Abstract
Animal cloning through somatic cell nuclear transfer is very inefficient, probably due to insufficient reprogramming of the donor nuclei, which in turn would cause the dysregulation of gene expression. X-Chromosome inactivation (XCI) is a multi-step epigenetic process utilized by mammals to achieve dosage compensation in females. Our aim was to determine if any dysregulation of X-linked genes, which would be indicative of unfaithful reprogramming of donor nuclei, was present in cloned pigs. Real time reverse transcription polymerase chain reaction (RT-PCR) was performed to quantify the transcript levels of five X-linked genes, XIST, TSIX, HPRT1, G6PD, ARAF1 and one autosomal gene, COL4A1 in major organs of neonatal deceased and surviving female cloned pigs and age-matched control pigs from conventional breeding. Aberrant expression level of these genes was prevalent in the neonatal deceased clones, while it was only moderate in cloned pigs that survived after birth. These results suggest a correlation between the viability of the clones and the normality of their gene expression and provide a possible explanation for the death of a large portion of cloned animals around birth.
Keywords: Gene expression, Porcine, X chromosome, nuclear transfer
Introduction
Since the creation of Dolly, the first mammal cloned from a differentiated adult somatic cell, production of live offspring through nuclear transfer (NT) of somatic cells has been successful in a variety of species, ranging from small laboratory mice to large domestic animals (Betthauser et al. 2000; Cibelli et al. 1998; Galli et al. 2003; Kato et al. 1998; Kubota et al. 2000; Onishi et al. 2000; Polejaeva et al. 2000; Senda et al. 2004; Shin et al. 2002; Wilmut et al. 1997). Somatic cell nuclear transfer revealed the extraordinary ability of the oocyte to reprogram the differentiated somatic genome to a totipotent state (Eggan et al. 2004; Humpherys et al. 2002). However, little is known about this reprogramming process, and the overall success rate of animal cloning using somatic cells remains very low across different animal species, likely due to insufficient epigenetic reprogramming. Faulty or incomplete epigenetic reprogramming has been observed in different stages of development (Cezar et al. 2003; Kang et al. 2001b; Mann et al. 2003). Even in cloned animals that survive to term, different aspects of epigenetic abnormalities are evident, although the majority of these animals appear to be healthy and normal (Archer et al. 2003; Li et al. 2004; Xue et al. 2002).
Pigs have been regarded as promising donors for xenotransplantation and attempts have been made to genetically modify the porcine genome through somatic cell NT in order to make the porcine organs immunologically more compatible to human (Dai et al. 2002; Kolber-Simonds et al. 2004; Lai et al. 2002; Ramsoondar et al. 2003). The production of pigs through NT has been particularly difficult, probably due to the inefficiencies of in vitro oocyte maturation and embryo culture, and the necessity for at least four good-quality embryos to establish a pregnancy. Consequently, few cloned offspring are available for comparison of gene expression to control pigs from conventional reproduction. To date, there has been only one report examining epigenetic reprogramming in full-term cloned pigs; Archer et al. found abnormal CpG methylation in a repetitive region in adult clones. Despite the availability of various studies on cloned mice and bovine calves, epigenetic reprogramming patterns found in one species may not be the same in others. For instance, the abnormal DNA demethylation patterns observed in cloned bovine and mouse embryos were absent in cloned porcine embryos (Kang et al. 2001b).
The mammalian X chromosome contains more than 1,000 genes and many are house-keeping genes (Ross et al. 2005). X chromosome inactivation (XCI) is a biological process utilized by mammals to ensure equal expression of X-linked genes between males and females (Lyon 1961). The inactive X chromosomes in the donor cell requires reactivation/reprogramming during NT and provides a very good marker for reprogramming studies. Interestingly, aberrant patterns of X chromosome inactivation were discovered in bovine clones (Xue et al. 2002), while in cloned mice both relatively normal (Eggan et al. 2000) and abnormal XCI patterns (Nolen et al. 2005; Senda et al. 2004) have been reported. In order to investigate the normality of XCI in cloned pigs, we selected five X-linked genes and one ubiquitous autosomal gene to compare their expression levels in major organs of cloned piglets and age-matched controls by quantitative real time RT-PCR. These genes are: 1) the untranslated X inactivation-specific transcript (XIST), which resides in the x-inactivation center (XIC) and serves as the master regulatory switch for X chromosome inactivation (Penny et al. 1996); 2) the noncoding antisense mRNA of XIST, TSIX (the reserve spelling of XIST), which regulates both imprinted and random XCI through modification of chromatin structure in the mouse (Lee 2000; Sado et al. 2005); 3) hypoxanthine guanine phosphoribosyltransferase 1 (HPRT1), a housekeeping gene on X-chromosome and its deficiency causes Lesch-Nyhan Syndrome in humans resulting symptoms such as gouty arthritis and kidney/bladder stones; 4) glucose-6-phosphate dehydrogenase (G6PD), an enzyme involved in the hexose monophosphate pathway and its deficiency in the embryonic tissues impairs the development of the placenta, causing the death of the embryo (Longo et al. 2002); 5) V-raf murine sarcoma 3611 viral oncogene homolog 1 (ARAF1), a member of the raf proto-oncogene superfamily, which encodes cytoplasmic protein serine/threonine kinases and plays important roles in cell growth and development (Lee et al. 1994); and 6) Alpha-1 type IV collagen (COL4A1), located on porcine Chromosome 11, when mutated causes vascular defects in mice followed by Porcencephaly in some mutants (Gould et al. 2005). We found severe dysregulated expression of these genes in newborn deceased cloned piglets and moderate dysregulation in the surviving clones when compared to their respective age-matched controls.
Materials and Methods
Piglets and sample collection
All pigs in this study were females and fell in two age groups. Group 1 included six cloned pigs that died at or shortly after birth, and three controls from conventional reproduction that were healthy at birth and sacrificed for tissue collection. Four of the six NT-derived piglets were transgenic with one allele of the porcine α1,3-galactosyltransferase (α1,3GT) gene experimentally disrupted in the donor cells. The experimental protocols for knocking out this gene and generating these piglets have been described previously (Dai et al. 2002). The gestation period of these pigs ranged from 115 to 121 days. Minor phenotypic abnormalities, such as curled toes, were observed in three of the six cloned newborn pigs. These phenotypes have been observed occasionally in cloned piglets, and are not necessarily lethal. Piglets with these kinds of minor abnormalities have survived post-weaning and appeared to develop normally. For sample collection, deceased piglets were weighed, and tissues from major organs (heart, lung, liver, kidney, brain, spleen) were collected. Group 2 included seven cloned pigs derived from the same donor cell line and five age-matched controls. All pigs in Group 2 were euthanized at one month of age for the collection of tissues from the same six major organs. Tissues were snap frozen in liquid nitrogen and stored at −80°C until analysis. Animal handling and experimentation were in accordance with the National Research Council's publication “Guide for the Care and Use of Laboratory Animals” and approved by Institutional Animal Care and Use Committees at University of Missouri-Columbia and an equivalent organization at PPL Therapeutics, Inc. (currently Revivicor, Inc., Blacksburg, VA).
