Abstract
Differentiation of myocytes is impaired in patients with mytonic dystrophy type 1, DM1. CUG repeat binding protein, CUGBP1, is a key regulator of translation of proteins that are involved in muscle development and differentiation. In this paper, we present evidence that RNA-binding activity of CUGBP1 and its interactions with initiation translation complex eIF2 are differentially regulated during myogenesis by specific phosphorylation and that this regulation is altered in DM1. In normal myoblasts, Akt kinase phosphorylates CUGBP1 at Ser28 and increases interactions of CUGBP1 with cyclin D1 mRNA. During differentiation, CUGBP1 is phosphorylated by cyclinD3-cdk4/6 at Ser302, which increases CUGBP1 binding with p21 and C/EBPβ mRNAs. While cyclin D3 and cdk4 are elevated in normal myotubes; DM1 differentiating cells do not increase these proteins. In normal myotubes, CUGBP1 interacts with cyclin D3/cdk4/6 and eIF2; however, interactions of CUGBP1 with eIF2 are reduced in DM1 differentiating cells and correlate with impaired muscle differentiation in DM1. Ectopic expression of cyclin D3 in DM1 cells increases the CUGBP1-eIF2 complex, corrects expression of differentiation markers, myogenin and desmin, and enhances fusion of DM1 myoblasts. Thus, normalization of cyclin D3 might be a therapeutic approach to correct differentiation of skeletal muscle in DM1 patients.
Keywords: Myotonic Dystrophy 1, CUG repeats, CUGBP1-eIF2 complex, cyclin D3, differentiation
Introduction
Impaired differentiation of skeletal muscle is one of the critical disorders in DM1 patients. A number of publications showed that expression of the mutant DMPK mRNA with long CUG repeats alters biological activities of CUGBP1 in DM1 patients [1–6]. CUGBP1 is a multifunctional RNA-binding protein which regulates RNA processing at several stages including promotion of cap-dependent and cap-independent translation [7–16], RNA stability [17] and splicing [3,18–20]. Investigations of biological functions of CUGBP1 in mouse models showed that the un-scheduled elevation of CUGBP1 leads to a delay of muscle development and differentiation [6,21], muscular dystrophy [6,21,22] and myotonia [22]. Recent studies of the mechanisms which regulate expression of CUGBP1 in normal myogenesis showed that levels of CUGBP1 mRNA are significantly increased during muscle differentiation and that this increase is mediated by activation of the CUGBP1 promoter [23]. It has been shown that myogenin/E2/p300 complex binds to the E-box within the CUGBP1 promoter and increases transcription of CUGBP1 mRNA during differentiation [23]. A search for CUGBP1 targets identified a number of mRNAs including several mRNAs which are important for muscle development and function such as a cdk inhibitor p21 [4], myocyte enhancer factor 2A [6], insulin receptor [18] and chloride ion channel [19]. We have shown that the elevation of the CUGBP1 mRNA levels during normal muscle differentiation in cell culture models leads to increase of CUGBP1 protein and its RNA-binding activity [4]. In normal myotubes, CUGBP1 contributes to the up-regulation of translation of a cdk inhibitor, p21, [4] and a myocyte enhancer factor 2, MEF2A, promoting differentiation [6]. In contrast to normal myotubes, CUGBP1 binding activity to p21 and MEF2A mRNAs is reduced in DM1 differentiating cells consistent with a reduction of p21 [4] and MEF2A proteins [6]. Despite large body of evidence for the role of CUGBP1 in DM1 pathology, little is known about mechanisms which regulate biological functions of CUGBP1 during myogenesis. We have initiated these studies with the goal to develop approaches to correct a delay of differentiation of DM1 myoblasts.
In this paper, we present evidence that two signal transduction pathways regulate CUGBP1 activity in normal muscle and that these pathways are altered in DM1 cells. We have found that CUGBP1 is phosphorylated by different kinases during myoblast proliferation and differentiation and that phosphorylation of CUGBP1 at different sites directs CUGBP1 to different mRNA targets. Moreover, cyclin D3-cdk4-mediated phosphorylation of CUGBP1 increases the interactions of CUGBP1 with eIF2 during normal myogenesis. We have found that cyclin D3-cdk4 pathway is reduced in DM1 cells and that the normalization of cyclin D3 expression in DM1 cells leads to the correction of differentiation.
Materials and Methods
Plasmids and purified proteins
Constructs containing wild type human CUGBP1 and CUGBP1 truncated mutants containing different number of RNA-binding domains (RBD) were described early [7]. CUGBP1 S28A, S28D, S302G and S302D mutants were generated by mutagenesis using wild type human CUGBP1 cloned in BS with a Quick Mutagenesis kit (Stratagene). The sequences of the mutant forward primers are as follows: (1) S28A – 5′-GTT CCA AGG ACC TGG GCT GAA AAG GAC TTG -3′; (2) S28D – 5′-TCC AAG GAC CTG GGA TGA AAA GGA CTTG G-3′; S302G - 5′-CA CCT AGC TCT AGC AGC GGT AAT TCT GTC AAC -3′; S302D - 5′-CCT AGC TCT AGC AGC GAT AAT TCT GTC AAC-3′. Resulting constructs were verified by sequencing; and the mutant CUGBP1 cDNAs were re-cloned into pMAL vector. The expression of wild type and mutant CUGBP1-MBP proteins was induced by IPTG according to the manufacturer’s protocol (New England Biolabs) and the CUGBP1 proteins were purified by affinity chromatography on amylose resin. The purity of proteins was examined by electrophoresis coomassie staining.
Kinase assay
Equal amounts of wild type and mutant CUGBP1 proteins were incubated with the purified cyclin D3-cdk6 complex or with the purified Akt for 30 min in the presence of 50μCi of 32P-γATP. After the phosphorylation, proteins were separated by PAGE, transferred on the nitrocellulose membrane and exposed to the X-ray film. Membranes were stained with coomassie blue to verify loading of CUGBP1 proteins.
UV-cross link
Un-phosphorylated wild type and mutant CUGBP1 proteins and phosphorylated CUGBP1 proteins were incubated with radioactive RNA probes labeled with 32P-γATP. C/EBPβ and p21 RNA oligomers were synthesized in the Oligo’s Etc. The sequences of p21 and C/EBPβRNAs used in these experiments were previously described [4, 7]. The sequence of cyclin D1 RNA probe is as follows: 5′-CCC AGC CAG GAC CCA CAG CCC UCC CCA GCU GCC CAG GAA GAG CCC CAG CC-3′. Proteins (0.1–1 μg) were incubated at room temperature for 30 min in the mix containing cold non-specific competitor (total tRNA from HeLa cells) (100 ng/μl), radioactive RNA probe (50,000–100,000 cpm), 10 mM tris-HCl, pH 7.5; 50mM KCl, 5% glycerol and 5 mM DTT. Incubation mixtures were treated with UV light, separated by PAGE, transferred onto nitrocellulose membrane and exposed to the X-ray film. The membranes were stained with coomassie blue to verify protein loading.
EMSA
Equal amounts (5 μg) of cytoplasmic proteins extracted from mouse and human myoblasts and myotubes were incubated with 75,000 cpm of radioactive C/EBPβRNA probe under conditions described previously [4]. RNA-protein complexes were separated by native 5% PAGE.
Western blotting and immunoprecipitation (IP)
C2C12 myoblasts were grown as described previously in our publication [23]. Myotube formation was induced with fusion medium lacking FBS and containing insulin and horse serum. Myotubes were maintained for 5 days with daily change of fusion medium. Human primary myoblasts derived from control and DM1 patients were grown as described previously [4,6]. Cytoplasmic protein extracts were prepared as described [4] and applied for Western blotting. Antibodies to cyclin D1, cdc2, PCNA, CUGBP1, myogenin, cdk4, cyclin D3, cdk6, C/EBPβ, ph-Akt, eIF2α, eIF2β, GRP78, CRT (Santa Cruz Biotechnologies) and antibodies to β-actin (Sigma) were used according to the manufacturers’ protocols.
2D-Western approach
To examine phosphorylation status of CUGBP1, cytoplasmic extracts from C2C12 cells were separated by 2D techniques as described in our previous paper [14]. The proteins were transferred on the nitrocellulose membrane and probed with antibodies to CUGBP1. For these experiments, we performed overnight incubation with primary antibodies in cold room and 4 hours incubation with secondary antibodies at room temperature.
