Summary
BACKGROUND
Epigenetic regulation by diverse classes of small RNAs is mediated by the highly conserved Argonaute/Piwi family of proteins. While Argonautes are broadly expressed, the Piwi subfamily primarily functions in the germ line. Piwi proteins are associated with germline-specific ribonucleoprotein (RNP) granules in Drosophila, zebrafish and mouse. Depending on the species and on the specific family member, Piwis play important roles in either spermatogenesis and/or in maintaining germ cell and stem cell totipotency. Piwis bind to a newly discovered class of small RNAs, called piRNAs. C. elegans contains a large set of Argonaute/Piwi related proteins, including two closely related to piwi, called prg-1 and prg-2. The function of prg-1 and prg-2, and whether piRNAs exist in C. elegans, is unknown.
RESULTS
Here, we demonstrate that the Piwi-like protein PRG-1 is localized to P granules in germ cells entering spermatogenesis, and is required for successful spermatogenesis. Loss of prg-1 causes a marked reduction in expression of a subset of mRNAs expressed during spermatogenesis, and prg-1 mutant sperm exhibit extensive defects in activation and fertilization. Moreover, prg-1 activity is required for the presence of the small RNAs called 21U-RNAs.
CONCLUSION
Our data suggest that PRG-1 promotes expression, processing, or stability of 21U-RNAs, which, in turn or in concert with PRG-1, promote proper expression of spermatogenesis transcripts.
Keywords: C. elegans, germ line, piwi, 21U-RNA, spermatogenesis
Introduction
Piwi proteins bind to and are required for the accumulation of small RNAs known as piRNAs (reviewed in [1]). piRNAs are derived from a few long precursor transcripts expressed from heterochromatic or non-coding regions of the genome. The biogenesis and function of piRNAs remains mysterious, in part because piRNA sequence and Piwi function differ between species. For instance, in Drosophila, most piRNA sequences correspond to transposons, and the Piwi proteins act primarily to suppress transposon activity in the germ line [2, 3]. In mice, the three Piwi-related proteins Miwi, Mili, and Miwi2 primarily function in spermatogenesis, and only Miwi2 and Mili have an apparent role in transposon silencing [4–7]. Mouse piRNAs fall into two groups, one expressed pre-meiotically that contains transposon-associated sequences, and one expressed meiotically that consists of unique sequences [8–10]. This second group of piRNAs does not have sequence complementarity to coding or 3′ UTR sequences, and their function in spermatogenesis remains unclear.
C. elegans contains a large set of Argonaute/Piwi related proteins, including two genes closely related to piwi, called prg-1 and prg-2 (prg = piwi-related gene [11]). prg-1 mutants display a temperature-sensitive defect in fertility that is currently uncharacterized [12]. Although piRNAs have not been identified in C. elegans, a class of small RNAs called 21U-RNAs was recently discovered [13]. 21U-RNAs bear several similarities to piRNAs: they have a uridine at the 5′ position, their sequences are diverse, nonconserved, and do not obviously match mRNAs or transposons, and the loci encoding 21U-RNAs are tightly clustered in the genome [13]. However, 21U-RNAs are shorter than piRNAs (21 nt rather than 26–31 nt), and appear to be individually transcribed, rather than derived from a long precursor transcript [13], so their relationship to piRNAs or other small RNA species in other organisms is uncertain. In this report, we investigate the function of PRG-1 and PRG-2 in the germ line of C. elegans and define both a set of mRNAs and a class of small RNAs that require PRG-1 for their expression.