RNA preparation and reverse transcription (RT)
Total RNA from dissected tissues was extracted using the RNeasy mini kit (Qiagen, Valencia, CA). RNA was then treated with the DNA-free Kit (Ambion, Austin, TX) to remove any possible DNA contamination. The complete removal of DNA contamination was confirmed by the absence of amplification of PCR products after 40 cycles using treated RNA as a template in the absence of RT. RNA was then quantified by absorbance at 260/280 nm using a spectrophotometer. Before RT, RNA quality was examined by ratios of A260/A280 (all above 1.85 and mostly above 1.90) and by gel electrophoresis for the presence of two clear ribosomal RNA bands. Only RNA samples that did not show signs of degradation were used in this study. For RT, 1 μg of total RNA was added to a 20 μl-reaction mixture containing random hexamers and reverse transcriptase (New England Biolabs, Beverly, MA). The reaction mixture was incubated at 25°C for 10 min followed by RT at 42°C for 1 h. The reverse transcriptase was then inactivated at 95°C for 5 min.
Validation of the comparative CT method for quantitative real time RT-PCR
Quantitative real time RT-PCR was performed using an ABI Prism 7000 Sequence Detection System (Applied Biosystems, Inc., Foster City, CA). Reactions were performed in triplicates in 96-well optical reaction plates. Each reaction contained cDNA reverse transcribed from 10 ng total RNA, 1X SYBR Green PCR master mix and 0.3 μM of each specific primers, which were designed using Primer Express software (ABI) and summarized in Table 1. The Tm for each primer pair was between 59 and 60°C and the thermal cycling conditions were as follows: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 15s at 95°C for denaturation and 1 min at 60°C for annealing and extension. The fluorescence intensity for each well of the PCR plate was automatically determined after every elongation step. The specificity of the PCR reaction was confirmed by a single peak in the dissociation curve (Fig. 1a) and also by a single band in agarose gel electrophoreses (data not shown). Data were acquired and analyzed by ABI prism SDS software (ABI). The 18S ribosomal RNA was used as an endogenous control throughout this study. The calibrator, which was included in every real time plate, was a mixture of RNA from all six organs.
Table 1.
Primers for real time RT-PCR
| Gene | Primer sequences (5′ to 3′) | Product size
(bp) |
Amplification efficiency |
|---|---|---|---|
| XIST | F: GAAGAGATGCTCCAGGCCAAT | 87 | 1.92 |
| R: AGGTGTTGCTGGCTGATGCT | |||
|
| |||
| HPRT11 | F: CGTCTTGCTCGAGATGTGATG | 98 | 1.91 |
| R: TCCAGCAGGTCAGCAAAGAA | |||
|
| |||
| G6PD | F: CCTCCTGCAGATGCTGTGTCT | 112 | 1.91 |
| R: CGCCTGCACCTCTGAGATG | |||
|
| |||
| ARAF1 | F: CGGGATGGCATGAGTGTCTAC | 108 | 1.98 |
| R: GACTGTCTTTCGCCCCTTGA | |||
|
| |||
| COL4A1 | F: CAGCAACGAACCCCAGAAAT | 120 | 1.89 |
| R: CAGCAAGAAGAGGCCAACAAG | |||
|
| |||
| 18S rRNA | F: GCCCGAAGCGTTTACTTTGA | 93 | 1.96 |
| R: CCGCGGTCCTATTCCATTATT | |||
F = Forward primer, R = Reverse primer.
Fig. 1.
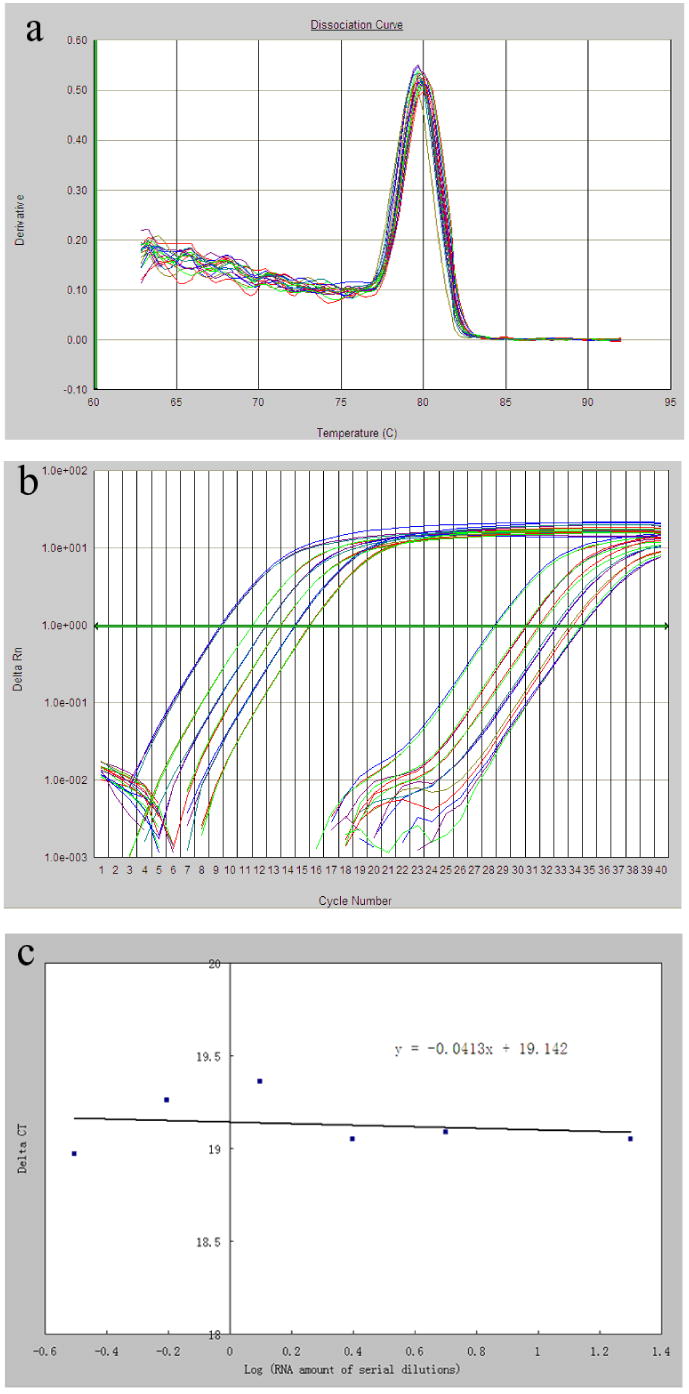
Primer validation for quantitative real time RT-PCR using the comparative CT method. a) A representative dissociation curve with a single peak, indicating the specificity of the amplification for the target gene (G6PD). b) A representative amplification plot of real time RT-PCR for the endogenous control, 18S rRNA, and the target gene (G6PD). The two sets of six curves for each gene (left set for 18S rRNA, right set for G6PD) represent serial cDNA dilutions of 20, 5, 2.5, 1.25, 0.625, 0.3125 ng (from left to right), respectively. The horizontal line in the middle was the threshold established for the calculation of the CT value. c) A representative linear regression between ΔCT, i.e. CT(18S rRNA)- CT(target gene) and the log value of cDNA amount. The absolute value of the slope (0.0413) was smaller than 0.1, demonstrating equal amplification efficiencies between the 18s rRNA (endogenous control) and the target gene (G6PD) within the cDNA dilutions tested.