Transfections
Normal and DM1 myoblasts were grown in the 4x chamber slides at 60% density. The cells were transfected with a pAdTrack-cyclin D3 plasmid expressing cyclin D3 and GFP from independent promoters using FuGene protocol (Roche) according to the manufacturer’s recommendations. For nucleofection, cells were grown in 10 cm dishes at 60–70% density. Nucleofection was performed according to the protocol from Amaxa and transfected cells were plated in 2–4 x chamber slides or 10 cm dishes at 80% density. In 24 hrs after transfection, growth myoblast medium was replaced with fusion medium and cells were maintained in the fusion medium for two-three days with daily medium change. Fixed cells were subjected to immunofluorescent analysis with antibodies to desmin at 1:20 dilution and secondary anti-rabbit antibodies labeled with TRITC (Santa Cruz). Nuclei were identified by staining with DAPI. For myogenin detection, proteins were extracted from the transfected cells grown in 10 cm dishes and myogenin was examined by Western blotting assay with antibodies from Santa Cruz Biotechnologies.
Results
Akt phosphorylates CUGBP1 at Ser28 and increases interactions of CUGBP1 with cyclin D1 mRNA
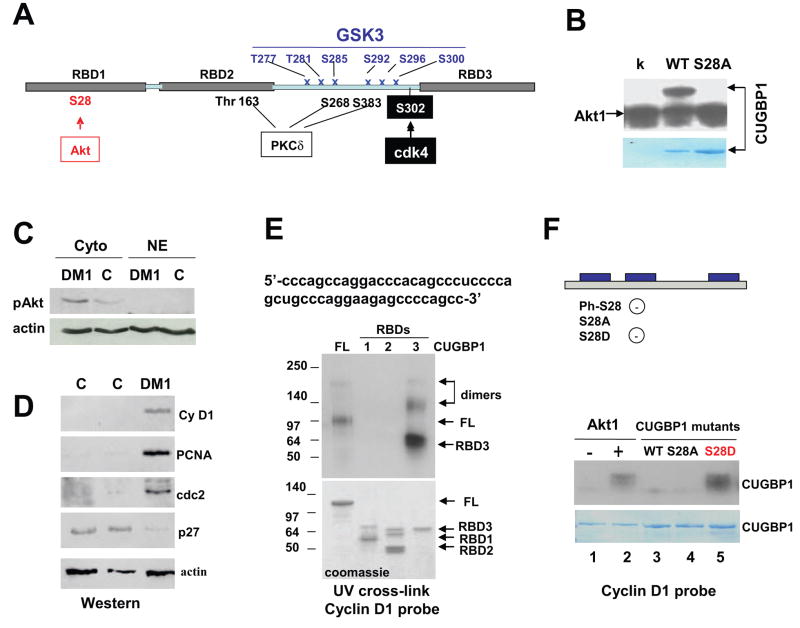
The transition of proliferating myoblasts to differentiation status involves alterations of several signal transduction pathways which might control activities of CUGBP1. Our previous data and data from other groups suggested that binding activity of CUGBP1 and formation of CUGBP1-protein complexes are regulated by phosphorylation of CUGBP1 [3, 8, 10, 14]. Particularly, we have found that cyclinD3/cdk4/6 complexes phosphorylate CUGBP1 at Ser302 in vitro and in vivo [14]. To identify other potential phosphorylation sites within CUGBP1, we have applied the Motif Scan Graphic Program. This program predicted multiple potential phosphorylation sites within the polylinker region of CUGBP1. These sites include consensuses for GSK3 (T277, T281, S285, S292, S300), three sites for PKC delta (Thr163, S268, S383) and a potential site for Akt kinase in the position 28 (Ser28) (Fig. 1A). In addition, our previous work identified a site for cyclinD3/cdk4/6 in the position S302 [14]. To examine if GSK3, PKC delta and Akt might phosphorylate CUGBP1, the purified fusion MBP-CUGBP1 was incubated with these kinases in the kinase buffer containing 32P-γATP. This kinase assay indicated that Akt1 strongly phosphorylates CUGBP1 in vitro (Fig. 1B), while GSK3 and PKC delta did not show detectable phosphorylation of CUGBP1 in this in vitro assay (data not shown). These results showed that CUGBP1 is a substrate for Akt kinase in vitro. Failure of GSK3 and PKC delta to phosphorylate CUGBP1 in vitro suggests that these kinases might require additional factors which are absent in the in vitro kinase assay. The Motif Scan Graphic Program predicted a potential Akt site at S28 (Fig. 1A). To examine if Ser28 is a specific site for Akt kinase, we have mutated S28 to alanine and examined if Akt is able to phosphorylate this mutant. As seen in Fig. 1B, replacement of S28 with alanine eliminates Akt-dependent phosphorylation of CUGBP1. Thus, these data demonstrate that Akt phosphorylates CUGBP1 at Ser28 in vitro.
Figure 1. Akt phosphorylates CUGBP1 at Ser28.
A. Prediction of specific phosphorylation sites within CUGBP1 by the Motif Scan Program. Three RNA-binding domains (RBDs) are shown by gray boxes. A site for cdk4-mediated phosphorylation of CUGBP1 (S302) was previously identified [14]. B. Akt phosphorylates the purified wild type CUGBP1 at Ser28. WT CUGBP1 and S28A-CUGBP1 mutant were incubated with Akt. Replacement of S28 with alanine eliminates Akt-dependent phosphorylation of CUGBP1. Lower band shows auto-phosphorylation of Akt. K (kinase only) - control with Akt kinase without addition of CUGBP1. Bottom image shows a coomassie stain of the membrane to verify loading of CUGBP1 C. Akt pathway is activated in DM1 cells. Cytoplasmic and nuclear extracts from cultured control and DM1 myoblasts were examined by Western blotting with antibodies to ph-473-Akt. The membrane was re-probed with antibodies toβ-actin. D. Cyclin D1, cdc2 and PCNA protein levels are increased in DM1 muscle; while expression of p27 is reduced. Western blotting was performed with protein extracts isolated from skeletal muscle samples of two controls and from skeletal muscle of DM1 patient with antibodies to cyclin D1, PCNA, cdc2 and p27. The membrane was re-probed with antibodies to β-actin. E. CUGBP1 binds to the 5′ region of cyclin D1 mRNA. Sequence of the 5’ region of cyclin D1 mRNA (riboprobe used in UV-cross link) is shown on the top. Full-length (FL) and truncated CUGBP1 proteins containing RBD1, RBD2 and RBD3 respectively were examined in UV cross link with the cyclin D1 probe. Positions of protein markers are shown on the left. Arrows show positions of the full-length CUGBP1 and truncated CUGBP1 proteins. Note that CUGBP1 and RBD3 protein form dimers. The membrane was exposed to the X-ray for 16 hrs. Bottom image shows a coomassie stain of the gel loaded with the corresponding proteins. F. Akt-mediated phosphorylation of CUGBP1 increases its interactions with cyclin D1 mRNA. Top. Diagram shows CUGBP1-S28 mutants which were used in these studies. CUGBP1-S28D mutant is mimicking phosphorylated state of CUGBP1. Generation of a negatively charged CUGBP1 by phosphorylation of wild type CUGBP1 (ph-S28) and by replacement of Ser28 with aspartate is also shown. Bottom. Purified CUGBP1-MBP proteins were tested in UV cross link with the cyclin D1 RNA probe. Lane 1; Un-phosphorylated WT CUGBP1, lane 2; CUGBP1 phosphorylated by Akt; lane 3; unphosphorylated wild type CUGBP1; lane 4; CUGBP1-S28A mutant; lane 5; CUGBP1-S28D mutant. The membrane was exposed to the X-ray film for 2 hrs. Coomassie staining of the membrane after UV-cross link shows identical loading of the proteins.
Given the fact that CUGBP1 is the substrate for Akt, we have examined if Akt pathway is altered in cells from DM1 patients. Cytoplasmic and nuclear proteins from cultured DM1 myoblasts were loaded on the SDS gel and probed with antibodies to active ph-473-Akt. As one can see in Fig. 1C, the amounts of active Akt are higher in cytoplasm of DM1 cells compared to those in control cells. Since one of functions of Akt is to promote cell cycle progression, we have examined expression of cell cycle proteins in skeletal muscle of DM1 patients, which contain elevated levels of CUGBP1 and have increased rate of proliferation [4,24]. Total protein extracts were isolated from skeletal muscle of control and DM1 patients and examined by Western blotting with antibodies to cell cycle proteins. These studies showed that PCNA, cdc2 and cyclin D1 are up-regulated in DM1 muscle; while expression of the inhibitor of proliferation p27 is reduced (Fig. 1D). This pattern of expression suggests that the a ctivation of Akt in DM1 cells might cause the increased levels of the cell cycle proteins. We have next tested this hypothesis by measuring the Akt-CUGBP1 pathway in DM1 cells. Since cyclin D1 is a strong promoter of cell proliferation, we have focused our further studies on the binding of CUGBP1 to the cyclin D1 mRNA. To test if CUGBP1 binds to the cyclin D1 mRNA, we have analyzed the nucleotide sequence of cyclin D1 mRNA and found that the 5′-region of cyclin D1 mRNA contains several GC-islands to which CUGBP1 binds (Fig. 1E, upper). Using RNA probe containing this sequence, we have examined interactions of purified CUGBP1 with the 5′ region of cyclin D1 mRNA. In these experiments, we used the purified full-length CUGBP1 and purified CUGBP1 truncated proteins containing RNA-binding domains (RBD) 1, 2 and 3. As shown in Fig. 1E, the full-length CUGBP1 binds to the 5′ region of cyclin D1 RNA. The purified proteins containing only RBD1 and RBD2 do not bind to this sequence, while RBD3 binds to the cyclin D1 riboprobe. Coomassie staining showed approximately identical loading of the proteins (Fig. 1E, bottom image). Thus, these studies revealed that CUGBP1 binds to the 5′ region of cyclin D1 mRNA and that this binding is mediated by RBD3.
