Abstract
Resveratrol (Resv), a natural occurring phytolexin present in grapes and other foods, possesses chemopreventive effects revealed by its striking modulation of diverse cellular events associated with tumor initiation, promotion and progression. Catechol estrogens generated in the metabolism of estrogens are oxidized to catechol quinones that react with DNA to form predominantly depurinating estrogen-DNA adducts. This event can generate the mutations responsible for cancer initiation. In this regard, Resv acts as both an antioxidant and an inducer of the phase II enzyme NAD(P)H:quinone oxidoreductase 1 (NQO1). In this report, we present the effects of Resv on the metabolism of estrogens in normal breast epithelial cells (MCF-10F) treated with 4-hydroxyestradiol (4-OHE2) or estradiol-3,4-quinone (E2-3,4-Q). Resv induced NQO1 in a dose- and time-dependent manner, but did not affect the expression of catechol-O-methyltransferase. Ultraperformance liquid chromatography/tandem mass spectrometry was used to determine the effects of Resv on estrogen metabolism. Preincubation of the cells with Resv for 48 h decreased the formation of depurinating estrogen-DNA adducts from 4-OHE2 or E2-3,4-Q and increased formation of methoxycatechol estrogens. When Resv was also present with the 4-OHE2 or E2-3,4-Q, even greater increases in methoxycatechol estrogens were observed, and the DNA adducts were undetectable. We conclude that Resv can protect breast cells from carcinogenic estrogen metabolites, suggesting that it could be used in breast cancer prevention.
Introduction
Resveratrol (3,5,4′-trihydroxystilbene, Resv), which is present in red wine and grapes, has been widely studied for its antioxidant and biological effects [1]. It has been shown to induce anticancer effects by modulating several cellular pathways related to cancer initiation, promotion and progression [2–4]. Resv inhibits cell proliferation and induces apoptosis to suppress and prevent a variety of tumors [5]. Resv stops the proliferation of human breast cancer MCF-7 cells and non-transformed MCF-10F cells treated with estrogens [6–8]. The molecular mechanisms underlying these activities are not completely understood.
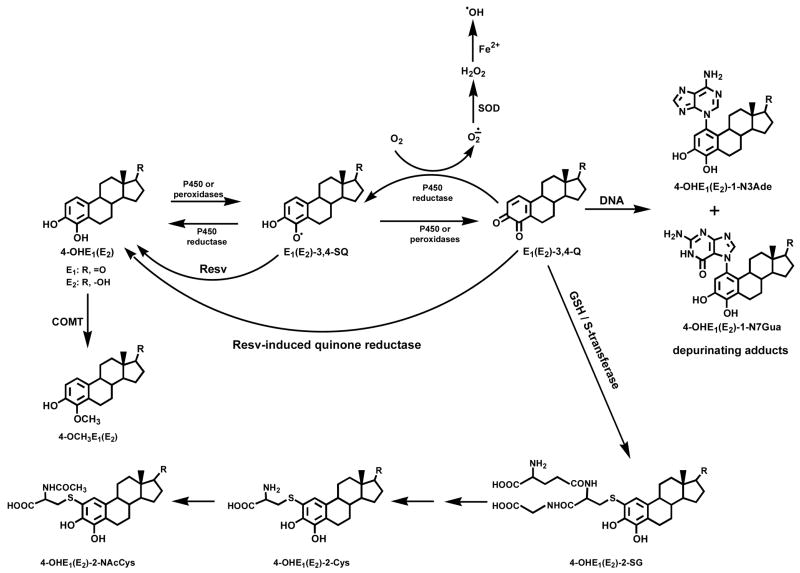
Estrogens are metabolized to catechol estrogens and then to catechol estrogen quinones, which can act as endogenous carcinogens [9–12] (Figure 1). These quinones react with DNA to form predominantly the depurinating 4-hydroxyestrone(estradiol) [OHE1(E2)-1-N3Ade] and 4-OHE1(E2)-1-N7Gua adducts [9–13]. Error-prone repair of the abasic sites left in the DNA by the depurination is thought to generate the mutations that are critical for cell proliferation and cancer initiation [14–16]. Low catechol-O-methyltransferase (COMT) activity and increased formation of DNA adducts mediated through the catechol estrogen quinones may be contributory factors in the initiation of breast cancer [17,18]. In fact, Gaikwad et. al. have shown that in healthy women the levels of estrogen-DNA adducts in urine are low and the levels of methoxylated estrogen metabolites and conjugates are high [11]. In contrast, higher levels of DNA adducts and lower levels of methoxylated estrogen metabolites and conjugates are present in the urine of high-risk women and women with breast cancer. Based on this information, it could be argued that lowering the depurinating DNA adduct levels may lead to reduction in the