In order to use the comparative CT method for quantification, also known as the ΔΔCT method, each pair of primers was validated for equal amplification efficiency to primers of the endogenous reference (18S rRNA) at a wide range of cDNA concentrations. Briefly, serial dilutions of cDNA from mixed RNA samples were made, real time RT-PCR reactions for both the 18S rRNA and each target gene was performed in triplicates at each cDNA dilution (Fig. 1b). CT, the threshold cycle, was determined after setting the threshold in the linear amplification phase of the PCR reaction and ΔCT for a particular gene was defined as CT(target gene) - CT(18S rRNA). As shown in Fig. 1c, a linear regression with a slope (absolute value) of less than 0.1 was obtained for each of the gene of interest, indicating that 18S rRNA and target genes had similar amplification efficiency which were close to 2.0 (Table 1), thus validating the quantitative nature of the RT-PCR reaction using the ΔΔCT method. The relative expression level of a target gene in a particular sample was calculated as: 2−ΔΔCT, where ΔΔCT = ΔCT(sample) - ΔCT(calibrator). The amplification efficiency was determined by the following formula: e=10-1/slope, where the slope is from the linear regression of the each gene's CT values over the log values of serial dilutions of cDNA from 20, 5, 2.5, 1.25, 0.625, 0.3125 ng mixed RNA.
Distinguishing transcripts of XIST and TSIX
Because XIST and TSIX are antisense mRNA for each other, a regular RT reaction, in which random hexamers are used for reverse transcription, would convert both transcripts to the same double-stranded cDNA molecule. Therefore, strand-specific primers were used during RT and this was followed by real time PCR to determine mathematically the ratio of XIST and TSIX. The gene-specific RT primers for XIST and TSIX were 5′-AGGTGTTGCTGGCTGATGCT-3′; and 5′- GAAGAGATGCTCCAGGCCAAT-3′, respectively. These primers were subjected to reverse-phase cartridge purification and RT was conducted using Endofree RT kit (Ambion, Austin, TX) per manufacture's instruction to reduce the endogenous priming by fragmented oligonucleotides.
Statistical analysis
All statistical analyses were performed using SAS/STAT software (Version 8.2, Cary, NC). All data were first examined for distribution normality and variance equality, the latter of which was tested by the folded form of the F statistic (F′) (Steel and Torrie 1980). Data with normal distribution and equal variance were subjected to pooled t-tests. Normally distributed data with unequal variance were subjected to the Cochran t-test. Data that were not normally distributed but have equal variance were tested using the Wilcoxon Rank Sum test. Data that were neither normally distributed nor equal in variance were log-transformed first and then tested as described above depending on their distribution and variance status. A p value of less than 0.05 was considered statistically significant.
Results
The mRNA levels of a total of five genes on the porcine X chromosome and one on the autosome were examined in our study and the results are summarized in Fig. 2 to Fig. 7. Comparisons of expression for each gene were made between controls and clones in a particular organ. For the neonate group, four comparisons were found to be significantly different between controls and clones among the 30 comparisons made. These include HPRT1 expression in lung, brain and spleen, and COL4A1 gene expression in kidney (Figs. 2a and 3a). While the other genes studied appear to have normal expression levels in all six organs. Interestingly, newborn deceased clones had lower levels of expression in all significant differences detected. When we compared the expression of each gene between controls and clones in the surviving group, five comparisons were found to be significantly different. All of these were due to up-regulation in gene expression in the clones, including the expression of XIST in heart, lung and spleen, and HPRT1 in lung and liver (Figs. 3b and 4b).
Fig. 2.
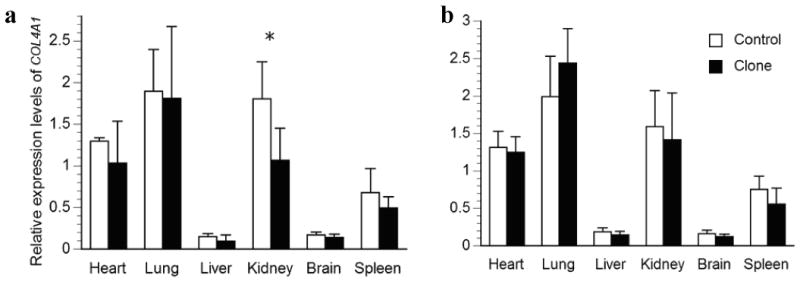
Relative transcription levels for the autosomal COL4A1 in major organs of a) deceased newborn clones or b) one-month-old surviving cloned piglets and their age-matched controls. The average transcription levels of each age group in the control piglets were normalized to 1. Bars with asterisks (*) are significantly different between clones and controls.
Fig. 7.