We have next examined if phosphorylation of CUGPB1 by Akt might regulate its interactions with cyclin D1 mRNA. In addition to S28A mutant, we also generated a phosphomimetic mutant in which Ser28 was replaced with aspartate (S28D), mimicking the phosphorylation status of CUGBP1. Wild type CUGBP1 and mutant S28A and S28D CUGBP1 proteins were purified to homogeneity, as determined by commassie staining, and their interactions with cyclin D1 RNA were examined by UV-cross link (Fig. 1F). UV-cross link showed that un-phosphorylated CUGBP1 binds to cyclin D1 RNA with low affinity; however, phosphorylation of CUGBP1 by Akt significantly increases interaction of CUGBP1 with cyclin D1 mRNA (Fig. 1F). Examination of S28A and S28D mutants showed that the mutation of Ser28 to alanine reduces interaction of CUGBP1 with cyclin D1 mRNA. In contrast, the S28D mutant binds stronger to cyclin D1 mRNA and this binding is identical to that of wild type CUGBP1 phosphorylated by Akt. Note that time of exposure of the membranes shown in Fig. 1E and 1F were different: 16 hrs exposure is shown in the Fig. 1E and 2 hrs exposure is shown in Fig. 1F. This suggest that, although the region of RBD3 mediates interaction of full-length CUGBP1 with cyclin D1 RNA, the binding of un-phosphorylated RDB3 to the cyclin D1 riboprobe is weaker than the binding of phosphorylated full-length CUGBP1. Taken together, these data show that phosphorylation of CUGBP1 by Akt at Ser28 significantly increases its binding to the cyclin D1 mRNA.
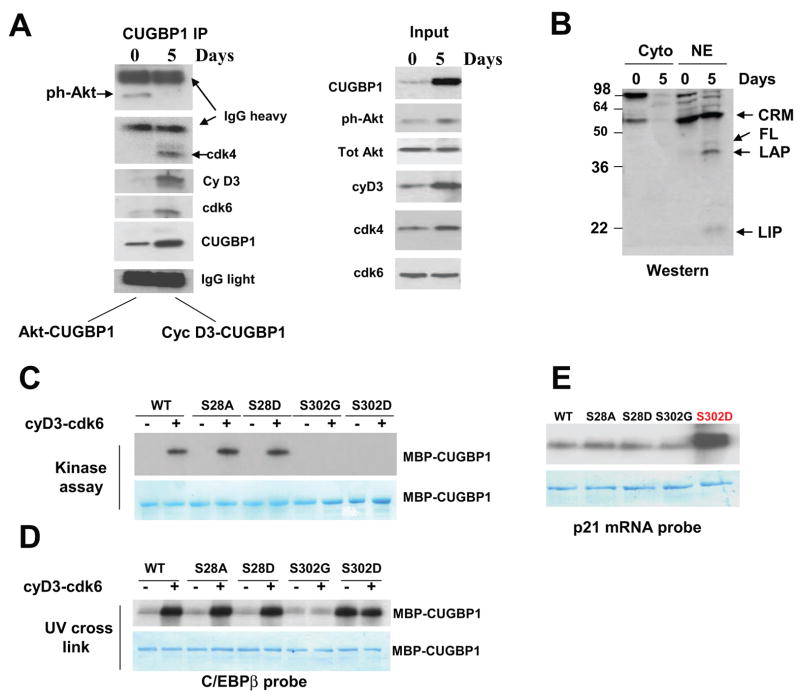
Figure 2. Cyclin D3-cdk4/6 increases binding of CUGBP1 to C/EBPβ and p21 mRNAs in differentiated myotubes.
A. CUGBP1 is associated with “active” Akt in proliferating myoblasts, while in differentiated myotubes CUGBP1 is associated with cyclinD3/cdk4/6. Left image. Co-IP-Western blotting. CUGBP1 was immunoprecipitated from proliferating and differentiated C2C12 cultured cells. “0” and “5” show days of culturing cells in fusion medium. These IPs were probed with antibodies to ph-Akt, cyclin D3, CUGBP1, cdk4 and cdk6. Light and heavy chains of IgGs are shown and revealed that equal amounts of CUGBP1 antibodies which were used for CUGBP1 precipitation from myoblasts and from myotubes. Right image (input) shows Western blotting of the cytoplasmic extracts used for the CUGBP1 IPs. B. The levels of CUGBP1 translational target, C/EBPβ, are increased in normal differentiated muscle cells. The cytoplasmic and nuclear extracts were analyzed by Western blotting with antibodies to C/EBPβ. The full-length and truncated C/EBPβ proteins (LAP and LIP) are shown by arrows. CRM; cross-reactive molecule. C. Cyclin D3-cdk6 phosphorylates CUGBP1 at S302. WT CUGBP1 and CUGBP1 mutants (shown on the top) were incubated with cyclinD3/cdk6 complex and 32P-γATP in in vitro kinase assay. The membrane was stained with coomassie to show loading of CUGBP1 proteins (bottom image). D. Phosphorylation of CUGBP1 by cyclinD3/cdk6 increases its binding to C/EBPβ mRNA. UV cross link was performed with un-phosphorylated CUGBP1 and CUGBP1 phosphorylated by cyclin D3/cdk6 using the C/EBPβ mRNA probe. Coomassie blue shows equal amounts of CUGBP1 used in this experiment. E. A phosphomimetic CUGBP1-S302D strongly interacts with the 5′ region of p21 mRNA. UV cross link was performed with non-phosphorylated wild type and mutant CUGBP1 proteins. Coomassie staining on the bottom shows equal amounts of purified CUGBP1-MBP used in these experiments.
Interactions of CUGBP1 with Akt and cyclin D3/cdk4/6 are dependent on differentiation stages of myogenesis
We have previously found that cyclin D3-cdk4/6 regulate activity of CUGBP1 in differentiated settings such as quiescent livers [14]. Therefore, we have next examined whether Akt1 and cdk4 are involved in the regulation of CUGBP1 during transition of myoblasts to myotubes. For this goal, we have used mouse C2C12 myoblasts and myotubes. CUGBP1 was precipitated from cytoplasmic extracts of proliferating mouse myoblasts and differentiated myotubes and the presence of ph-Akt and cyclin D3/cdk4/6 in the CUGBP1-IPs was examined by Western blotting. Fig. 2A demonstrates that, in C2C12 myoblasts, the phosphorylated (“active”) Akt is present in the CUGBP1-IP, while cyclin D3/cdk4/6 are undetectable. In contrast, ph-Akt is not detectable in CUGBP1-IP from myotubes, but the association of CUGBP1 with cdk4, cdk6 and cyclin D3 is dramatically increased. The right panel of the Fig. 2A (input) shows levels of the proteins in cytoplasmic extracts used for the IP. Although levels of CUGBP1, cyclin D3 and ph-Akt are increased during differentiation, the interactions of CUGBP1 with ph-Akt are reduced; while interaction of CUGBP1 with cyclin D3 is increased. These data show that different kinases are associated with CUGBP1 in proliferating myoblasts and in differentiated myotubes and that the association of CUGBP1 with ph-Akt does not depend on the levels of the proteins, but rather depends on the stage of cell differentiation.
To determine if the cyclin D3-cdk4 activates translational functions of CUGBP1 in differentiated C2C12 cells, we have examined expression of C/EBPβ since previous studies showed that the phosphorylation of CUGBP1 by cdk4-cyclin D3 enhances the ability of CUGBP1 to increase translation of C/EBPβ isoforms FL, LAP and LIP [14]. We found that these C/EBPβ isoforms are increased in nuclear extracts of C2C12 myotubes (Fig. 2B). The elevation of C/EBPβ isoforms correlates with the increased association of CUGBP1 with eIF2 and with increased phosphorylation of CUGBP1 by cyclin D3/cdk4/6 (see below).