risk of developing breast cancer. This could be achieved by methylation of 4-OHE1(E2) by COMT, reduction of estrogen quinones by quinone oxidoreductase 1 or 2 (NQO1 or NQO2 [19,20]), reduction of estrogen semiquinones by an antioxidant [21,22] and/or conjugation of estrogen quinones with glutathione or derivatives [9,11]. In a preliminary in vitro study, we tested the ability of four antioxidant dietary supplements, N-acetylcysteine, Resv, melatonin and reduced lipoic acid to inhibit formation of estrogen-DNA adducts [22]. Resv inhibited best the formation of depurinating adducts when enzyme-oxidized 4-OHE2 was reacted with DNA. Only the antioxidant effects of Resv, presumably the reduction of E2-3,4-semiquinone to 4-OHE2, were evaluated in this study. The in vivo preventive effects of Resv are a combination of its antioxidant properties and its ability to induce NQO1 [23,24] and modulate CYP1B1 [24,25] (Figure 1).
Figure 1.
Mechanisms by which Resv can prevent estrogen-initiated breast cancer.
The effects of Resv on the metabolism of estrogens in general, and on the formation of depurinating DNA adducts in particular, have never been investigated. In this article we show that Resv, indeed, completely inhibits estrogen-DNA adduct formation in human breast epithelial cells (MCF-10F) through its antioxidant properties, as well as induction of the phase II enzyme NQO1. This study is the first report on the metabolism of E2-3,4-Q and 4-OHE2 by MCF-10F cells in the presence of Resv, and it provides evidence that Resv may be a new hope for prevention of breast cancer.
Materials and Methods
Chemicals and Reagents
4-OHE2 and all standards were synthesized in our laboratory, as previously described [26–29]. For quinone treatments, the E2-3,4-Q was freshly synthesized as previously described [10]. Resv and all other chemicals were purchased from Sigma (St. Louis, MO).
Cell culture and treatment
MCF-10F cells were obtained from the ATCC (Rockville, MD) and cultured in DMEM and Ham’s F12 media (Mediatech, Inc.) with 20 ng/ml epidermal growth factor, 0.01 mg/ml insulin, 500 ng/ml hydrocortisone, 5% horse serum and 100 μg/ml penicillin/streptomycin mixture. Estrogen-free medium was prepared in phenol red-free DME/F12 medium with charcoal-stripped FBS (HyClone, Logan, UT). Cell viability was determined by the MTT [3,(4,5-dimethylthiazole-2-yl)-2,5-diphenyltrazolium bromide] assay [30].
The MCF-10F cells (0.75 × 105 cells) were seeded for 48 h in estrogen-containing medium. The medium was changed to estrogen-free medium and the cells were grown for another 72 h. To see the effect of Resv on NQO1 and COMT induction, cells were first pre-incubated for 48 h with 25 μM Resv, washed with PBS and, after adding fresh medium, treated with E2-3,4-Q or 4-OHE2 (1, 10 or 30 μM) for 24 h. To check the combined induction and antioxidant effects, cells were pre-incubated with 25 μM Resv for 48 h and then again with fresh 25 μM Resv after changing medium and while treating with 4-OHE2 or E2-3,4-Q (1, 10 or 30 μM). To examine the dose-dependency of the Resv effects, cells were treated with 1, 10 or 30 μM Resv, while keeping the 4-OHE2 constant at 10 μM. To keep the concentration of DMSO (0.001%) the same in all experiments, different stock solutions of 4-OHE2 (1–30 mM) and Resv (0.1–30 mM) were prepared. Media from four T-150 flasks of MCF-10F cells treated with an equal amount of organic solvent were used as controls.