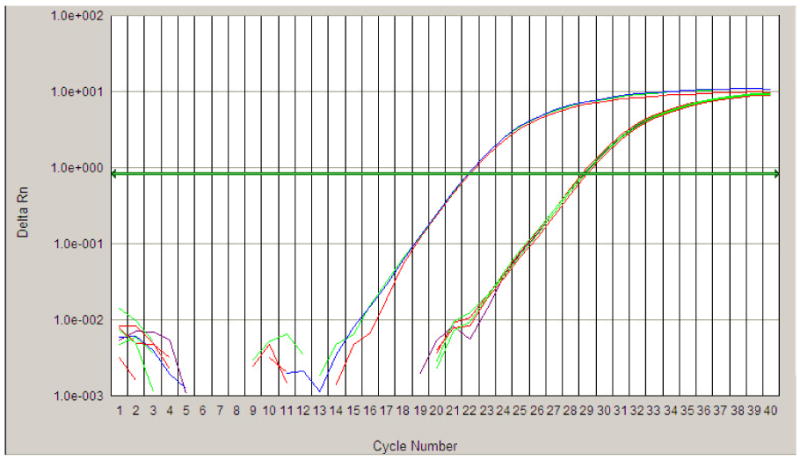
A quantitative real time amplification plot for the determination of the TSIX transcripts. The series of curves on the left represent triplicate amplifications of XIST using the XIST-specific primer. The series of curves on the right represent triplicate amplifications TSIX using either the TSIX-specific primer or no primers. The amplification curves for TSIX with and without primers overlapped, indicating a lack of TSIX transcripts in the samples. The same amount of total RNA was used in all amplifications in this figure.
Fig. 3.
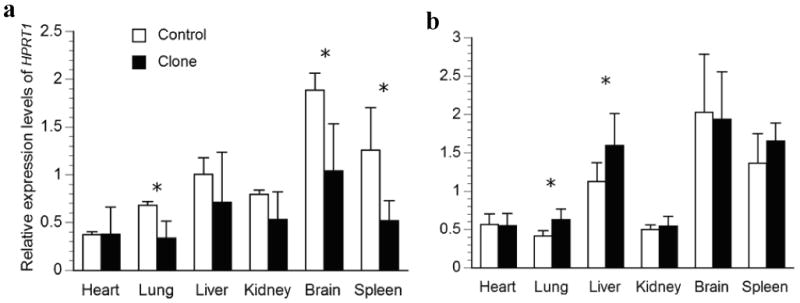
Relative transcription levels for HPRT1 in major organs of a) deceased newborn; b) one-month-old surviving cloned piglets and their age-matched controls. The average transcription levels of each age group in the control piglets were normalized to 1. Bars with asterisks (*) are significantly different between clones and controls.
Fig. 4.
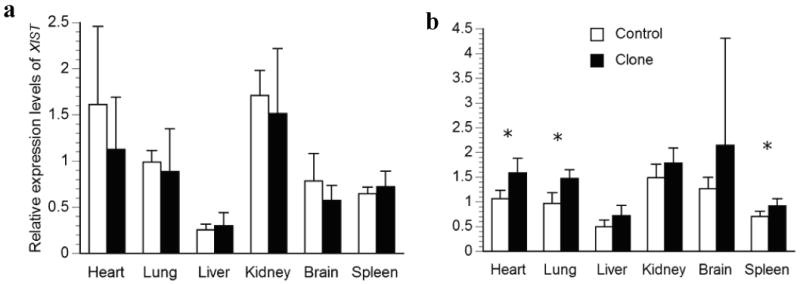
Relative transcription levels for XIST in major organs of a) deceased newborn; b) one-month-old surviving cloned piglets and their age-matched controls. The average transcription levels of each age group in the control piglets were normalized to 1. Bars with asterisks (*) are significantly different between clones and controls.
Relatively large variations in the expression levels of the genes studied were observed among different organs in newborns and this trend continued in one-month old pigs, regardless of their reproduction method. Interestingly, the most dramatic differences were found for the autosomal COL4A1 gene, which were high in lung and kidney but very low in liver and brain in both aged groups. For the X-linked genes, the liver seemed to have the lowest XIST levels, while the brain had the highest HPRT1 levels.
In addition, we tested homogeneity of variance and found six comparisons to have unequal variance in the clones when compared to the controls. These include COL4A1 expression in heart (Fig. 2a), HPRT1 expression in heart and kidney (Fig. 3a), XIST expression in brain (Fig. 4b), and G6PD expression in heart and kidney (Fig. 6a). Interestingly, larger variances were found in the clones for five out of six of these heterogeneous variances. When clones and controls were compared in groups, these large variances could mask dysregulation of individual clones and result in a lack of significant differences. Therefore, we used 2 standard deviations (2SD) of the expression level of controls as a cut-off to compare the cloned pigs individually, which gives us 95% confidence. Indeed, four genes, XIST, HPRT1, COL4A1, and G6PD, had more than 30% of parameters that varied more than 2SD of the control's values in the newborn deceased clones, while the remaining two genes studied were less variable (Table 2). On average, we found 36% of the 180 parameters (5 genes in each of 6 organs from 6 pigs = 180) determined in cloned newborn pigs that was dysregulated by at least 2SD of the control's values (Table 2). While in the live clones, the dysregulation was only moderate, 19% on average (Table 2). Interestingly, XIST appears to be the most deregulated gene in both the diseased newborn clones as well as clones at one-month of age, when these animals were analyzed individually.
Fig. 6.
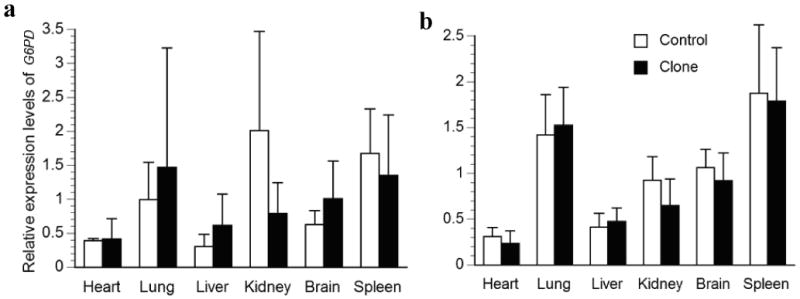
Relative transcription levels for G6PD in major organs of a) deceased newborn; b) one-month-old surviving cloned piglets and their age-matched controls. The average transcription levels of each age group in the control piglets were normalized to 1. Bars with asterisks (*) are significantly different between clones and controls.
Table 2.