Phosphorylation of CUGBP1 by cyclinD3/cdk4 increases interactions of CUGBP1 with C/EBPβ and p21 mRNAs
Since the phosphorylation of CUGBP1 by Akt increases its RNA-binding affinity to cyclin D1 (Fig. 1E), we asked if phosphorylation of CUGBP1 by cyclinD3/cdk4/6 at Ser302 might also control its RNA-binding activity. To address this question, we have used purified electrophoretically homogenous wild type CUGBP1 and several CUGBP1 mutants. In one of the mutants, Ser302 was replaced with glycine (S302G) resulting in the expression of CUGBP1 un-phosphorylatable at Ser302 [14]. We have also generated another mutant, S302D, which mimics cdk4-mediated phosphorylation of CUGBP1. We have first examined phosphorylation of WT CUGBP1 and CUGBP1 mutants by cyclin D3-cdk6 in in vitro kinase assay. The complex of cyclin D3/cdk6 has been used because the complex of cyclin D3 with cdk4 is not formed efficiently in vitro. Fig. 2C shows that cyclin D3-cdk6 phosphorylates WT, S28A and S28D mutants; while phosphorylation of S302G and S302D mutants is very weak or not detectable. These data confirmed our previous findings that S302 is a key amino acid for the phosphorylation by cdk4/cdk6 [14]. We have next examined whether phosphorylation of CUGBP1 at Ser302 regulates its RNA-binding affinity toward C/EBPβ mRNA. CUGBP1 and CUGBP1 mutants were phosphorylated by cold ATP with baculovirus expressed cyclin D3-cdk6 as shown in Fig. 2C and examined for the ability to interact with C/EBPβ. As seen in Fig. 2D, the phosphorylation of WT CUGBP1, S28A and S28D mutants by cyclin D3/cdk6 complex significantly increases CUGBP1 binding to C/EBPβ mRNA, while the binding of un-phosphorylated CUGBP1 to C/EBPβ mRNA is weaker. A different result was observed for S302G and S302D mutants which are no longer under cdk4/cdk6 control and the pre-incubation with cyclin D3-cdk6 did not change their interactions with the C/EBPβprobe. However, the mutation of S302 to aspartate has increased the interactions with C/EBPβ mRNA. We have next examined the interaction of WT CUGBP1 and CUGBP1 mutants with p21 mRNA since our previous data showed that CUGBP1 regulates translation of p21 during differentiation of C2C12 cells. UV-cross linking studies showed that S302D mutant (mimicking CUGBP1 phosphorylation at Ser302) binds to the p21 mRNA much stronger than WT CUGBP1 and other CUGBP1 mutants (Fig. 2E). Taken together, these investigations revealed that phosphorylation of CUGBP1 at Ser302 and the mutation of Ser302 to aspartate significantly increase interactions of CUGBP1 with C/EBPβ and p21 mRNAs.
Cyclin D3-cdk4-mediated phosphorylation of CUGBP1 increases formation of the translational CUGBP1-eIF2 complexes during normal muscle differentiation
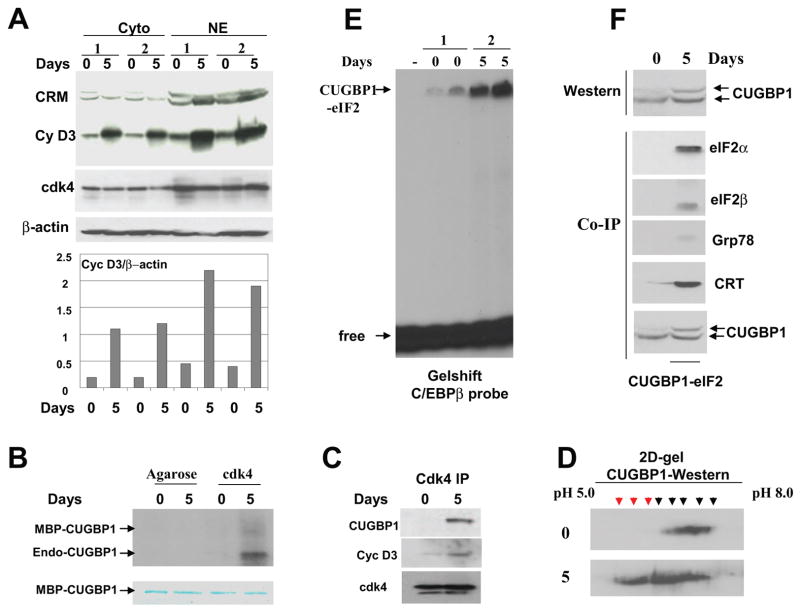
In differentiated settings, the translational activity of CUGBP1 is mediated by its interactions with translation initiation factor eIF2 and by forming a high MW complex CUGBP1-eIF2 which contains eIF2 alpha and beta, Grp78 and CRT [14]. Therefore, we have examined if differentiated myotubes utilize the same pathway. In contrasts to cyclins D1 and D2 which promote cell proliferation, cyclin D3 is highly expressed in certain differentiating settings such as differentiating myotubes and hepatocytes [14, 25–28] and supports differentiation and growth arrest of these cells. Previous studies of the cyclin D3 in differentiated myotubes have been focused on the nuclear accumulation of cyclin D3 and on nuclear functions of this protein [25–28]. Since our data suggested that cyclin D3 also has biological functions in cytoplasm, we have first determined expression of cyclin D3 in cytoplasm of proliferating and differentiated C2C12 cells and compared these levels with levels of nuclear cyclin D3. Examination of two sets of differentiated cells showed that the levels of cyclin D3 are increased during differentiation in both cytoplasm and nuclei (Fig. 3A). Although the amounts of cyclin D3 in cytoplasm are 2-fold lower than in nuclei (as ratios to β-actin), cyclin D3 is abundant and easy detectable in cytoplasm. Western blotting with antibodies to cdk4 also determined cdk4 in both cellular compartments with slightly higher concentrations in nuclei. Most important, the elevation of cyclin D3 during differentiation of myotubes is observed in cytoplasm suggesting that cyclin D3 plays a role in the regulation of cytoplasmic functions during differentiation. One of the possible functions of cyclin D3-cdk4 in cytoplasm of differentiated myotubes is the regulation of biological activities of CUGBP1.
Figure 3. The phosphorylation of CUGBP1 by cyclin D3/cdk4 during differentiation of C2C12 cells correlates with increased amounts of the CUGBP1-eIF2 complex.
A. Cyclin D3 levels are increased in cytoplasm and in nuclei of C2C12 myotubes. Western blotting of cytoplasmic and nuclear extracts isolated from two sets of C2C12 cells subjected to differentiation (days 0 and 5) was performed with antibodies against cyclin D3 and cdk4. The membrane was re-probed with Abs to β-actin as control. Cross reactive molecule (CRM) is shown and serves as an internal control for the loading. Bar graphs show protein levels of cyclin D3 as ratios to β-actin. B. Cdk4 phosphorylates CUGBP1 in normal myotubes. Cdk4 was precipitated from myoblasts (0) and myotubes (5) and examined in kinase assay with MBP-CUGBP1 as a substrate. Arrows show the positions of MBP-CUGBP1 and endogenous CUGBP1 interacting with cdk4 in myotubes. Phosphorylation of the endogenous CUGBP1 is more efficient relatively to the MBP-CUGBP1 purified from bacteria. Bottom part is a commassie stain of the membrane which shows the equal amounts of MBP-CUGBP1 used in this assay. C. CUGBP1 and cyclin D3 are abundant in cdk4 IPs from C2C12 myotubes. Western blotting was performed with cdk4 IPs using antibodies to CUGBP1 and cyclin D3. D. Phosphorylation of CUGBP1 is increased in differentiated myotubes. 2D-gel-electrophoresis-Western blotting of protein extracts from C2C12 myoblasts (top) and myotubes (bottom) with antibodies to CUGBP1. The 2D-We stern was performed as described in Materials and Methods. Phosphorylated CUGBP1 isoforms with similar mobility in myoblasts and in myotubes are shown by black arrows. Additional hyper-phosphorylated isoforms of CUGBP1 in myotubes are shown by red arrows. E. The amounts of the high molecular weight complex migrating in the position of CUGBP1-eIF2 are increased in differentiated myotubes. EMSA assay was performed with two sets of cytoplasmic extracts from mouse C2C12 myoblasts and myotubes using C/EBPβ RNA as a probe. The high molecular weight complex binding with C/EBPβ probe (putative CUGBP1-eIF2) is shown by arrow. F. The translational CUGBP1-eIF2 complex is increased in differentiated myotubes. Top. Amounts of CUGBP1 and phosphorylation of CUGBP1 are increased during normal differentiation. Western blotting was performed with extracts isolated from myoblasts (0) and from myotubes (5). Bottom. The amounts of CUGBP1-eIF2 complex are increased in differentiated C2C12 cells. CUGBP1 was immunoprecipitated from cytoplasms of myoblasts (0) and from cytoplasm of myotubes (5 days), and these IPs were probed with antibodies to the components of the CUGBP1-eIF2 complex (shown on the right).