Analysis of estrogen-DNA adducts
Following the treatments, the media were harvested, supplemented with 2 mM ascorbic acid (to prevent possible decomposition of the compounds) and processed immediately. Sample preparation and analysis by ultraperformance liquid chromatography/tandem mass spectrometry (UPLC-MS/MS) were carried out as follows:
i. Sample preparation
Culture media from four flasks (40 mL) were processed by passing through Varian C8 Certify II cartridges (Varian, Harbor City, CA). The cartridges were pre-equilibrated by sequentially passing 1 ml of methanol, distilled water, and potassium phosphate buffer (100 mM, pH 8) through them. Culture media were adjusted with 1 ml of 1 M potassium phosphate buffer to pH 8.0 and passed through the cartridges. After washing with 200 μl of the phosphate buffer, the analytes were eluted with 1 ml of elution buffer [methanol:acetonitrile:water: trifluoroacetic acid (8:1:1:0.1)] and processed as previously described [18].
ii. UPLC-MS/MS
Samples were analyzed on an Acquity UPLC system connected to a MicroMass QuattroMicro triple quadrupole mass spectrometer (Waters, Milford, MA) [11]. The 10-μl injections were carried out on a Waters Acquity UPLC BEHC18 column (1.7 μm, 1 × 100 mm). The instrument was operated in the positive electrospray ionization mode. All aspects of system operation, data acquisition and processing were controlled using QuanLynx v4.0 software (Waters). The column was eluted starting with 5% acetonitrile in water (0.1% formic acid) for 1 min at a flow rate of 150 μl/min; then a gradient to 55% acetonitrile in 10 min was run. Ionization was achieved using the following settings: capillary voltage 3 kV; cone voltage 15–40 V; source block temperature 100 °C; desolvation temperature 200 °C, with a nitrogen flow of 400 l/h. Nitrogen was used as both the desolvation and auxiliary gas. Argon was used as the collision gas. Three-point calibration curves were run for each standard. Triplicate samples were analyzed for each data point.
The UPLC-MS/MS limit of detection for the estrogen derivatives was in the range of 0.002 to 0.06 pmole, which was well below the experimental values obtained for all the compounds, and the standard error for this analysis, calculated from triplicate analyses was 0.00002–0.0066 pmole.
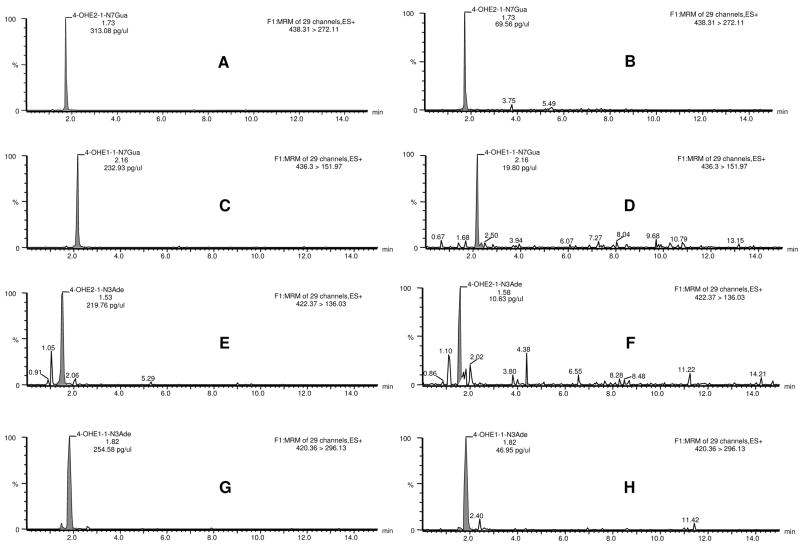
UPLC-MS/MS both identified and quantified the estrogen derivatives (Figure 2). For example, in one experiment the adducts were detected as single peaks and pretreatment of the cells with Resv decreased the level of 4-OHE2-1-N7Gua from 313 to 70 pg/μl (Figure 2A and B), 4-OHE1-1-N7Gua from 233 to 20 pg/μl (Figure 2C and D), 4-OHE2-1-N3Ade from 220 to 11 pg/μl (Figure 2E and F) and 4-OHE1-1-N3Ade from 255 to 47 pg/μl (Figure 2G and H).
Figure 2.