Results
Disruption of prg-1 causes temperature-sensitive sterility
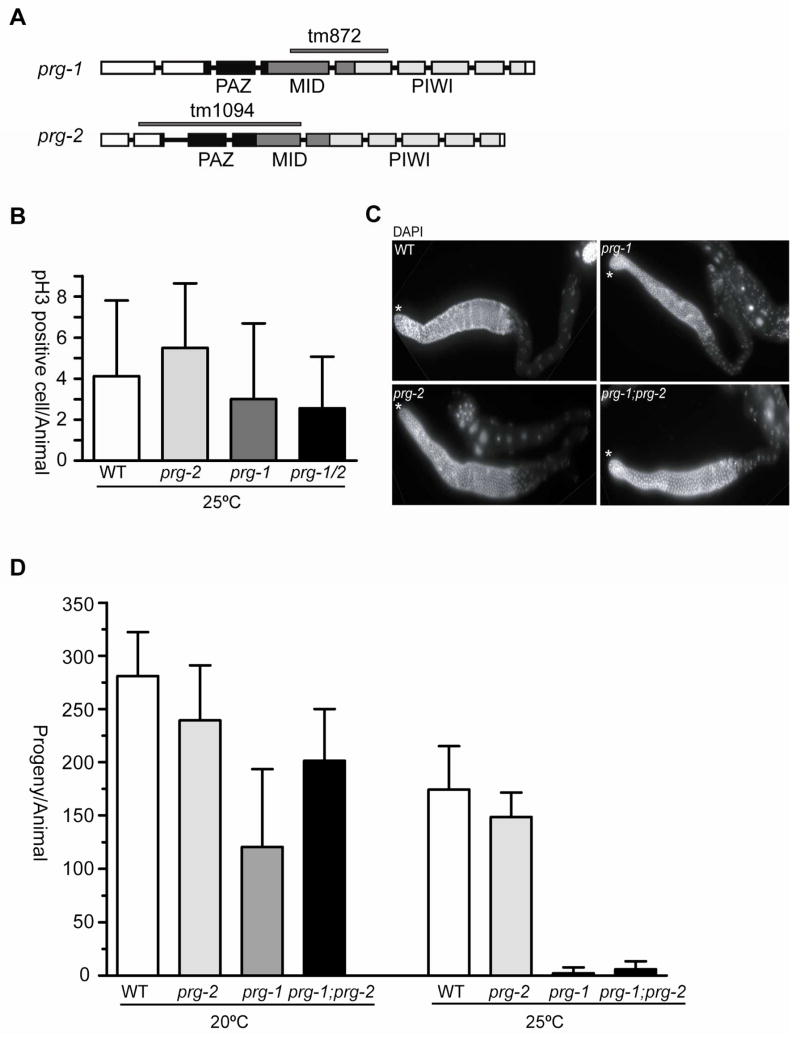
The key functional motifs of Argonaute/Piwi proteins include an N-terminal PAZ domain that binds the 3′ end of a small RNA, and a C-terminal PIWI domain that has RNAse H catalytic activity (Figure 1a). The PIWI domain also coordinates binding of the 5′ phosphate of the small RNA, along with a basic pocket domain called the MID domain [14]. To study the function of the C. elegans piwi-related genes prg-1 and prg-2, we obtained deletion mutants [11]. prg-1(tm872) deletes 640 bp encoding most of the MID domain and part of the PIWI domain, and prg-2(tm1094) removes 1065 bp, which includes the entire PAZ domain. Both prg-1(tm872) and prg-2(tm1094) therefore should be null or loss-of-function alleles.
Figure 1. Disruption of prg-1 causes reduction in brood size and temperature-sensitive sterility.
A) Schematic of prg-1 and prg-2 genomic structure. Box, exon; connecting line, intron. Functional domains (PAZ, MID and PIWI) of PRG proteins are indicated below. The solid lines above indicate the deletions in prg-1(tm872) and prg-2(tm1094). B) The average number of germ cells per gonad positive for phosphorylated Ser10 of histone H3 at 25°C is graphed. The X-axis indicates genotype; the Y-axis represents mean number of pH3+ cells. N>10 gonads. C) Dissected gonad arms from different genotypes stained with DAPI. Asterisk marks distal end of gonad. D) prg-1 mutants have a temperature sensitive sterile phenotype. Brood size of wild-type or mutant worms was determined at 20°C or 25 °C. The X-axis indicates corresponding genotypes; the Y-axis is the mean value of total number of progeny per animal. Greater than eight animals per trial, average of three trials. Error bars indicate standard deviation.
A previous report demonstrated that treating animals with RNAi to reduce prg-1 and prg-2 levels resulted in decreased germline stem cell proliferation [11]. We therefore first examined prg-1(tm872), prg-2(tm1094), and prg-1(tm872); prg-2(tm1094) mutants for defects in proliferation by monitoring mitotic germ cell number with phosphoS10 histone H3 staining, but did not see any statistically significant differences from wild type. Moreover, both DIC microscopy and DAPI staining of adult prg-1, prg-2, and prg-1; prg-2 double mutant gonads revealed essentially normal germ cell number and development (Figure 1b, c). However, as previously reported, prg-1(tm872) mutants exhibit temperature sensitive sterility: they have reduced brood size at 20°C and are essentially sterile at 25°C (<5 progeny/animal; [12]) (Figure 1d). prg-2(tm1094) single mutants do not display any obvious defects compared to wild type, and do not enhance or otherwise affect prg-1(tm872) sterility at 25°C.