To determine if cyclin D3/cdk4 is able to phosphorylate CUGBP1 in differentiated C2C12 cells, cdk4 was immunoprecipitated from proliferating (day 0) and differentiated (day 5) C2C12 cells and used in a kinase assay with MBP-CUGBP1 substrate. Fig. 3B shows that cdk4 phosphorylates CUGBP1. Interestingly, we have also detected the phosphorylation of an additional protein which is co-precipitated with cdk4. Since the size of this protein is 50 kDa, we suggested that this protein might be the endogenous CUGBP1. To test this suggestion and to determine amounts of cyclin D3 in cdk4 IPs, we have performed Western blotting of cdk4 IPs with antibodies to CUGBP1 and cyclin D3. As one can see in Fig. 3C, both CUGBP1 and cyclin D3 are abundant in cdk4-IPs from differentiated myotubes; while only small amounts of cyclin D3 are detected in cdk4-IPs from proliferating myoblasts. Taken together, these studies showed that cyclin D3-cdk4 phosphorylates CUGBP1 in differentiated C2C12 cells.
To examine phosphorylation status of CUGBP1 during myogenesis we used 2D gel electrophoresis–Western blotting approach. Cytoplasmic proteins from proliferating (day 0) and differentiated (day 5) C2C12 were separated by 2D technique, transferred on the filter and probed with antibodies to CUGBP1. As can be seen in Fig. 3D, differentiated C2C12 cells contained three additional phosphorylated isoforms of CUGBP1. These isoforms represent different phosphorylated forms of CUGBP1 because treatment of the protein extracts with alkaline phosphatase resulted in the shift of the signals to the alkaline region of the gel (data not shown). Thus, CUGBP1 phosphorylation is increased during normal differentiation.
We have next examined if the interaction of CUGBP1 with eIF2 is increased during muscle differentiation. For this goal, we have used two approaches: EMSA with the C/EBPβ probe and Co-immunoprecipitation analysis. Cytoplasmic proteins from myoblasts and myotubes were incubated with C/EBPβRNA probe containing the CUGBP1-binding site. The CUGBP1-eIF2 complex contains 8 protein components and, under conditions of EMSA, migrates close to the top of the gel because of high molecular weight of whole complex [14]. Fig. 3E shows that the RNA binding activity of the high molecular weight complex interacting with the C/EBPβ probe is significantly increased in myotubes compared with the activity of this complex observed in myoblasts. The high molecular weight complex binding with C/EBPβ in C2C12 myotubes migrates in the position typical for CUGBP1-eIF2 complex. However, the analysis of this complex by the traditional supershift assay is quite difficult, since the position of this complex in the EMSA gel is very close to the top of the gel. Therefore, we have determined if the interaction of CUGBP1 with eIF2 is increased during normal differentiation using a second approach: IP-Western analysis. CUGBP1 was immunoprecipitated from C2C12 cells and the CUGBP1-IPs were probed with antibodies to the known components of the CUGBP1-eIF2 complex. Upper image of the Fig. 3F shows amounts of CUGBP1 in the lysates used for the Co-IP. As one can see, eIF2 alpha and beta, CRT and Grp78 are associated with CUGBP1 in myotubes; while these proteins do not bind or bind very weakly (CRT) to CUGBP1 in proliferating myoblasts. Consistent with the results of 2D-Western, the additional isoform of CUGBP1 is detectable in myotubes by regular Western blotting. Taken together, these studies showed that CUGBP1 interacts with cdk4 and cyclin D3 in differentiated myotubes and that the interactions of CUGBP1 with eIF2 complex are also increased in normal differentiated myotubes.
Misregulation of Akt and cyclinD3/cdk pathways in DM1 myogenesis
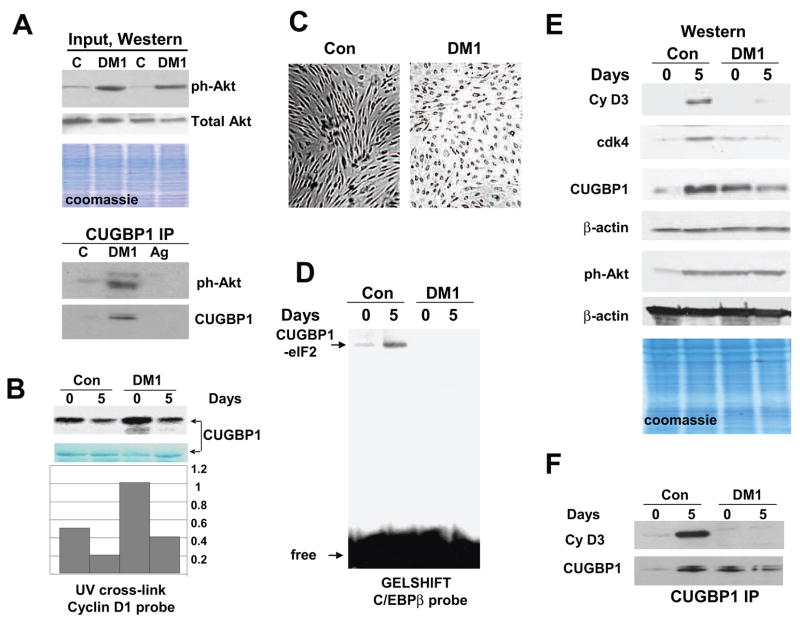
Given established pathways which regulate CUGBP1 activities during normal differentiation of mouse myotubes, we have examined these pathways in human normal myoblasts and in myoblasts from DM1 patients. In mouse myoblasts, CUGBP1 is phosphorylated by Akt kinase which increases interactions of CUGBP1 with cyclin D1 RNA. We have found that protein levels of PCNA, cdc2 and cyclin D1, which is a strong stimulator of proliferation, are increased in DM1 patients (Fig. 1D). Examination of DM1 cells also revealed that Akt pathway is activated in cytoplasm of DM1 cells. These data suggested that DM1 cells might have misregulated Akt-CUGBP1 pathway. To test this hypothesis, we have applied primary myoblast cell lines derived from control and DM1 patients. Consistent with data shown in Fig. 1C, Western blotting with ph-Akt revealed that Akt is activated in DM1 cell lines; while total levels of Akt are not changed (Fig. 4A, upper image, input). We have next examined if Akt interacts with CUGBP1 in DM1 myoblasts. CUGBP1 was precipitated from control and DM1 myoblasts and ph-Akt was measured in CUGBP1-IPs. As shown in Fig. 4A, the interaction of CUGBP1 with “active” Akt is significantly higher in DM1 myoblasts relatively to the interaction in control myoblasts. Thus, the elevation of ph-Akt in DM1 proliferating cells correlates with the increased interaction of active Akt with CUGBP1. These data are consistent with the hypothesis that the translation of cyclin D1 is increased in DM1 cells by Akt-mediated activation of interaction of CUGBP1 with cyclin D1 RNA. To test this suggestion, we have examined if the incubation of bacterially expressed CUGBP1 with proteins from DM1 cells might enhance interactions of CUGBP1 with cyclin D1 probe. For this goal, we incubated bacterially expressed, purified CUGBP1 with cytoplasmic extracts from control and DM1 cells at day 0 and 5 after initiation of differentiation and examined the RNA binding activity of the phosphorylated CUGBP1 towards cyclin D1 mRNA. Fig. 4B shows that the incubation of CUGBP1 with proteins from DM1 myoblasts increases the interaction of CUGBP1 with cyclin D1 mRNA much stronger than incubation with proteins from myoblasts from normal patients. Protein extracts from differentiating cells have much lower capability to increase interactions of CUGBP1 with cyclin D1 mRNA. Nevertheless, densitometric calculations showed that the activation of CUGBP1 towards cyclin D1 by proteins from DM1 cells at day 5 is stronger than that observed in experiments with control cells at day 5 (Fig. 4B, Bar graphs). This pattern of the activation of CUGBP1 towards cyclin D1 mRNA is consistent with the hypothesis that the increase of Akt-CUGBP1 pathway in DM1 cells supports high level of cyclin D1.