UPLC-MS/MS chromatograms of depurinating DNA adducts: 4-OHE2-1-N7Gua (A, B), 4-OHE1-1-N7Gua (C, D), 4-OHE2-1-N3Ade (E, F) and 4-OHE1-1-N3Ade (G, H) in medium from MCF-10F cells treated with 10 μM 4-OHE2 alone (A, C, E and G) or with 10 μM 4-OHE2 plus 10 μM Resv (B, D, F and H), respectively.
Immunoblotting
Comprehensive analysis of the expression patterns of the protective phase II enzymes (COMT and NQO1) from control and treated MCF-10F cells at different time points was conducted by immunoblotting. Whole cell lysates were prepared by suspending cells in RIPA buffer with protease inhibitor and lysed by three freeze-thaw cycles. After removing cellular debris by centrifugation, protein concentrations were determined by using a BCA protein assay kit (Pierce Biotechnology, Rockford, IL). The proteins were separated by SDS-PAGE and transferred to PVD membranes for immunodetection as previously described [17]. Dilutions of the primary anti-COMT (Chemicon International, Temecula, CA) and NQO1 (Abcam, Temecula, CA) antibodies were made in blocking solution (5% non-fat dry milk in PBS). The blots were incubated for 3 h with primary antibody and for 1 h with secondary antibody coupled to horseradish peroxidase at room temperature. After each step, blots were washed with PBST (PBS and 0.1% Tween-20), incubated with ECL solution (Amersham Biotech, Piscataway, NJ) for 1 min, and visualized with radiographic film. The intensities of the bands were quantified by Alpha DigiDoc 1201 (Alpha Innotech, San Leandro, CA).
Determination of NQO1 activity by UPLC-MS/MS
Cellular NQO1 (25 μg of whole cell lysate protein) from control and 25 μM Resv-treated MCF-10F cells was prepared. Triplicate enzyme reactions were carried out in a final volume of 100 μl of 0.1 M sodium phosphate, pH 7. We used freshly synthesized E2-3,4-Q as the substrate and 25 μM NADH as the co-factor in the buffer system containing 0.7 mg/ml BSA. Recombinant NQO1 protein (10 units, Sigma, St. Louis, MO) in the reaction system served as the positive control. Inhibition studies were carried out with inclusion of a potent inhibitor of NQO1, dicumarol (10 μM), in the assay mixture. Control assays were carried out without enzyme or cofactor. The reaction mixture was pre-incubated for 3 min at room temperature prior to the addition of substrate. The reaction was then initiated by adding 5 μl of 100 μM E2-3,4-Q in acetonitrile and terminated after 27 min at 37 °C by adding 100 μl methanol. Following centrifugation to precipitate proteins, the supernatant was passed through a 5,000 M.W. cut-off filter (Millipore, Billerica, MA), and 100 μl of each sample was analyzed for the product, 4-OHE2, by UPLC-MS/MS.
Determination of COMT activity by UPLC-MS/MS
Cellular COMT from control and 25 μM Resv (0–72 h)-treated MCF-10F cells was prepared as a cytosolic extract. Enzyme reactions were carried out in a final volume of 250 μl of 0.1 M sodium phosphate, pH 7.8, containing 1 mM DTT, 5 mM MgCl2, 100 μg cytosolic protein, 100 μM 4-OHE2, and 200 μM S-adenosylmethionine (SAM, saturating). The incubation mixture, except for the substrate, was pre-incubated for 3 min at 37 °C. Then, the reaction was initiated by adding 5 μl of 4-OHE2 in DMSO and terminated after 27 min by adding 25 μl of 4 M perchloric acid. Following centrifugation to precipitate proteins, the supernatant (245 μl) was passed through a 5,000 M.W. cut-off filter (Millipore, Billerica, MA), and 10 μl of each sample was analyzed by UPLC-MS/MS. Each analysis was conducted in triplicate.
Statistical Analysis
The statistical significance of the results was determined by Student’s t-test and ANOVA analysis by using SAS and GraphPad Prism 4.0 software. P<0.05 was considered significant. All data were obtained from triplicate experiments.