The sterility of prg-1(tm872) mutants arises from a defect in spermatogenesis
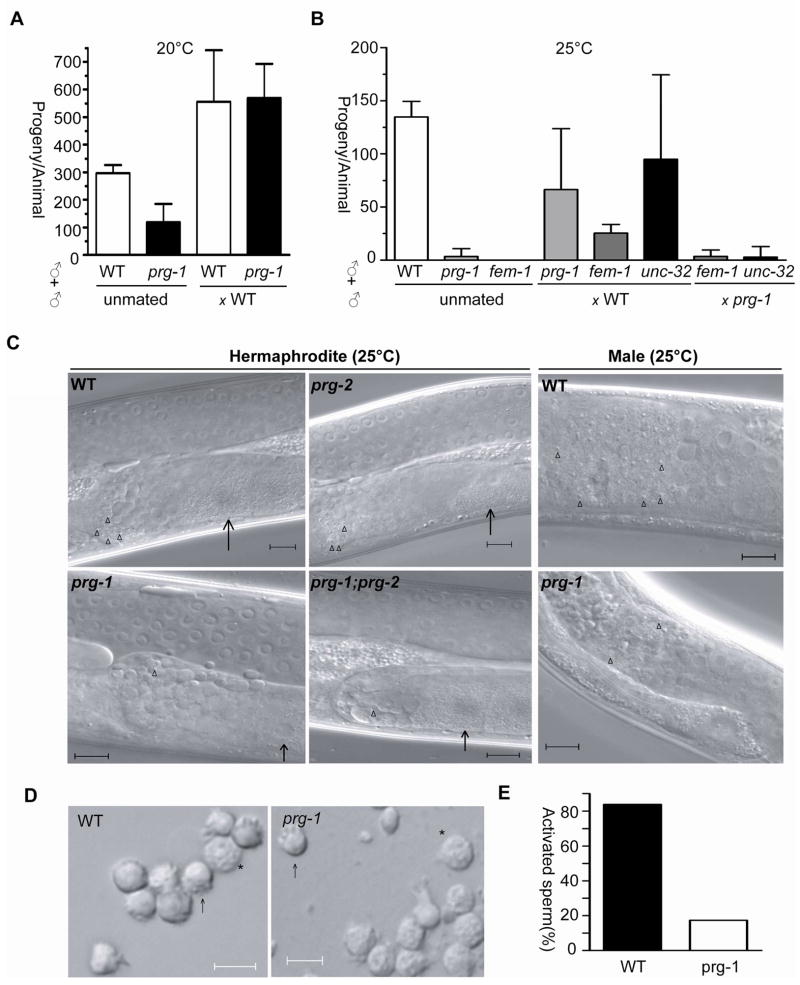
The germ line of adult C. elegans hermaphrodites contains both sperm and oocytes. Sperm are made only during the fourth stage (L4) of larval development, and oocytes are subsequently produced throughout early adulthood. In males, sperm production persists into adulthood. Even though hermaphrodites are self-fertile, male sperm are preferentially used over self-sperm for oocyte fertilization if mating occurs. To determine whether the sterility of prg-1 mutants arises from a defect in oogenesis or in spermatogenesis, we tested whether providing wild type sperm to prg-1 hermaphrodites by mating would rescue the brood size defects at 20°C and 25°C. After mating to wild type males, prg-1(tm872) hermaphrodites exhibited substantially improved fertility at both temperatures; in particular, at 20°C the brood size increased to the same extent as seen in mated wild type hermaphrodites (Figure 2a). This observation suggests that prg-1(tm872) mutant germ cells do not have major impairments in proliferation or oogenesis, and that the sterility at 25°C arises from a defect in spermatogenesis.
Figure 2. prg-1(tm872) mutants exhibit defects in spermatogenesis.
A) Wild type or prg-1(tm872) L4 hermaphrodites were either left unmated or mated to wild type males at 20°C, and the number of progeny counted. X-axis = genotypes, Y-axis = mean progeny per animal. N ≥16. Error bars indicate standard deviation. B) Wild type or prg-1(tm872) males were mated with prg-1, fem-1 or unc-32 mutant hermaphrodites at 25°C, and the number of progeny counted (cross-progeny only for unc-32). X-axis = genotypes, Y-axis = mean progeny per animal. Greater than eight animals per trial, two or three trials averaged. Error bars indicate standard deviation. C) Young adult hermaphrodites or males were examined by microscopy under DIC. Arrows indicate the first oocyte produced after completing spermatogenesis in hermaphrodites. Triangles indicate spermatids. Scale bar: 10μm. D) Image of pronase-treated spermatids in wild type and prg-1 mutants. Scale bar: 5μm. Arrow: activated spermatids; asterisk: unactivated spermatids. E) Quantification of spermatid activation, N>137 spermatids.
prg-1 mutant males also exhibit a temperature-sensitive defect in spermatogenesis. prg-1(tm872) males raised at 20°C are capable of producing cross progeny when mated to a self-fertile hermaphrodite, but not at 25°C (Figure 2b). At 25°C, prg-1 males could generate only ~2 cross progeny in 10% of matings, while wild type males generated ~100 cross progeny in 83% of matings. prg-1 males are physically capable of mating, as we could visualize sperm from the prg-1 male in a fem-1 mutant hermaphrodite, which lacks self-sperm. Although mating occurred, prg-1 males could not rescue the infertility of fem-1 mutants or produce cross progeny after mating to self-fertile unc-32 hermaphrodites at 25°C.
During spermatogenesis, undifferentiated germ cells undergo meiotic divisions with incomplete cytokinesis. Most cellular contents are dumped into a shared residual body, from which the haploid spermatids eventually bud. These spermatids then undergo further morphological changes to become activated, ameboid spermatozoa that are competent for oocyte fertilization. Microscopic examination of spermatogenesis revealed that prg-1 mutant hermaphrodites and males raised at 25°C possessed differentiated spermatocytes but relatively few mature spermatids (Figure 2c). Because some spermatids were present, we tested whether they were deficient in activation by treatment with pronase, which stimulates pseudopod formation [15]. prg-1 mutant sperm almost always failed to produce normal pseudopodia (17% in prg-1 compared to 84% in wild type; Figure 2d–e). Taken together, our data suggest that a defect at very late stages in spermatogenesis is the principal reason for the reduced fertility in prg-1(tm872) hermaphrodites and males at both 20°C and 25°C.