Percentage of gene expression dysregulated by at least 2SD of controls' expression
| XIST | HPRT11 | G6PD | ARAF1 | COL4A1 | |
|---|---|---|---|---|---|
| Dead Clones (n=6) | 39% | 72% | 31% | 3% | 36% |
| Live Clones (n=7) | 45% | 17% | 12% | 17% | 2% |
To distinguish the XIST and TSIX transcripts, primers designed in the non-overlapping regions of these transcripts are necessary, as performed in the mouse and human (Lee, Migeon). Due to the lack of complete sequence information, we were unable to design such primers. Using strand-specific RT reactions should allow us to convert only the interested mRNA to cDNA. However, we were still able to the detect positive band by gel electrophoresis (data not shown), suggesting endogenous priming was not completely eliminated despite the use of a system to reduce it. In order to circumvent this problem, we combined strand-specific RT and real time PCR in order to mathematically calculate the ratios of XIST and TSIX expression. We conducted these reactions in RNA extracts from all tissues and similar results were observed. Therefore, we pooled RNA (20 ng) from all six organs for strand-specific RT and real time PCR and only presented results from the pooled sample (Fig. 7). We found that the CT values of RT-PCR with TSIX strand-specific primer and RT-PCR without primers (endogenous priming only) were the same. This indicated that there was no TSIX expression in these pig tissues at these ages.
Discussion
Abnormal expression of genes on the X-chromosome have been reported in cloned bovine embryos and full-term calves as well as in cloned mouse embryos (Wrenzycki et al. 2002; Xue et al. 2002). This is possibly due to the insufficient reprogramming of the donor cells and failure to reactivate/inactivate the X chromosome during embryonic development (Nolen et al. 2005). The present study is the first to investigate gene transcription levels in full-term piglets produced by somatic cell NT. Specifically, expression of five X-linked genes and one autosomal gene was studied. We found that while expression of these genes in surviving clones was largely within the normal range, abnormal expression was prevalent in deceased newborns.
Although porcine embryos from somatic cell nuclear transfer acquire a typical methylation pattern as control embryos and have normal telomere length (Jeon et al. 2005; Kang et al. 2001b), they display sporatic dysregulation of genes that are critical to embryo development (Lee et al. 2004; Miyazaki et al. 2005). This is consistent with the widely reported abnormal gene expression in mouse and bovine NT embryos (Bortvin et al. 2003; Daniels et al. 2000; Han et al. 2003; Mann et al. 2003; Wrenzycki et al. 2002; Wrenzycki et al. 2001). Different from the cloned pig embryos, however, bovine and murine cloned embryos also have abnormal DNA methylation patterns (Kang et al. 2001a), suggesting species differences in DNA methylation reprogramming. The wide-spread gene dysregulation in cloned embryos, regardless of species, may be a major contributor to the substantial loss of embryos throught pregenency. Because XCI occurs during early embryonic development, the abnormalities of X-linked gene expression observed in full-term piglets in the present study possibly persisted throughout fetal development, suggesting tolerence of these animals to dysregulated gene expression.
In the present study, we found that comparing clones to controls in groups is insufficient to discover the extent of expression anomalies. This is primarily due to the large degree of gene expression variation in the cloned animals, despite the fact that many of them were genetically identical to each other, for which a smaller variance would be expected. Using the 2SD of the normal expression as the cutoff, we found 65 out of 180 (36%) parameters in deceased newborns were abnormal vs. only 4 out of 30 (13%) parameters when clones and controls were compared in groups. In fact, the expression of all but one gene (ARAF1) in organs of individual deceased clones was found to exceed this threshold and was likely abnormal (Table 2). Similar expression abnormalities were found in DNA methylation in cloned pigs and deceased cloned bovine calves (Archer et al. 2003; Li et al., 2004; Yang et al., 2005). The same 2SD comparisons in the surviving clones revealed much less dysregulation (Table 2). This strongly suggests a correlation between the normality of gene expression and viability of the cloned piglets. Our finding was in accordance with a recent study in which the expression of several imprinted genes were found to be relatively normal in bovine clones that survived to adulthood compared to those that died at/after birth (Yang et al. 2005). Interestingly, all the above-mentioned studies, together with ours, reported more variation of gene expression in the cloned group, suggesting the universal random nature of nuclear reprogramming in different species.
We also noticed large variations in the expression levels of the same genes among different organs, regardless of how the animals were reproduced. This suggests that the expression of X-linked genes is not only regulated by XCI but also by their own individual promoters which may be subjected to tissue-specific stimulation/inhibition. The relatively consistent recapitulation of tissue specificity of gene expression in the cloned animals indicates that organ specific functions may be reprogrammed, despite the lack of complete reprogramming of XCI.
Finally, TSIX has been shown to repress XIST expression in cis and to regulate both imprinted and random X inactivation in mice (Lee 2000; Lee and Lu 1999). Unlike mice where TSIX expression is limited to undifferentiated cells, TSIX was reported to be present in human embryonic cell lines, fetal somatic cells and placental tissues, skin fibroblasts of 2-month old females (Chow et al. 2003; Migeon et al. 2001; Migeon et al. 2002), and even in adult bovine organs (Farazmand et al. 2004). These studies used TSIX-specific or strand-specific RT-PCR to distinguish the antisense transcripts of TSIX from XIST. Due to limited sequence information at the porcine XIST/TSIX locus, we were unable to design primers in non-overlapping regions of TSIX and XIST. The novel combination of strand-specific RT and quantitative real time PCR, however, allowed us to assertively conclude that TSIX was absent in our samples from newborn and one-month old pigs.
In summary, we document for the first time that cloned, deceased newborn pigs have prevalent abnormalities in the levels of X-linked genes, while clones that survived to one-month of age tend to have only moderate levels of expression abnormalities. Pigs do not express TSIX in major organs after birth.
Fig. 5.
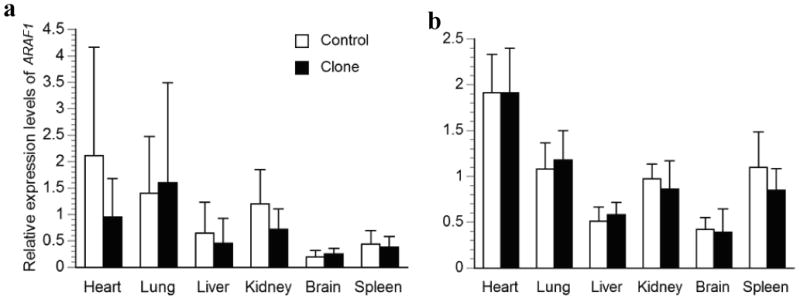
Relative transcription levels for ARAF1 in major organs of a) deceased newborn; b) one-month-old surviving cloned piglets and their age-matched controls. The average transcription levels of each age group in the control piglets were normalized to 1. Bars with asterisks (*) are significantly different between clones and controls.