Results
Effect of Resv on NQO1 and COMT expression and activity
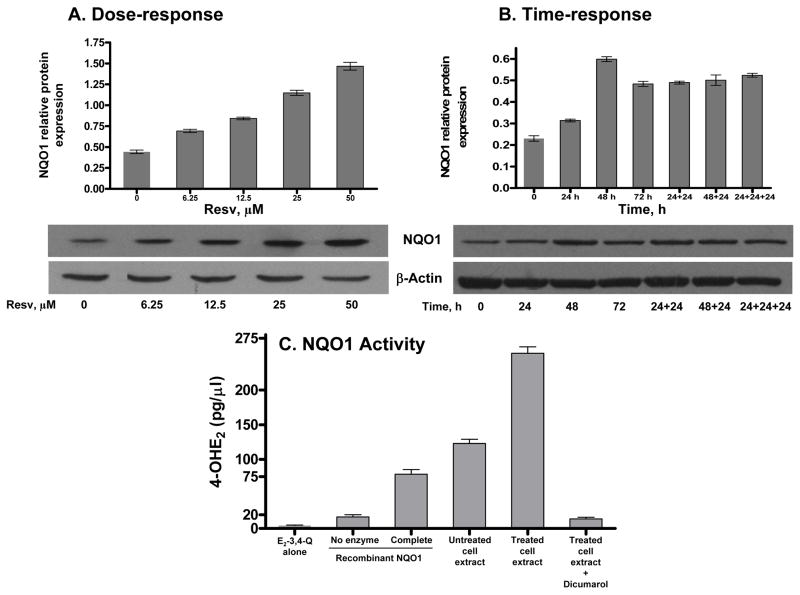
To see the effect of Resv on NQO1, MCF-10F cells were treated with various concentrations of Resv (0–50 μM) for 48 h and the level of NQO1 protein was determined by immunoblotting. Densitometric analyses showed that Resv dose- and time-dependently induced the expression of NQO1 (Figure 3). Resv (0–50 μM) induced NQO1 protein 2–3 fold (Figure 3A). A time course study using 25 μM Resv for 24–72 h demonstrated that induction of NQO1 protein occurred by 24 h and peaked at 48 h, while its level fell slightly by 72 h (Figure 3B). Treatment of the cells with 25 μM Resv 2 or 3 times for 24 h each time or for 48 h + 24 h, resulted in approximately the same amount of NQO1 protein (Figure 3B). The catalytic activity of cellular NQO1 in control and Resv-treated cells was determined by an in vitro enzyme activity assay using UPLC-MS/MS. The levels of the reaction product, 4-OHE2, in treated cells were significantly greater than in the untreated cells (p< 0.05), as determined by ANOVA (Figure 3C). The level of 4-OHE2 in reaction mixtures containing recombinant NQO1 protein, the positive control, was 78.5±11.5 pg/μl, whereas the cellular proteins from 25 μM Resv-treated cells showed a 2-fold increase in enzymatic activity compared with that from control cells (4-OHE2= 123±10.3 pg/μl). The reduction of E2-3,4-Q to 4-OHE2 by NQO1 was subsequently confirmed by adding its potent inhibitor dicumarol (10 μM) to the assay mixture, which almost completely inhibited the formation of 4-OHE2. Control assays carried out without enzyme or cofactor had lower levels of 4-OHE2 (4.3±1.2 pg/μl). In the presence of NADH, the level was slightly higher, 16.7 ± 5.7 pg/μl. The increased NQO1 activity in Resv-treated cells corresponded well with the induction in NQO1 protein.
Figure 3.
Induction of NQO1 expression and activity by Resv. Each lane contains 25 μg of cell lysate. Intensity of the bands was quantified and normalized as described in Methods and Materials (n=3). (A) NQO1 expression in MCF-10F cells treated with increasing concentrations of Resv for 48 h. (B) Cells were treated with one 25 μM dose of Resv for various times (24–72 h) or with multiple 25 μM doses of Resv for the indicated time periods. (C) Cellular NQO1 (25 μg) enzymatic activity was determined by UPLC-MS/MS. The levels of the reaction product, 4-OHE2, in treated cells were significantly different from those in the untreated cells, p< 0.05 as determined by ANOVA.