PRG-1 is localized to P granules during spermatogenesis
To determine the distribution and subcellular localization of PRG-1 during germline development, we generated two transgenic lines expressing full-length PRG-1 fused to GFP under the control of the pie-1 promoter, which is permissive for germline expression [16]. These lines showed identical spatial and temporal expression patterns. GFP was detectable in the cytoplasm but was greatly enriched in perinuclear granular structures in pachytene germ cells in males and in L4 hermaphrodites during spermatogenesis (Figure 3a). PRG-1 was preferentially expressed during spermatogenesis, since expression decreases over two-fold in hermaphrodite adults after the switch from spermatogenesis to oogenesis, and the granular localization completely disappeared (Figure 3b, c). This transgenic expression pattern likely reflects endogenous expression because the pie-1-driven PRG-1:GFP transgene partially rescues the prg-1 mutant sterile phenotype at 25°C from two to 40 progeny (n=11). Moreover, we have generated transgenic strains expressing PRG-1:GFP under the control of its own promoter that exhibited identical spatial, temporal, and subcellular localization to pie-1- driven PRG-1:GFP (data not shown).
Figure 3. PRG-1 localizes to P granules during spermatogenesis.
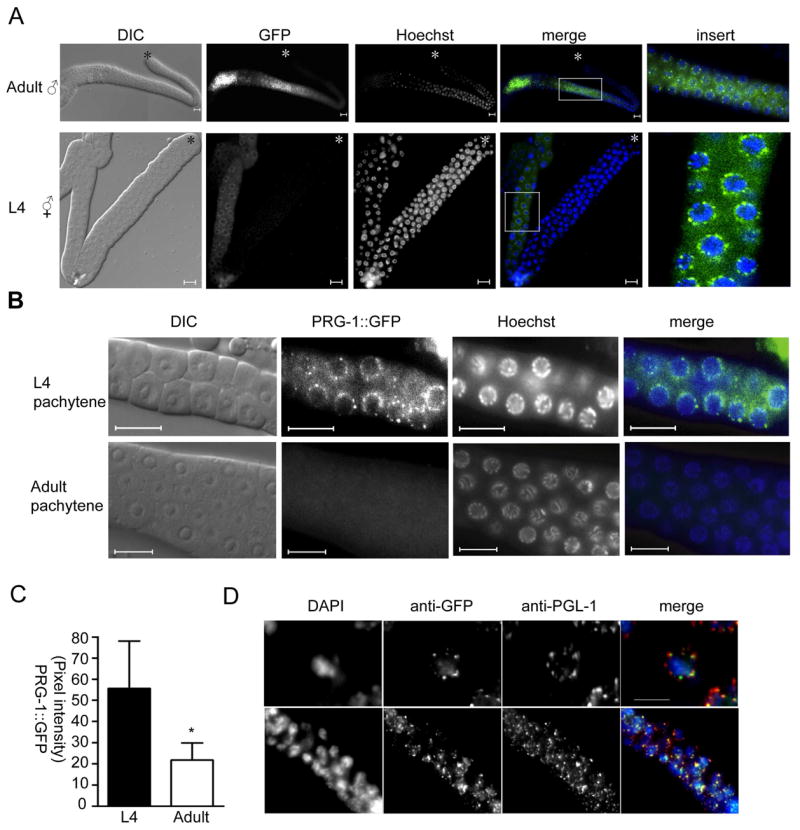
A) Live animals expressing the PRG-1:GFP transgene were exposed to Hoechst dye, and then gonads directly dissected and mounted for viewing. DIC and fluorescent images were collected. Scale bar: 10μm. B) The pachytene region of L4 and adult gonads from PRG-1::GFP transgenic animals are shown. Green, PRG-1::GFP; blue, Hoechst; scale bar, 10μm. C) PRG-1:GFP intensity was measured from equivalent exposures of pachytene germ cells from 10 animals each at L4 and adult stages. Error bar indicates standard deviation. * = Student’s t-test, P<0.01 D) L4 hermaphrodite gonads were stained with anti-GFP (green) and anti-PGL-1 (red). Upper panels: 1000X magnification; scale bar, 5 μm; lower panels: 400X magnification.
C. elegans germ cells possess RNP granules called P granules. Similar to germ granules in other species, P granules likely regulate mRNA stability and/or translation, perhaps to correctly localize germline determinants or suppress somatic fates. To determine whether the perinuclear granular structures containing PRG-1:GFP are P granules, we co-stained the dissected gonads of PRG-1:GFP animals with antibodies to GFP and the constitutive P granule component PGL-1 [17]. We found extensive co-localization between PGL-1 and PRG-1:GFP (Figure 3d), indicating that PRG-1 was indeed localized to P granules. This pattern was consistent with the localization of Piwi family proteins to germ granules in Drosophila, zebrafish and mammals [2, 18–20].
