 102: 12477–12482, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rorsman P, Trube G: Glucose dependent K+-channels in pancreatic beta-cells are regulated by intracellular ATP. Pflugers Arch 405: 304–309, 1985 [DOI] [PubMed] [Google Scholar]
- 26.Arkell RM, Cadman M, Marsland T, Southwell A, Thaung C, Davies JR, Clay T, Beechey CV, Evans EP, Strivens MA, Brown SD, Denny P: Genetic, physical, and phenotypic characterization of the Del(13)Svea36H mouse. Mamm Genome 12: 687–694, 2001 [DOI] [PubMed] [Google Scholar]
- 27.Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PAJ, van de Wetering M, Verbeek S, Lamers WH, Kruisbeek AM, Cumanoparallel A, Clevers H: Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature 380: 711–714, 1996 [DOI] [PubMed] [Google Scholar]
- 28.Ya J, Schilham MW, de Boer PA, Moorman AF, Clevers H, Lamers WH: Sox4-deficiency syndrome in mice is an animal model for common trunk. Circ Res 83: 986–994, 1998 [DOI] [PubMed] [Google Scholar]
- 29.Lioubinski O, Muller M, Wegner M, Sander M: Expression of Sox transcription factors in the developing mouse pancreas. Dev Dyn 227: 402–408, 2003 [DOI] [PubMed] [Google Scholar]
- 30.Eliasson L, Renstrom E, Ammala C, Berggren PO, Bertorello AM, Bokvist K, Chibalin A, Deeney JT, Flatt PR, Gabel J, Gromada J, Larsson O, Lindstrom P, Rhodes CJ, Rorsman P: PKC-dependent stimulation of exocytosis by sulfonylureas in pancreatic beta cells. Science 271: 813–815, 1996 [DOI] [PubMed] [Google Scholar]
- 31.Kulkarni RN, Brüning J, Winnay JN, Postic C, Magnuson MA, Kahn CR: Tissue-specific knockout of the insulin receptor in pancreatic beta cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell 96: 329–339, 1999 [DOI] [PubMed] [Google Scholar]
- 32.Martinez SC, Cras-Meneur C, Bernal-Mizrachi E, Permutt MA: Glucose regulates Foxo1 through insulin receptor signaling in the pancreatic islet β-cell. Diabetes 55: 1581–1591, 2006 [DOI] [PubMed] [Google Scholar]
- 33.Ashcroft F, Rorsman P: Type 2 diabetes mellitus: not quite exciting enough? Hum Mol Genet 13: R21–R31, 2004 [DOI] [PubMed] [Google Scholar]
- 34.Wegner M: From head to toes: the multiple facets of Sox proteins. Nucleic Acid Res 27: 1409–1420, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamachi Y, Uchikawa M, Kondoh H: Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet 16: 182–187, 2000 [DOI] [PubMed] [Google Scholar]
- 36.Wissmuller S, Kosian T, Wolf M, Finzsch M, Wegner M: The high-mobility-group domain of Sox proteins interacts with DNA-binding domains of many transcription factors. Nucleic Acid Res 34: 1735–1744, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinner D, Kordich JJ, Spence JR, Opoka R, Rankin S, Lin SC, Jonatan D, Zorn AM, Wells JM: Sox17 and Sox4 differentially regulate {beta}-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol 27): 7802–7815, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith U: TCF7L2 and type 2 diabetes: we WNT to know. Diabetologia 50: 5–7, 2007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.