Acknowledgments
The authors would like to thank Lan Yang for the technical support and helpful discussion; Marina Julian and Sadie L. Smith for the critical reading of the manuscript, support from the NIH via the National Center for Research Resources (RSP R01 RR013438)
Footnotes
Publisher's Disclaimer: This PDF receipt will only be used as the basis for generating PubMed Central (PMC) documents. PMC documents will be made available for review after conversion (approx. 2-3 weeks time). Any corrections that need to be made will be done at that time. No materials will be released to PMC without the approval of an author. Only the PMC documents will appear on PubMed Central -- this PDF Receipt will not appear on PubMed Central.
References
- Archer GS, Dindot S, Friend TH, Walker S, Zaunbrecher G, Lawhorn B, Piedrahita JA. Hierarchical Phenotypic and Epigenetic Variation in Cloned Swine. Biol Reprod. 2003;69(2):430–436. doi: 10.1095/biolreprod.103.016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betthauser J, Forsberg E, Augenstein M, Childs L, Eilertsen K, Enos J, Forsythe T, Golueke P, Jurgella G, Koppang R, Lesmeister T, Mallon K, Mell G, Misica P, Pace M, Pfister-Genskow M, Strelchenko N, Voelker G, Watt S, Thompson S, Bishop M. Production of cloned pigs from in vitro systems. Nat Biotechnol. 2000;18(10):1055–1059. doi: 10.1038/80242. [DOI] [PubMed] [Google Scholar]
- Bortvin A, Eggan K, Skaletsky H, Akutsu H, Berry DL, Yanagimachi R, Page DC, Jaenisch R. Incomplete reactivation of Oct4-related genes in mouse embryos cloned from somatic nuclei. Development. 2003;130(8):1673–1680. doi: 10.1242/dev.00366. [DOI] [PubMed] [Google Scholar]
- Cezar GG, Bartolomei MS, Forsberg EJ, First NL, Bishop MD, Eilertsen KJ. Genome-wide epigenetic alterations in cloned bovine fetuses. Biol Reprod. 2003;68(3):1009–1014. doi: 10.1095/biolreprod.102.010181. [DOI] [PubMed] [Google Scholar]
- Chow JC, Hall LL, Clemson CM, Lawrence JB, Brown CJ. Characterization of expression at the human XIST locus in somatic, embryonal carcinoma, and transgenic cell lines. Genomics. 2003;82(3):309–322. doi: 10.1016/s0888-7543(03)00170-8. [DOI] [PubMed] [Google Scholar]
- Cibelli JB, Stice SL, Golueke PJ, Kane JJ, Jerry J, Blackwell C, Ponce de Leon FA, Robl JM. Cloned transgenic calves produced from nonquiescent fetal fibroblasts. Science. 1998;280(5367):1256–1258. doi: 10.1126/science.280.5367.1256. [DOI] [PubMed] [Google Scholar]
- Dai Y, Vaught TD, Boone J, Chen SH, Phelps CJ, Ball S, Monahan JA, Jobst PM, McCreath KJ, Lamborn AE, Cowell-Lucero JL, Wells KD, Colman A, Polejaeva IA, Ayares DL. Targeted disruption of the alpha1,3-galactosyltransferase gene in cloned pigs. Nat Biotechnol. 2002;20(3):251–255. doi: 10.1038/nbt0302-251. [DOI] [PubMed] [Google Scholar]
- Daniels R, Hall V, Trounson AO. Analysis of Gene Transcription in Bovine Nuclear Transfer Embryos Reconstructed with Granulosa Cell Nuclei. Biol Reprod. 2000;63(4):1034–1040. doi: 10.1095/biolreprod63.4.1034. [DOI] [PubMed] [Google Scholar]
- Eggan K, Akutsu H, Hochedlinger K, Rideout W, 3rd, Yanagimachi R, Jaenisch R. X-Chromosome inactivation in cloned mouse embryos. Science. 2000;290(5496):1578–1581. doi: 10.1126/science.290.5496.1578. [DOI] [PubMed] [Google Scholar]
- Eggan K, Baldwin K, Tackett M, Osborne J, Gogos J, Chess A, Axel R, Jaenisch R. Mice cloned from olfactory sensory neurons. Nature. 2004;428(6978):44–49. doi: 10.1038/nature02375. [DOI] [PubMed] [Google Scholar]
- Farazmand A, Basrur PK, Stranzinger G, Graphodatskaya D, Reyes ER, King WA. Expression of XIST sense and antisense in bovine fetal organs and cell cultures. Chromosome Res. 2004;12(3):275–283. doi: 10.1023/b:chro.0000021864.66235.81. [DOI] [PubMed] [Google Scholar]
- Galli C, Lagutina I, Crotti G, Colleoni S, Turini P, Ponderato N, Duchi R, Lazzari G. Pregnancy: a cloned horse born to its dam twin. Nature. 2003;424(6949):635. doi: 10.1038/424635a. [DOI] [PubMed] [Google Scholar]
- Gould DB, Phalan FC, Breedveld GJ, van Mil SE, Smith RS, Schimenti JC, Aguglia U, van der Knaap MS, Heutink P, John SW. Mutations in Col4a1 cause perinatal cerebral hemorrhage and porencephaly. Science. 2005;308(5725):1167–1171. doi: 10.1126/science.1109418. [DOI] [PubMed] [Google Scholar]
- Han DW, Song SJ, Uhum SJ, Do JT, Kim NH, Chung KS, Lee HT. Expression of IGF2 and IGF receptor mRNA in bovine nuclear transferred embryos. Zygote. 2003;11(3):245–252. doi: 10.1017/s0967199403002296. [DOI] [PubMed] [Google Scholar]
- Humpherys D, Eggan K, Akutsu H, Friedman A, Hochedlinger K, Yanagimachi R, Lander ES, Golub TR, Jaenisch R. Abnormal gene expression in cloned mice derived from embryonic stem cell and cumulus cell nuclei. Proc Natl Acad Sci U S A. 2002;99(20):12889–12894. doi: 10.1073/pnas.192433399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon HY, Hyun SH, Lee GS, Kim HS, Kim S, Jeong YW, Kang SK, Lee BC, Han JY, Ahn C, Hwang WS. The analysis of telomere length and telomerase activity in cloned pigs and cows. Mol Reprod Dev. 2005;71(3):315–320. doi: 10.1002/mrd.20279. [DOI] [PubMed] [Google Scholar]
- Kang YK, Koo DB, Park JS, Choi YH, Chung AS, Lee KK, Han YM. Aberrant methylation of donor genome in cloned bovine embryos. Nat Genet. 2001a;28(2):173–177. doi: 10.1038/88903. [DOI] [PubMed] [Google Scholar]