prg-1 mutant males exhibit reduced levels of spermatogenesis-expressed transcripts
To further explore the function of PRG-1 during spermatogenesis, we analyzed prg-1-dependent gene expression. We performed microarray analysis comparing dissected gonads from wild type and prg-1 mutant males grown at 25°C, in triplicate on biologically independent preparations. Examination of differentially expressed genes identified several telling trends (Supplemental Table 1). First, 529 transcripts are down-regulated (≥1.5×, Z-test, p<0.05) in prg-1 mutants, while only 48 are up-regulated, suggesting that PRG-1 is primarily involved in promoting transcript accumulation. Of the down-regulated set, 63% are preferentially expressed during spermatogenesis, based on previous microarray experiments [21], compared to 10% of the up-regulated set. Moreover, prg-1-regulated genes displayed a very strong preference for residence on chromosome IV, as well as a striking exclusion from chromosome X, consistent with the known distribution of genes expressed during spermatogenesis [21, 22]. Consistent with the prg-1 mutant phenotype, the down-regulated genes included several that are known to act relatively late during spermatogenesis. For instance, spe-15 acts during budding of spermatocytes from the residual body, and spe-11, spe-41, and spe-42 all act during spermatid activation [23].
Notably, over 1000 genes known to be expressed during spermatogenesis are not significantly affected by loss of prg-1. This set includes an extremely abundant and highly variable set of genes encoding major sperm protein (MSP). Fourteen probes on the array detecting different msp transcripts had an average fold regulation of 0.95±0.13. The general stability of most spermatogenesis transcripts, and most especially the stability of msp transcripts, in prg-1 mutants argues against the likelihood that the gene expression changes are a downstream consequence of disrupted spermatogenesis, and suggests a more direct role for the differentially regulated genes in causing the mutant phenotype.
prg-1 mutants fail to express 21U-RNAs
We hypothesized that PRG-1 might function in the production of a small RNA species related to piRNAs, based on the role of Piwi-like proteins in other species [1]. A recently described class of small C. elegans RNAs called 21U-RNAs resembles piRNAs in two respects: they have a uridine at the 5′ position, and the corresponding 21U-RNA genes are tightly clustered in the genome [13]. We noted that the 21U-RNA gene clusters are all located on chromosome IV, near regions that encode many sperm-enriched transcripts, and that 21U-RNAs were present at slightly higher frequency in a population containing males [13]. These observations suggested to us that 21U-RNA expression might be regulated by PRG-1.
We therefore isolated RNA from wild type and a set of mutant hermaphrodites with various germline defects. We enriched for RNAs <200bp and used northern analysis to examine the expression profile of the 21U-RNA 21ur-1 [13]. 21ur-1 RNA can be detected in wild type hermaphrodites at L3, L4, and adult stages, but not in prg-1 mutants at those stages (Figure 5a). 21ur-1 was still expressed in prg-2 mutants, but not in prg-1; prg-2 mutants or in glp-1 mutants, which lack germ cells (Figure 5b). A fem-1(lf) mutant lacking sperm but containing oocytes displayed reduced levels of 21ur-1, while a fem-3(gf) mutant that contains sperm and no oocytes had normal levels. The effect of loss of prg-1 activity is specific, as expression of the miRNA let-7 was unaffected. Thus, 21ur-1 displays the expected characteristics of a small RNA expressed during spermatogenesis that requires PRG-1 for its expression. Another 21U-RNA, 21ur-1353, also depended upon prg-1 for its expression (data not shown), indicating that the expression of 21U-RNAs generally require PRG-1.
Figure 5. prg-1 mutants fail to express 21U RNAs.
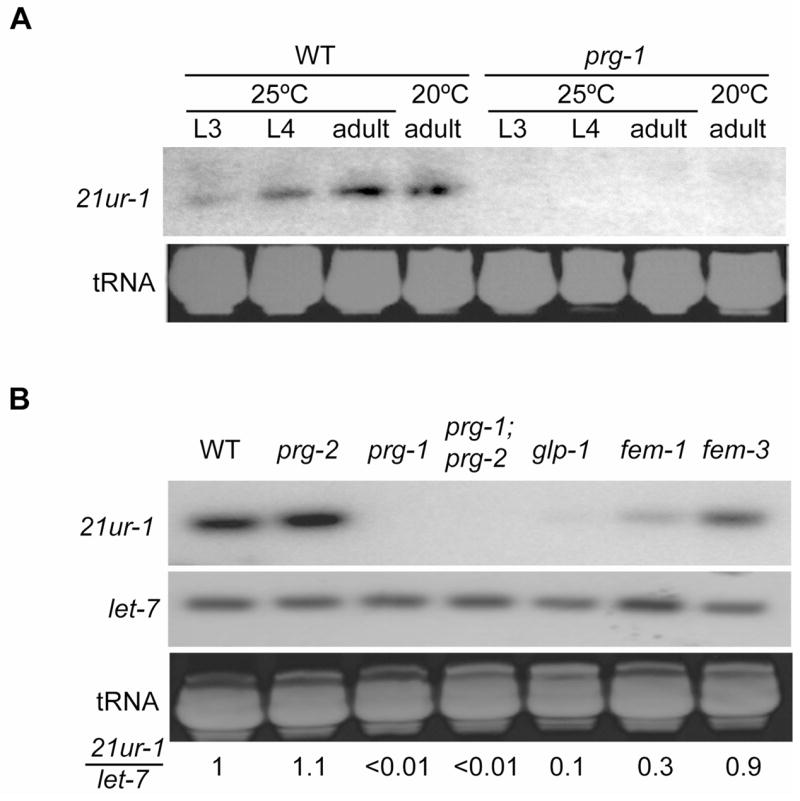
A) Temporal expression of 21ur-1 in wild-type and prg-1(tm872) worms at 20°C and 25°C. B) 21ur-1 RNA expression in mutants with defects in germline development: glp-1 has almost no germ cells; fem-1 has oocytes only, and fem-3 has sperm only. Numbers indicate the relative intensity ratio of 21ur-1 to let-7, after comparison to wild type, which was set to one. Ethidium bromide staining of tRNA shows similar loading levels.