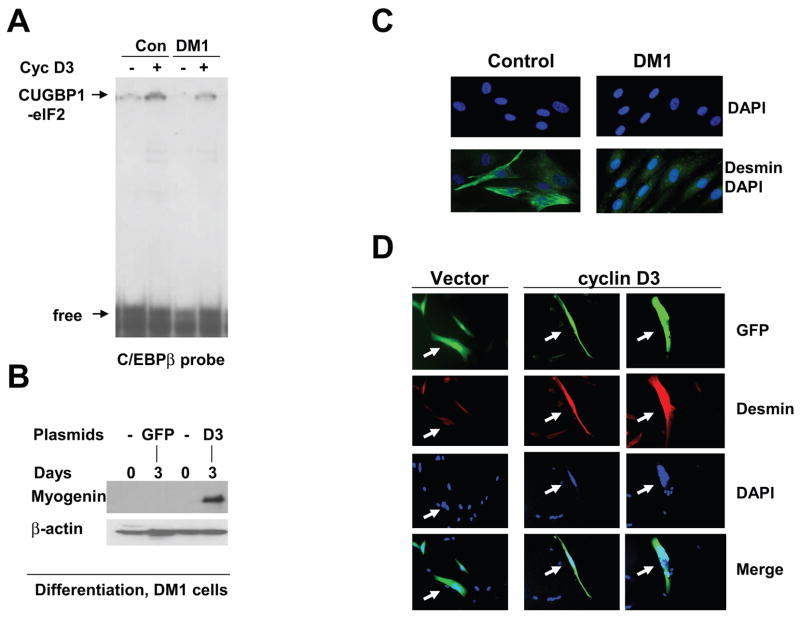
Figure 4. Mis-regulation of Akt-CUGBP1 and cyclinD3/cdk-CUGBP1 pathways in DM1 myogenesis.
A. Input. The levels of active ph-Akt and total Akt in cytoplasm used for the IP. Western blotting of cytoplasmic extracts from control and DM1 myoblasts was performed with ph-Akt antibodies and with antibodies to total Akt. Cells from two control and two DM1 patients were analyzed. Bottom image shows coomassie staining of the membrane. CUGBP1-IP: Interactions of “active” Akt with CUGBP1 are increased in DM1 myoblasts. CUGBP1 was precipitated from control and DM1 myoblasts and ph-Akt was detected by Western blotting. Ag; mock agarose control. B. Incubation of bacterially expressed CUGBP1 with cytoplasmic proteins from DM1 cells increases interactions of CUGBP1 with cyclin D1 mRNA. CUGBP1 was incubated with cytoplasmic proteins from control and DM1 cells in the presence of 1mM ATP and examined by UV cross-link with cyclin D1 probe. The filter was stained with coomassie. Bar graphs show the ratio of CUGBP1 signals on UV cross link film to signals of CUGBP1 detected by coomassie stain. C. Delay of differentiation of DM1 cells. Bright field microscopy shows cells differentiating for 5 days. Cells were derived from control and DM1 patient. D. Amounts of the CUGBP1-eIF2 complex are reduced in DM1 differentiating cells. Gelshift was performed with cytoplasmic protein extracts isolated from control and DM1 proliferating and differentiating cells using C/EBPβ probe. Positions of the CUGBP1-eIF2 complex and free probe are shown by arrows. E. Mis-regulation of Akt and cyclin D3/cdk4 pathways in DM1 myogenesis. Cyclin D3 and cdk4 levels are reduced in DM1 differentiating cells while levels of active Akt remain high during proliferation and differentiation of DM1 muscle cells. Cytoplasmic proteins from control and DM1 proliferating and differentiating cells were analyzed by Western blotting with antibodies to cyclin D3, cdk4, CUGBP1 and ph-Akt. The membranes were re-probed with antibodies to β-actin as a loading control. Bottom image shows a coomassie stain of the membrane. F. Cyclin D3 does not interact with CUGBP1 in DM1 cells during differentiation. CUGBP1 was immunoprecipitated from control and DM1 cytoplasm. The IPs were probed with antibodies to cyclin D3 and to CUGBP1.
Since CUGBP1 displays its translational activities in differentiated cells via interactions with eIF2, we have examined if the formation of the CUGBP1-eIF2 complex is increased in control human myotubes and if the amounts of this complex are affected in DM1 patients. Myoblasts from normal and DM1 patients were subjected to differentiation, cytoplasmic proteins were isolated from myoblasts and myotubes and the formation of CUGBP1-eIF2 complex was examined by EMSA assay. Parallel plates were monitored to detect morphological changes typical for differentiation. In agreement with previous observations, DM1 cells show a delay of differentiation, while control cells are forming elongated myotubes (Fig. 4C). Gelshift analysis indicated that the amounts of the high molecular weight CUGBP1-eIF2 complex are low in proliferating myoblasts from control patients and from patients with DM1; however, the amounts of this complex are significantly increased during differentiation of control myoblasts. In contrast, the CUGBP1-eIF2 complex is not increased in DM1 differentiating muscle cells (Fig. 4D). Since cyclin D3-cdk4 phosphorylates CUGBP1 in vitro [14] and because cyclin D3 and cdk4 are associated with CUGBP1 in differentiated mouse C2C12 cells (Fig. 2), we suggested that the lack of the CUGBP1-eIF2 complexes in DM1 differentiating cells might be associated with reduced levels of cyclin D3 and cdk4. To test this suggestion, we have examined the expression of cyclin D3, cdk4 and CUGBP1 in control and DM1 cells during differentiation. As can be seen in Fig. 4E, cyclin D3 levels are increased in differentiated cells from control patients, but DM1 cells do not increase cyclin D3. Examination of cdk4 levels showed that there is also no increase of cdk4 in DM1 differentiating cells. The lack of cyclin D3/cdk4 elevation in DM1 differentiating myoblasts might be the cause for a failure to form the CUGBP1-eIF2 complex. Examination of ph-Akt showed that the amounts of active Akt are increased in normal cells during differentiation. In contrast, the activation of Akt was observed in DM1 cells before differentiation and it is not significantly changed during differentiation. Protein levels of CUGBP1 are elevated during differentiation of normal human myoblasts [4]. Interestingly, DM1 cells already have elevated levels of CUGBP1 in myoblasts. In contrast to normal myotubes, cytoplasmic levels of CUGBP1 in DM1 differentiating cells are not increased but reduced compared to levels in proliferating DM1 cells (Fig. 4E).
The lack of elevation of cyclin D3 in DM1 cells after initiation of differentiation suggests that the cyclin D3-CUGBP1 pathway is not activated in DM1 cells. To test this suggestion, CUGBP1 was immunoprecipitated from control and DM1 cells and cyclin D3 was examined in these IPs. As can be seen in Fig. 4F, cyclin D3 is strongly associated with CUGBP1 in normal differentiated myotubes; however, no interaction of cyclin D3 with CUGBP1 is observed in DM1 cells. These data clearly show that cyclin D3-CUGBP1 pathway is inhibited in DM1 differentiating cells.
Ectopic expression of cyclin D3 corrects differentiation of DM1 myocytes
Based on our data described above, we hypothesized that the lack of cyclin D3 elevation might be a major cause of the impaired differentiation of DM1 cells. To test this hypothesis, we transfected pAdTrack-cyclin D3 plasmid (which, in addition to cyclin D3, also expresses GFP protein from an independent CMV promoter) into DM1 myoblasts and initiated differentiation of the cells by addition of fusion media. To determine if the transfected cyclin D3 activates CUGBP1-eIF2 complex, we have performed EMSA assay with cytoplasmic extracts from transfected cells. Fig. 5A shows that transfection of cyclin D3 into DM1 cells increases amounts of the CUGBP1-eIF2 complex. Three approaches were used for the examination of differentiation of DM1 cells transfected with cyclin D3: immunostaining of desmin, examination of cell fusion and the quantification of myogenin by Western blot assay. First, the expression of desmin was determined using immunostaining of control and DM1 cells after two days of differentiation. Fig. 5C shows that the levels of desmin are dramatically increased in normal cells in 2 days after initiation of differentiation. However, DM1 cells showed no or very weak increase of desmin at day 2 of differentiation. The lack of increase of desmin is consistent with a delay of differentiation in DM1 patients [4]. DM1 cells were transfected with a pAdTrack-cyclin D3 plasmid and the transfected cells were analyzed by IF with antibodies to desmin. These studies showed that expression of desmin is dramatically increased in cells transfected with cyclin D3 at day 3 after initiation of differentiation, while un-transfected cells have very weak signal for desmin (Fig. 5D). These un-transfected cells serve as a good control since these cells have been treated the same way as transfected cells, but they do not increase expression of desmin. The elevation of desmin was observed in each examined cyclinD3-positive cell. Thus, these data show that normalization of cyclin D3 levels during differentiation of DM1 cells rescues early steps of the differentiation.