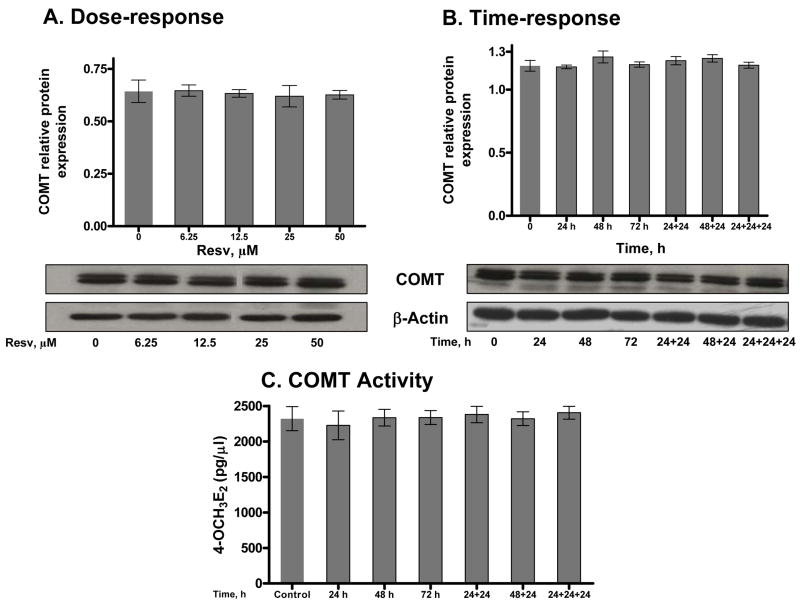
Although the levels of methoxy catechol estrogens increased in the culture medium when Resv was present, it did not change the COMT protein level in a time- or dose-dependent manner (Figure 4A and 4B). To investigate the activity of COMT in Resv-treated and untreated MCF-10F cells, and to validate the findings from our immunoblotting experiments, catalytic activity was measured by LC/MS using 4-OHE2 as the substrate (Figure 4C). No change in COMT activity was observed. This is not consistent with earlier reports in which COMT enzyme levels were not changed, but catalytic activity was affected [31].
Figure 4.
Effect of Resv on COMT expression and activity. Each lane contained 25 μg of cell lysate. Intensity of the bands was quantified and normalized as described in Methods and Materials (n=3). (A) COMT expression in MCF-10F cells treated with increasing concentrations of Resv for 48 h. (B) Cells were treated with one 25 μM dose of Resv for various times (24–72 h) or with multiple 25 μM doses of Resv for the indicated time periods. (C) Cellular COMT (100 μg) enzymatic activity was determined by UPLC-MS/MS as described in Methods and Materials. The levels of the reaction product, 4-OCH3E2, were similar in Resv treated and untreated cells, p< 0.05 as determined by ANOVA.
Taken together, our results clearly demonstrate that Resv induces NQO1 protein in a time and dose dependent manner, whereas COMT protein and activity remain unaffected.
Treatment of cells
MCF-10F cell treatments with either 4-OHE2 or E2-3,4-Q were divided into three groups. First, the control group was treated for 24 h with 4-OHE2 or E2-3,4-Q alone. In the second group, to ascertain the contribution of induced NQO1, cells were pretreated with 25 μM Resv for 48 h, followed by 4-OHE2 or E2-3,4-Q for 24 h. In the third group, pretreatment with 25 μM Resv was followed by treatment with 4-OHE2 or E2-3,4-Q plus fresh 25 μM Resv to determine its antioxidant effect.