Discussion
Based on our data, we propose that PRG-1 functions in a manner analogous to mammalian Miwi. Both PRG-1 and Miwi proteins achieve peak expression during meiosis ([4]; this work). Like prg-1, miwi mutations cause a relatively late defect at the onset of spermiogenesis (the transformation of round spermatids to mature sperm), whereas the other mammalian Piwi-like proteins, mili and miwi2, act at an earlier stage, in prophase of meiosis I. Moreover, Miwi-associated piRNAs do not have additional sequence matches in the genome and do not correspond to transposons [10, 24], similar to the 21U-RNAs regulated by PRG-1.
This model of PRG-1 function differs from Drosophila, where Piwi, along with related proteins Ago3 and Aubergine, regulate piRNAs to silence transposable elements through an amplification mechanism [2, 25]. 21U-RNAs do not show sequence similarity to transposons and do not have the 10-bp sequence overlap with each other that is characteristic of the Ago3/Aubergine-mediated amplification mechanism [13]. Additionally, our data does not support the notion that PRG-1 and PRG-2 function together to amplify 21U-RNAs, as 21U-RNAs are still present in a prg-2 mutant, and the prg-1 mutant phenotype is not affected by loss of prg-2.
Intriguingly, the genomic sites of 21U-RNA loci clusters are likely conserved between C. elegans and the related species C. briggsae, based on the conserved location of a consensus sequence found upstream of C. elegans 21U-RNAs, which is also found clustered on the syntenic chromosome in C. briggsae. However, in C. briggsae, the sequences downstream of this consensus sequence that likely correspond to 21U-RNAs are not conserved [13], suggesting that the sequence of individual 21U-RNAs is not critical for their function. The location of 21U-RNA loci on chromosome IV and the high density of spermatogenesis-enriched genes on the same chromosome initially suggested that 21U-RNAs might affect expression of these genes in cis. However, many of the spermatogenesis-enriched genes on IV were unaffected in prg-1 mutants, including several msp genes. Moreover, prg-1-regulated genes are located on chromosomes other than IV, and even those on IV are not strongly enriched in the regions from which 21U-RNAs are produced (only about two-fold). These results suggest that PRG-1 and/or 21U-RNAs do not globally or uniformly alter the expression state of the entire chromosome. However, the fact that genes enriched during spermatogenesis are found on the same chromosome as 21U-RNAs suggests that some advantage might exist for the coordinated evolution of a subset of spermatogenesis-enriched genes and 21U-RNA genes.
A temperature-sensitive sterile phenotype, similar to that seen in prg-1 mutants, is a common phenotype of other genes encoding P granule-associated proteins, including PGL-1, GLH-1, MEG-1, and DEPS-1 [17, 26–28]. This commonality suggests that the underlying function of P granules is inherently sensitive to temperature. Two reports indicate that P granules might have a role in RNAi-related processes, as mutations in genes encoding the two P granule proteins PGL-1 and DEPS-1, cause defects in germline RNAi [28, 29]. P granule localization of PRG-1, combined with its requirement for 21U-RNA expression, also suggests that at least one function of P granules is small RNA-mediated regulation of gene expression. However, in contrast to RNAi-based mechanisms, PRG-1 appears to function to promote transcript accumulation, rather than transcript degradation. Thus, multiple, independent small RNA pathways might converge at P granules.
The localization of PRG-1 to P granules on the outside of the nuclear envelope suggests that it does not directly function in the transcriptional activation of spermatogenesis-expressed genes or 21U-RNAs, but we cannot rule out the possibility that PRG-1 is present in the nucleus below detectable levels. Indeed, Drosophila PIWI is found in the nucleus, where it promotes transcriptionally permissive histone modifications, as well as the expression of a nuclear piRNA that silences a master regulatory locus [3]. Alternatively, PRG-1 in P granules could play a role in 21U-RNA processing, as the PIWI domain of PRG-1 retains key residues for “slicing” activity [13]. Although 21U-RNAs appear to be individually transcribed [12], whether or where the nascent 21U-RNA transcript undergoes processing to produce a 21-nt final product is unknown. Finally, 21U-RNAs could associate with PRG-1 in an effector complex of unknown function, perhaps at P granules.