Figure 5. Increase of the levels of cyclin D3 in DM1 myoblasts corrects early steps of differentiation.
A. Ectopic expression of cyclin D3 in DM1 cells activates the CUGBP1-eIF2 pathway. Control and DM1 cells were transfected with pAdTrack-cyclin D3 plasmid, cytoplasmic extracts were isolated and used for EMSA with the C/EBPβ probe. Positions of the CUGBP1-eIF2 complex and free probe are shown by arrows. B. Ectopic expression of cyclin D3 increases protein levels of myogenin in DM1 differentiating cells. DM1 myoblasts transfected with GFP or cyclin D3/GFP were subjected to differentiation in fusion medium for three days. Myogenin levels were determined by Western blotting. The membrane was re-probed with antibodies to β-actin. C. DM1 cells do not increase expression of desmin at early step of differentiation. Immunofluorescent analysis with antibodies to desmin was performed using control and DM1 muscle cells differentiating for two days in fusion medium. Nuclei were stained with DAPI. C. Ectopic expression of cyclin D3 in DM1 cells increases expression of desmin during differentiation. DM1 myoblasts were transfected with pAdTrack-cyclin D3 plasmid expressing cyclin D3 and GFP from different promoters (green), differentiation was initiated by fusion media and cells were subjected to IF with antibodies to desmin (red). Nuclei were stained with DAPI. The picture shows representative analysis of 45 cells transfected with cyclin D3 and 41 cells transfected with an empty vector. Arrows show the transfected cells.
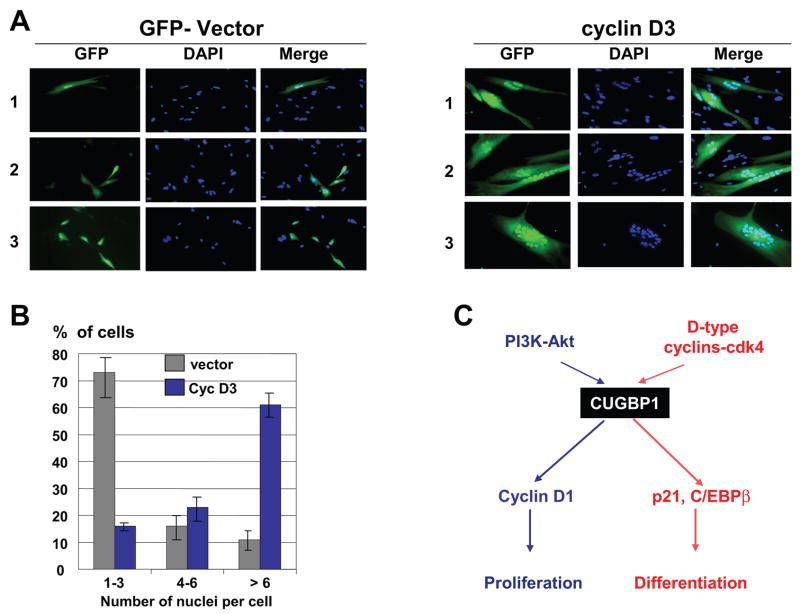
To further examine if ectopic expression of cyclin D3 corrects differentiation of DM1 cells, we have applied a second approach and have monitored the fusion of cells at day 3 after initiation of differentiation. Fig. 6A shows that the fusion of DM1 cells is dramatically increased after transfection with cyclin D3; while GFP alone did not affect the fusion of DM1 cells. Calculation of the efficiency of fusion by counting multi-nucleated cells showed that only 11% of DM1 cells transfected with empty vector contain more than 6 nuclei per cell upon maintenance in fusion medium for 3 days. However, up to 61% of DM1 cells transfected with cyclin D3 contain high number of nuclei per cell (6–25). In these experiments, we found a significant portion of DM1-cyclin D3 transfected cells which had more than 20 nuclei in the fused cells (Fig. 6A, right, field #3). This dramatic increase of the fusion of DM1 cells by the ectopic cyclin D3 is accompanied by the elevation of myogenin. As shown in Fig. 5B, myogenin is almost undetectable in DM1 cells transfected with an empty vector; however, in the presence of cyclin D3 the levels of myogenin are significantly increased upon differentiation. In summary, these studies showed that ectopic expression of cyclin D3 in DM1 cells leads to the activation of CUGBP1-eIF2 pathway (Fig. 5A) and to correction of differentiation of DM1 cells (Figs 5B,D; 6A,B).
Figure 6.
A. Ectopic expression of cyclin D3 dramatically increases fusion of DM1 cells during differentiation. Empty vector and pAdTrack-cyclin D3 plasmid were transfected into DM1 cells using Amaxa Transfector protocol. Fusion media was added on the next day after transfections and nuclei were stained with DAPI at day 3 after addition of fusion media. Three fields (numbered on the left) are shown for the control (vector) and cyclin D3 transfections. The field #3 of cyclin D3 transfection shows an example of cells containing more than 20 nuclei. These cells represent approximately 10% of green cells in cyclin D3 transfections and are not detectable in control DM1 cells. B. Bar graphs show a summary of three independent experiments examining the efficiency of DM1 myoblast fusion after ectopic expression of cyclin D3. Percentage of cells containing 1–3, 4–6 and more than 6 nuclei per cell was calculated. 200–300 transfected cells were used for these calculations. C. Diagram showing a hypothesis for the pathways which regulate CUGBP1 activity in normal myogenesis. These pathways are altered in DM1 cells and are involved in the development of DM1 pathology (see text).
Discussion
CUGBP1 plays a critical role in differentiation of skeletal muscle [4, 6]. DM1 mouse models have demonstrated that genetically un-programmed increase of CUGBP1 levels resulted in the development of muscular dystrophy and in a delay of muscle differentiation [6,21]. Examination of the recently developed new DM1 mouse model has revealed that a normalization of CUGBP1 levels results in reversing muscular dystrophy and myotonia [22]. These investigations showed that alterations in the regulation of CUGBP1 activities might be key events in the development of DM1 pathology. In order to establish approaches for the correction of activities of CUGBP1 in DM1 patients, we have initiated investigations of pathways which regulate the activities of CUGBP1 under normal conditions and mechanisms by which expanded CUG repeats alter these pathways. In this paper, we examined mechanisms which control interactions of CUGBP1 with mRNA targets during differentiation of normal myoblasts. Our data show that CUGBP1 is a subject of phosphorylation by different kinases and that specific phosphorylation of CUGBP1 increases its RNA-binding affinity toward different mRNAs. We found that Akt phosphorylates CUGBP1 at Ser28 and that S28-phosphorylated CUGBP1 has higher affinity to cyclin D1 RNA compared to interactions with C/EBPβ and p21 mRNAs. On the contrary, phosphorylation of CUGBP1 by cyclin D3-cdk4/6 increases its interactions with C/EBPβ and p21 mRNAs. Examination of protein-protein interactions of CUGBP1 revealed that, in proliferating myoblasts, CUGBP1 interacts with “active” Akt, while during differentiation of normal cells it is associated with cyclin D3/cdk4/6. These data suggested that differential phosphorylation of CUGBP1 might be responsible for control of specific interactions of CUGBP1 with certain mRNAs (Fig. 6C). In addition to alterations in RNA binding activity, the phosphorylation of CUGBP1 by cyclin D3-cdk4/6 also increases its interactions with translation initiation complex eIF2. Thus, the studies in normal mouse myoblasts showed that biological activities of CUGBP1 depend on interactions with different kinases and that these interactions are tightly regulated during differentiation. One of the key regulators of CUGBP1 activities in differentiated myotubes is cyclin D3. The elevation of cyclin D3 in differentiated myotubes has been previously published by several groups [25–27] and suggested that cyclin D3 might be a critical regulator of differentiation of myotubes. A recent paper by De Santa et al has shown that the elevation of cyclin D3 during myogenesis is mediated by Rb-dependent protein stabilization and that this elevation is required for myogenic differentiation [28]. While previous publications have been focused on nuclear functions of cyclin D3, our paper presented evidence for the cytoplasmic functions of cyclin D3 in differentiated myotubes. We have found that, in normal cells, a significant portion of cyclin D3-cdk4 is located in cytoplasm and these complexes phosphorylate CUGBP1. Examination of myoblasts from DM1 patients has identified two significant alterations. First, proliferating myoblasts from DM1 patients show much stronger interactions of CUGBP1 with active Akt than in normal proliferating myoblasts. Second, differentiation of normal human myoblasts is associated with elevation of cyclinD3-cdk4/6 which phosphorylates CUGBP1 and increases amounts of the CUGBP1-eIF2 complex; however, DM1 differentiating cells fail to increase cyclin D3 and have limited amounts of the CUGBP1-eIF2 complexes. The lack of elevation of cyclin D3 in DM1 differentiating cells seems to be a critical event leading to impaired myoblast fusion. We have found that normalization of cyclin D3 levels in DM1 cells corrects early steps of differentiation, as it is shown by measuring expression of desmin and myogenin, and dramatically increases fusion of DM1 cells leading to formation of multinucleated cells. In summary, data in this paper revealed an important role of phosphorylation of CUGBP1 in the regulation of differentiation of myoblasts and showed that DM1 cells lost a proper control of the phosphorylation dependent regulation of CUGBP1. Our data showing the correction of differentiation of DM1 cells by cyclin D3 suggest that the normalization of cyclin D3 levels in DM1 patients might be possible therapeutic approach for correction of a delayed differentiation of skeletal muscle in DM1 patients.