- Kang YK, Koo DB, Park JS, Choi YH, Kim HN, Chang WK, Lee KK, Han YM. Typical demethylation events in cloned pig embryos. Clues on species-specific differences in epigenetic reprogramming of a cloned donor genome. J Biol Chem. 2001b;276(43):39980–39984. doi: 10.1074/jbc.M106516200. [DOI] [PubMed] [Google Scholar]
- Kato Y, Tani T, Sotomaru Y, Kurokawa K, Kato J, Doguchi H, Yasue H, Tsunoda Y. Eight calves cloned from somatic cells of a single adult. Science. 1998;282(5396):2095–2098. doi: 10.1126/science.282.5396.2095. [DOI] [PubMed] [Google Scholar]
- Kolber-Simonds D, Lai L, Watt SR, Denaro M, Arn S, Augenstein ML, Betthauser J, Carter DB, Greenstein JL, Hao Y, Im GS, Liu Z, Mell GD, Murphy CN, Park KW, Rieke A, Ryan DJ, Sachs DH, Forsberg EJ, Prather RS, Hawley RJ. Production of alpha-1,3-galactosyltransferase null pigs by means of nuclear transfer with fibroblasts bearing loss of heterozygosity mutations. Proc Natl Acad Sci U S A. 2004;101(19):7335–7340. doi: 10.1073/pnas.0307819101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota C, Yamakuchi H, Todoroki J, Mizoshita K, Tabara N, Barber M, Yang X. Six cloned calves produced from adult fibroblast cells after long-term culture. Proc Natl Acad Sci U S A. 2000;97(3):990–995. doi: 10.1073/pnas.97.3.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L, Kolber-Simonds D, Park KW, Cheong HT, Greenstein JL, Im GS, Samuel M, Bonk A, Rieke A, Day BN, Murphy CN, Carter DB, Hawley RJ, Prather RS. Production of alpha-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science. 2002;295(5557):1089–1092. doi: 10.1126/science.1068228. [DOI] [PubMed] [Google Scholar]
- Lee JE, Beck TW, Brennscheidt U, DeGennaro LJ, Rapp UR. The complete sequence and promoter activity of the human A-raf-1 gene (ARAF1) Genomics. 1994;20(1):43–55. doi: 10.1006/geno.1994.1125. [DOI] [PubMed] [Google Scholar]
- Lee JT. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell. 2000;103(1):17–27. doi: 10.1016/s0092-8674(00)00101-x. [DOI] [PubMed] [Google Scholar]
- Lee JT, Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 1999;99(1):47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- Lee SH, Kim DY, Nam DH, Hyun SH, Lee GS, Kim HS, Lee CK, Kang SK, Lee BC, Hwang WS. Role of messenger RNA expression of platelet activating factor and its receptor in porcine in vitro-fertilized and cloned embryo development. Biol Reprod. 2004;71(3):919–925. doi: 10.1095/biolreprod.103.026138. [DOI] [PubMed] [Google Scholar]
- Li S, Li Y, Du W, Zhang L, Yu S, Dai Y, Zhao C, Li N. Aberrant Gene Expression in Cloned Bovine of Neonatal Death. Biol Reprod. 2004 doi: 10.1095/biolreprod.104.029462. biolreprod.104.029462. [DOI] [PubMed] [Google Scholar]
- Longo L, Vanegas OC, Patel M, Rosti V, Li H, Waka J, Merghoub T, Pandolfi PP, Notaro R, Manova K, Luzzatto L. Maternally transmitted severe glucose 6-phosphate dehydrogenase deficiency is an embryonic lethal. Embo J. 2002;21(16):4229–4239. doi: 10.1093/emboj/cdf426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Naturwissenschaften. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Mann MR, Chung YG, Nolen LD, Verona RI, Latham KE, Bartolomei MS. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol Reprod. 2003;69(3):902–914. doi: 10.1095/biolreprod.103.017293. [DOI] [PubMed] [Google Scholar]
- Migeon BR, Chowdhury AK, Dunston JA, McIntosh I. Identification of TSIX, encoding an RNA antisense to human XIST, reveals differences from its murine counterpart: implications for X inactivation. Am J Hum Genet. 2001;69(5):951–960. doi: 10.1086/324022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migeon BR, Lee CH, Chowdhury AK, Carpenter H. Species differences in TSIX/Tsix reveal the roles of these genes in X-chromosome inactivation. Am J Hum Genet. 2002;71(2):286–293. doi: 10.1086/341605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K, Tomii R, Kurome M, Ueda H, Hirakawa K, Ueno S, Hiruma K, Nagashima H. Evaluation of the quality of porcine somatic cell nuclear transfer embryo by gene transcription profiles. J Reprod Dev. 2005;51(1):123–131. doi: 10.1262/jrd.51.123. [DOI] [PubMed] [Google Scholar]
- Nolen LD, Gao S, Han Z, Mann MR, Gie Chung Y, Otte AP, Bartolomei MS, Latham KE. X chromosome reactivation and regulation in cloned embryos. Dev Biol. 2005;279(2):525–540. doi: 10.1016/j.ydbio.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Onishi A, Iwamoto M, Akita T, Mikawa S, Takeda K, Awata T, Hanada H, Perry AC. Pig cloning by microinjection of fetal fibroblast nuclei. Science. 2000;289(5482):1188–1190. doi: 10.1126/science.289.5482.1188. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379(6561):131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Polejaeva IA, Chen SH, Vaught TD, Page RL, Mullins J, Ball S, Dai Y, Boone J, Walker S, Ayares DL, Colman A, Campbell KH. Cloned pigs produced by nuclear transfer from adult somatic cells. Nature. 2000;407(6800):86–90. doi: 10.1038/35024082. [DOI] [PubMed] [Google Scholar]