To date, our experiments have established a link between the C. elegans Piwi, PRG-1, and a novel class of 21U-RNAs in the successful completion of spermatogenesis. Future work should address the precise role of 21U-RNAs in mediating PRG-1 function.
Experimental Procedures
Nematode strains and maintenance
C. elegans were grown on NGM as described in Brenner [30]. Strains used: as wild-type variety Bristol, strain N2; LGI, prg-1(tm872); LGIII, glp-1(e2141), unc-32(e189); LGIV, prg-2(tm1094), fem-1(hc17), fem-3(q20gf), BS585. Strains were maintained at 20°C, except for the temperature-sensitive mutants: glp-1, fem-1 and fem-3, which were maintained at 15°C and analyzed at 25°C. Both prg-1(tm872) and prg-2(tm1094) strains were outcrossed to N2 animals six times prior to analysis.
Brood size and fertility test
To measure hermaphrodite brood size, 8–10 self-fertilizing hermaphrodites were plated and permitted to lay eggs at 20°C for two hours before removal. Eggs were either kept at 20°C or shifted to 25°C. The brood counts were performed by placing L4 hermaphrodites singly on plates with daily transfer to a fresh plate until egg production ceased (about three days); the number of progeny on each plate was counted after all worms reached the L4 stage. To measure male fertility, L4 males were placed with fem-1(hc17) or unc-32(q20gf) hermaphrodites at 3:2 ratios and the hermaphrodites were cloned to new plates after 16–24 hr. The number of cross progeny was counted after reaching adulthood. Students’ t-test was used to assess statistical significance.
Sperm activation assay
Sperm activation assays were performed as described previously [15]. Briefly, spermatids were dissected from wild type and prg-1(tm872) males separated from hermaphrodites prior to sexual maturity, grown at 25°C, and analyzed in parallel. Dissections were performed in sperm medium (50 mM HEPES pH 7.8, 50 mM NaCl, 25 mM KCl, 5 mM CaCl2, 1 mM MgSO4) supplemented with 10 mg/ml PVP and 0.2mg/ml of pronase. The percentage of activated spermatids (N>100) was determined after 20 min.
Construction of PRG-1::GFP transgene
prg-1 genomic sequence from ATG to 215 bp downstream of the stop codon, which includes the 3′ untranslated region, was amplified by PCR from genomic DNA using primers 5′-GGGGACAACTTTGTACAAAAAAGTTGGCTTGGCATCTGGAAGTGGTCGCGG -3′ and 5′-GGGGACAACTTTGTACAAGAAAGTTGATGTCCAACTTCCTTACGAAAC-3′, which contain attB1 and attB2 sequences. The resulting PCR product was first cloned into pDONR201 vector (Invitrogen, Carlsbad, CA) using Gateway technology and sequence verified before recombining into pID3.02B [31]. This construct was transformed into worms by microparticle bombardment as described previously [32]. Expression of GFP was examined by DIC and fluorescence microscopy.
Immunofluorescence
Immunostaining of extruded gonads was carried out as in [33]. Briefly, gonads were dissected in M9 buffer, placed on slides, and immersed in liquid nitrogen, before cracking off the coverslip. The slide was placed in ice-cold methanol for 20′, followed by immersion in ice-cold acetone for 10 minutes. We used the following primary antibodies: goat anti-GFP (Rockland, Gilbertsville, PA) diluted 1:500 and mouse anti-PGL-1 (OIC4D; [17]) diluted 1:30. Samples were examined by epifluorescence microscopy and images collected using Zeiss Axiovision.
Small RNA northern blot analysis
Total RNA was isolated from L4 stage wild type, prg-1, prg-2, prg-1;prg-1, glp-1, fem-1 and fem-3 mutant worms. This RNA was enriched for species <200 bp using the mirVana miRNA Isolation Kit (Ambion, Austin, TX) according to the manufacturer’s instructions. All RNA samples were separated by electrophoresis using 15% polyacrylamide (19:1) gels cast in 7M urea and buffered with 20 mM MOPS/NaOH (pH7). After gel electrophoresis, RNAs were transferred to Hybond-N (Amersham, Piscataway, NJ) membrane and cross-linked to membrane by carbodiimide as described previously [34]. Northern hybridization and washes were carried out as in [35]. Probes used to detect RNA levels of 21ur-1 (5′-GCACGGTTAACGTACGTACCA-3′), 21ur-1353 (5′-TACCATCATGTGATGTTCTGA-3′) and let-7 (5′-ACTATACAACCTACTACCTCA-3′) were made using the StarFire Oligonucleotide Labeling System (IDT, Coralville, IA).
Gonad dissection and microarray analysis
Wild-type and prg-1 mutant males were collected by bleaching mated gravid hermaphrodites to collect eggs, which were then hatched in S-basal solution in the absence of food. Starved L1 larvae were cultured with food (bacterial strain OP50) at 25°C for 48 hours, then L4 males were picked to fresh plates and cultured at 25°C for another 16–24 hr before gonad dissection. Total RNA was isolated from 75 dissected male gonads as described previously [36], and amplified using MessageAmpII aRNA kit (Ambion). Amplified mRNA (2μg) was labeled with ULS-Cy3 or ULS-Cy5 dye (Kreatech, Amsterdam) and hybridized to a C. elegans whole genome DNA array from the Washington University Genome Science Center (St Louis, MO). Three independent wild type and prg-1 mutant samples were collected and hybridized. The fold differences from the three experiments were averaged, and a Z test [Z=(observed-expected)/SE] was performed. Genes with up- or down-regulation greater than 1.5 fold, P<0.05 in any given mutant were selected for inclusion. Chromosome bias was assessed using the Chi square test.