Acknowledgments
This work is supported by NIH grants, AR052791, AR49222, and AR44387 (LTT) and by grants from the Deutsche Gesellschaft fur Muskelkranke (BGHS). BGHG is member of the German network on muscular dystrophies (MD-NET, 01GM0601) funded by the German Ministry of Education and Research (BMBF, Bonn, Germany). MD-NET is partner of TREAT-NMD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Timchenko LT, Timchenko NA, Caskey CT, Roberts R. Novel proteins with binding specificity for DNA CTG repeats and RNA CUG repeats: implications for myotonic dystrophy. Hum Mol Genet. 1996;5:115–121. doi: 10.1093/hmg/5.1.115. [DOI] [PubMed] [Google Scholar]
- 2.Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 4.Timchenko NA, Iakova P, Cai ZJ, Smith JR, Timchenko LT. Molecular basis for impaired cell cycle withdrawal in myotonic dystrophy. Mol Cell Biol. 2001;21:6927–6938. doi: 10.1128/MCB.21.20.6927-6938.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Timchenko NA, Cai ZJ, Welm AL, Reddy S, Ashizawa T, Timchenko LT. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J Biol Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- 6.Timchenko NA, Patel R, Iakova P, Cai ZJ, Quan L, Timchenko LT. Overexpression of CUG triplet repeat binding protein, CUGBP1, inhibits myogenesis. J Biol Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- 7.Timchenko NA, Welm AL, Lu X, Timchenko LT. CUG repeat binding protein (CUGBP1) interacts with the 5′ region of C/EBPβ mRNA and regulates translation of C/EBPβ isoforms. Nucl Acids Res. 1999;27:4517–4525. doi: 10.1093/nar/27.22.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welm AL, Mackey SL, Timchenko LT, Darlington GJ, Timchenko NA. Translational induction of LIP during acute phase response leads to repression of C/EBP alpha mRNA. J Biol Chem. 2000;275:27406–27413. doi: 10.1074/jbc.M002343200. [DOI] [PubMed] [Google Scholar]
- 9.Baldwin BR, Timchenko NA, Zhanow CA. EGF receptor stimulation activates the RNA binding protein, CUGBP1, and increases expression of C/EBPβ-LIP in mammary epithelial cells. Mol Cell Biol. 2004;24:3682–3691. doi: 10.1128/MCB.24.9.3682-3691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Timchenko NA, Wang GL, Timchenko LT. CUG triplet repeat binding protein, CUGBP1, increases translation of C/EBPβ isoform, LIP, by interacting with the α and α subunits of eIF2. J Biol Chem. 2005;280:20549–20557. doi: 10.1074/jbc.M409563200. [DOI] [PubMed] [Google Scholar]
- 11.Bae EJ, Kim SG. Enhanced CCAAT/enhancer-binding protein beta-liver-enriched inhibitory protein production by Oltipraz, which accompanies CUG repeat-binding protein-1 (CUGBP1) RNA-binding protein activation, leads to inhibition of preadipocyte differentiation. Mol Pharmacol. 2005;68:660–669. doi: 10.1124/mol.105.012997. [DOI] [PubMed] [Google Scholar]
- 12.Guerzoni C, Bardini M, Mariani SA, Ferrari-Amorotti G, Neviani P, Panno ML, Zhang Y, Martinez R, Perrotti D, Calabretta B. Inducible activation of CEBPB, a gene negatively regulated by BCR/ABL, inhibits proliferation and promotes differentiation of BCR/ABL-expressing cells. Blood. 2006;107:4080–4089. doi: 10.1182/blood-2005-08-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karagiannides I, Thomou T, Tchkonia T, Pirtkalava Y, Kypreos KE, Cartwright A, Dalageorgou G, Lash TL, Farmer SR, Timchenko NA, Kirkland JL. Increased CUG triplet repeat binding protein-1 predisposes to impaired adipogenesis with aging. J Biol Chem. 2006;281:23025–23033. doi: 10.1074/jbc.M513187200. [DOI] [PubMed] [Google Scholar]
- 14.Timchenko LT, Salisbury E, Wang GL, Nguyen HD, Albrecht JH, Hershey JWB, Timchenko NA. Age-specific CUGBP1-eIF2 complex increases translation of C/EBPβ in old liver. J Biol Chem. 2006;281:32806–32819. doi: 10.1074/jbc.M605701200. [DOI] [PubMed] [Google Scholar]
- 15.Choi WT, Folsom MR, Azim MF, Meyer C, Kowarz E, Marchalek R, Timchenko NA, Naeem R, Lee DA. C/EBPβ suppression by interruption of CUGBP1 resulting from a complex rearrangement of MLL. Cancer Genetics Cytogen. 2007;177:108–114. doi: 10.1016/j.cancergencyto.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woeller C, Fox JT, Perry C, Stover PJ. A ferritin-responsive internal ribosome entry site regulates folate metabolism. J Biol Chem. 2007;282:29927–29935. doi: 10.1074/jbc.M706264200. [DOI] [PubMed] [Google Scholar]
- 17.Barreau C, Paillard L, Mereau A, Osborne BH. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 2005;88:515–525. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 18.Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 19.Charlet BN, Savkur RS, Singh G, Philips AV, Grice EA, Cooper TA. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. (2002) Mol Cell. 2002;10:43–45. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 20.Zhang W, Liu H, Han K, Grabowski PJ. Region-specific alternative splicing in the nervous system: implications for regulation by the RNA-binding protein NAPOR. RNA. 2002;8:671–685. doi: 10.1017/s1355838202027036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho TH, Bundman D, Armstong DL, Cooper TA. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum Mol Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- 22.Mahadevan MS, Yadava RS, Yu Q, Balijepalli S, Frenxel-McCardell CD, Bourne TD, Phillips LH. Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat Genet. 2006;38:1066–1070. doi: 10.1038/ng1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huichalaf CH, Sakai K, Wang GL, Timchenko NA, Timchenko LT. Regulation of the promoter of CUG triplet repeat binding protein, Cugbp1, during myogenesis. Gene. 2007;396:391–402. doi: 10.1016/j.gene.2007.04.022. [DOI] [PubMed] [Google Scholar]
- 24.Khajavi M, Tari AM, Patel NB, Tsuji K, Siwak DR, Meistrich ML, Terry NHA, Ashizawa T. Mitotic drive of expanded CTG repeats in myotonic dystrophy type 1 (DM1) Hum Mol Gen. 2001;10:855–863. doi: 10.1093/hmg/10.8.855. [DOI] [PubMed] [Google Scholar]
- 25.Bartkova J, Lukas J, Strauss M, Bartek J. Cyclin D3: requirement for G1/S transition and high abundance in quiescent tissues suggest a dual role in proliferation and differentiation. Oncogene. 1998;17:1027–1037. doi: 10.1038/sj.onc.1202016. [DOI] [PubMed] [Google Scholar]
- 26.Cenciarelli C, De Santa F, Puri PL, Mattei E, Ricci L, Bucci F, Felsani A, Caruso M. Critical Role played by cyclin D3 in the MyoD-mediated arrest of cell cycle during myoblasts differentiation. Mol Cell Biol. 1999;19:5203–5217. doi: 10.1128/mcb.19.7.5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarruf DA, Iankova I, Abella A, Assou S, Miard S, Fajas L. Cyclin D3 promotes adipogenesis through activation of peroxisome proliferator-activated receptor gamma. Mol Cell Biol. 2005;25:9985–9995. doi: 10.1128/MCB.25.22.9985-9995.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Santa F, Albini S, Mezzaroma E, Baron L, Felsani A, Caruso M. pRb-dependent cyclin D3 protein stabilization is required for myogenic differentiation. Mol Cell Biol. 2007;27:7248–65. doi: 10.1128/MCB.02199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]