- Ramsoondar JJ, Machaty Z, Costa C, Williams BL, Fodor WL, Bondioli KR. Production of alpha 1,3-galactosyltransferase-knockout cloned pigs expressing human alpha 1,2-fucosylosyltransferase. Biol Reprod. 2003;69(2):437–445. doi: 10.1095/biolreprod.102.014647. [DOI] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, Frankish A, Lovell FL, Howe KL, Ashurst JL, Fulton RS, Sudbrak R, Wen G, Jones MC, Hurles ME, Andrews TD, Scott CE, Searle S, Ramser J, Whittaker A, Deadman R, Carter NP, Hunt SE, Chen R, Cree A, Gunaratne P, Havlak P, Hodgson A, Metzker ML, Richards S, Scott G, Steffen D, Sodergren E, Wheeler DA, Worley KC, Ainscough R, Ambrose KD, Ansari-Lari MA, Aradhya S, Ashwell RI, Babbage AK, Bagguley CL, Ballabio A, Banerjee R, Barker GE, Barlow KF, Barrett IP, Bates KN, Beare DM, Beasley H, Beasley O, Beck A, Bethel G, Blechschmidt K, Brady N, Bray-Allen S, Bridgeman AM, Brown AJ, Brown MJ, Bonnin D, Bruford EA, Buhay C, Burch P, Burford D, Burgess J, Burrill W, Burton J, Bye JM, Carder C, Carrel L, Chako J, Chapman JC, Chavez D, Chen E, Chen G, Chen Y, Chen Z, Chinault C, Ciccodicola A, Clark SY, Clarke G, Clee CM, Clegg S, Clerc-Blankenburg K, Clifford K, Cobley V, Cole CG, Conquer JS, Corby N, Connor RE, David R, Davies J, Davis C, Davis J, Delgado O, Deshazo D, Dhami P, Ding Y, Dinh H, Dodsworth S, Draper H, Dugan-Rocha S, Dunham A, Dunn M, Durbin KJ, Dutta I, Eades T, Ellwood M, Emery-Cohen A, Errington H, Evans KL, Faulkner L, Francis F, Frankland J, Fraser AE, Galgoczy P, Gilbert J, Gill R, Glockner G, Gregory SG, Gribble S, Griffiths C, Grocock R, Gu Y, Gwilliam R, Hamilton C, Hart EA, Hawes A, Heath PD, Heitmann K, Hennig S, Hernandez J, Hinzmann B, Ho S, Hoffs M, Howden PJ, Huckle EJ, Hume J, Hunt PJ, Hunt AR, Isherwood J, Jacob L, Johnson D, Jones S, de Jong PJ, Joseph SS, Keenan S, Kelly S, Kershaw JK, Khan Z, Kioschis P, Klages S, Knights AJ, Kosiura A, Kovar-Smith C, Laird GK, Langford C, Lawlor S, Leversha M, Lewis L, Liu W, Lloyd C, Lloyd DM, Loulseged H, Loveland JE, Lovell JD, Lozado R, Lu J, Lyne R, Ma J, Maheshwari M, Matthews LH, McDowall J, McLaren S, McMurray A, Meidl P, Meitinger T, Milne S, Miner G, Mistry SL, Morgan M, Morris S, Muller I, Mullikin JC, Nguyen N, Nordsiek G, Nyakatura G, O'Dell CN, Okwuonu G, Palmer S, Pandian R, Parker D, Parrish J, Pasternak S, Patel D, Pearce AV, Pearson DM, Pelan SE, Perez L, Porter KM, Ramsey Y, Reichwald K, Rhodes S, Ridler KA, Schlessinger D, Schueler MG, Sehra HK, Shaw-Smith C, Shen H, Sheridan EM, Shownkeen R, Skuce CD, Smith ML, Sotheran EC, Steingruber HE, Steward CA, Storey R, Swann RM, Swarbreck D, Tabor PE, Taudien S, Taylor T, Teague B, Thomas K, Thorpe A, Timms K, Tracey A, Trevanion S, Tromans AC, d'Urso M, Verduzco D, Villasana D, Waldron L, Wall M, Wang Q, Warren J, Warry GL, Wei X, West A, Whitehead SL, Whiteley MN, Wilkinson JE, Willey DL, Williams G, Williams L, Williamson A, Williamson H, Wilming L, Woodmansey RL, Wray PW, Yen J, Zhang J, Zhou J, Zoghbi H, Zorilla S, Buck D, Reinhardt R, Poustka A, Rosenthal A, Lehrach H, Meindl A, Minx PJ, Hillier LW, Willard HF, Wilson RK, Waterston RH, Rice CM, Vaudin M, Coulson A, Nelson DL, Weinstock G, Sulston JE, Durbin R, Hubbard T, Gibbs RA, Beck S, Rogers J, Bentley DR. The DNA sequence of the human X chromosome. Nature. 2005;434(7031):325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sado T, Hoki Y, Sasaki H. Tsix silences Xist through modification of chromatin structure. Dev Cell. 2005;9(1):159–165. doi: 10.1016/j.devcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Senda S, Wakayama T, Yamazaki Y, Ohgane J, Hattori N, Tanaka S, Yanagimachi R, Shiota K. Skewed X-inactivation in cloned mice. Biochem Biophys Res Commun. 2004;321(1):38–44. doi: 10.1016/j.bbrc.2004.06.096. [DOI] [PubMed] [Google Scholar]
- Shin T, Kraemer D, Pryor J, Liu L, Rugila J, Howe L, Buck S, Murphy K, Lyons L, Westhusin M. A cat cloned by nuclear transplantation. Nature. 2002;415(6874):859. doi: 10.1038/nature723. [DOI] [PubMed] [Google Scholar]
- Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385(6619):810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- Wrenzycki C, Lucas-Hahn A, Herrmann D, Lemme E, Korsawe K, Niemann H. In vitro production and nuclear transfer affect dosage compensation of the X-linked gene transcripts G6PD, PGK, and Xist in preimplantation bovine embryos. Biol Reprod. 2002;66(1):127–134. doi: 10.1095/biolreprod66.1.127. [DOI] [PubMed] [Google Scholar]
- Wrenzycki C, Wells D, Herrmann D, Miller A, Oliver J, Tervit R, Niemann H. Nuclear transfer protocol affects messenger RNA expression patterns in cloned bovine blastocysts. Biol Reprod. 2001;65(1):309–317. doi: 10.1095/biolreprod65.1.309. [DOI] [PubMed] [Google Scholar]
- Xue F, Tian XC, Du F, Kubota C, Taneja M, Dinnyes A, Dai Y, Levine H, Pereira LV, Yang X. Aberrant patterns of X chromosome inactivation in bovine clones. Nat Genet. 2002;31(2):216–220. doi: 10.1038/ng900. [DOI] [PubMed] [Google Scholar]
- Yang L, Chavatte-Palmer P, Kubota C, O'Neill M, Hoagland T, Renard JP, Taneja M, Yang X, Tian XC. Expression of imprinted genes is aberrant in deceased newborn cloned calves and relatively normal in surviving adult clones. Mol Reprod Dev. 2005;71(4):431–438. doi: 10.1002/mrd.20311. [DOI] [PubMed] [Google Scholar]