Supplementary Material
Figure 4. prg-1- regulated genes exhibit the same chromosome bias as genes expressed during spermatogenesis.

A total of 1300 genes known to be expressed during spermatogenesis and the 529 genes downregulated by prg-1 were mapped to their chromosomal locations and the numbers compared to the expected number given the representation of each chromosome on the microarray. * = p<0.01 (Chi square).
Acknowledgments
The authors thank Haifan Lin for discussions, Aurora Esquela-Kerscher for technical advice, Susan Strome for anti-PGL-1 antibodies, and Shoshei Mitani and the CGC for strains. This work was supported by NIH GM65682 and the Pew Charitable Trust (VR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- 2.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 3.Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- 4.Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 5.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, Nakano T. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 6.Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 8.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 9.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 11.Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 13.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 14.Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- 15.Kelleher JF, Mandell MA, Moulder G, Hill KL, L’Hernault SW, Barstead R, Titus MA. Myosin VI is required for asymmetric segregation of cellular components during C. elegans spermatogenesis. Curr Biol. 2000;10:1489–1496. doi: 10.1016/s0960-9822(00)00828-9. [DOI] [PubMed] [Google Scholar]
- 16.Pellettieri J, Reinke V, Kim SK, Seydoux G. Coordinate activation of maternal protein degradation during the egg-to-embryo transition in C. elegans. Dev Cell. 2003;5:451–462. doi: 10.1016/s1534-5807(03)00231-4. [DOI] [PubMed] [Google Scholar]
- 17.Kawasaki I, Shim YH, Kirchner J, Kaminker J, Wood WB, Strome S. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell. 1998;94:635–645. doi: 10.1016/s0092-8674(00)81605-0. [DOI] [PubMed] [Google Scholar]
- 18.Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RH, Hannon GJ, Draper BW, Ketting RF. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 19.Megosh HB, Cox DN, Campbell C, Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol. 2006;16:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 20.Kotaja N, Lin H, Parvinen M, Sassone-Corsi P. Interplay of PIWI/Argonaute protein MIWI and kinesin KIF17b in chromatoid bodies of male germ cells. J Cell Sci. 2006;119:2819–2825. doi: 10.1242/jcs.03022. [DOI] [PubMed] [Google Scholar]
- 21.Reinke V, Gil IS, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–323. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- 22.Reinke V, Smith HE, Nance J, Wang J, Van Doren C, Begley R, Jones SJ, Davis EB, Scherer S, Ward S, Kim SK. A global profile of germline gene expression in C. elegans. Mol Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- 23.L’Hernault SW. Spermatogenesis. WormBook. 2006:1–14. doi: 10.1895/wormbook.1.85.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci U S A. 2006;103:13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 26.Kuznicki KA, Smith PA, Leung-Chiu WM, Estevez AO, Scott HC, Bennett KL. Combinatorial RNA interference indicates GLH-4 can compensate for GLH-1; these two P granule components are critical for fertility in C. elegans. Development. 2000;127:2907–2916. doi: 10.1242/dev.127.13.2907. [DOI] [PubMed] [Google Scholar]
- 27.Leacock SW, Reinke V. MEG-1 and MEG-2 are embryo-specific P-granule components required for germline development in Caenorhabditis elegans. Genetics. 2008;178:295–306. doi: 10.1534/genetics.107.080218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spike CA, Bader J, Reinke V, Strome S. DEPS-1 promotes P-granule assembly and RNA interference in C. elegans germ cells. Development. 2008;135:983–993. doi: 10.1242/dev.015552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robert VJ, Sijen T, van Wolfswinkel J, Plasterk RH. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev. 2005;19:782–787. doi: 10.1101/gad.332305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reese KJ, Dunn MA, Waddle JA, Seydoux G. Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol Cell. 2000;6:445–455. doi: 10.1016/s1097-2765(00)00043-5. [DOI] [PubMed] [Google Scholar]
- 32.Praitis V. Creation of transgenic lines using microparticle bombardment methods. Methods Mol Biol. 2006;351:93–107. doi: 10.1385/1-59745-151-7:93. [DOI] [PubMed] [Google Scholar]
- 33.Strome S, Wood WB. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell. 1983;35:15–25. doi: 10.1016/0092-8674(83)90203-9. [DOI] [PubMed] [Google Scholar]
- 34.Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007;35:e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- 36.Chi W, Reinke V. Promotion of oogenesis and embryogenesis in the C. elegans gonad by EFL-1/DPL-1 (E2F) does not require LIN-35 (pRB) Development. 2006;133:3147–3157. doi: 10.1242/dev.02490. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.