Effect of Resv on 4-OHE2 metabolism in MCF-10F cells
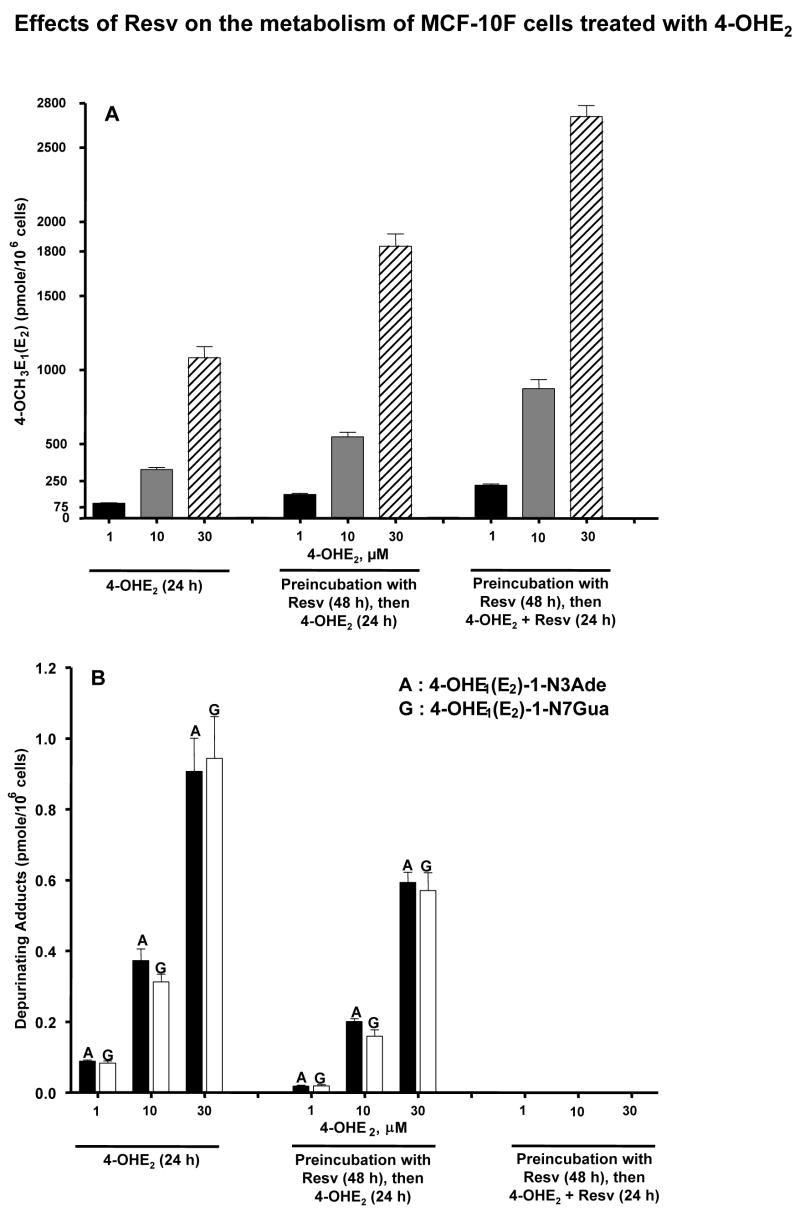
UPLC-MS/MS analysis of the cell culture media from the control group, which was treated with 1, 10 or 30 μM 4-OHE2, showed that 4-OHE2 is predominantly metabolized to 4-methoxycatechol estrogens, with small amounts of thiol conjugates and depurinating estrogen-DNA adducts in a dose-dependent manner (Table 1, Figure 5). Both adducts were present in the media at concentrations ranging from 10 pM to 470 pM, depending on the concentration of 4-OHE2 used to treat the cells.
Table 1.
Incubation of MCF-10F cells with 4-OHE2 in the presence or absence of 25 μM Resv1
| Detected Compounds2 | 4-OHE2 treatment | 48 h Resv Pre-incubation 4-OHE2 (24 h) | 48 h Resv Pre-incubation 4-OHE2 + Resv (24 h) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 10 | 30 | 1 | 10 | 30 | 1 | 10 | 30 | |
| pmol/106 cells | |||||||||
| 4-OHE1(E2) | 0.16 ± 0.02 | 0.91 ± 0.11 | 2.45 ± 0.51 | 0.22 ± 0.01 | 1.20 ± 0.19 | 2.97 ± 0.40 | 0.25 ± 0.03 | 1.48 ± 0.16 | 3.74 ± 0.79 |
| 4-OCH3E1(E2) | 102 ± 4 | 327 ± 27 | 1081 ± 131 | 158 ± 15 | 548 ± 52 | 1832 ± 146 | 223 ± 14 | 874 ± 103 | 2708 ± 130 |
| 4-OHE1(E2)-2-SG | 0.07 ± 0.01 | 0.55 ± 0.16 | 0.89 ± 0.13 | 0.16 ± 0.02 | 1.12 ± 0.16 | 1.25 ± 0.08 | 0.05 ± 0.01 | 0.46 ± 0.08 | 0.65 ± 0.13 |
| 4-OHE1(E2)-2-Cys | 0.33 ± 0.05 | 0.80 ± 0.18 | 3.53 ± 0.51 | 0.50 ± 0.05 | 1.31 ± 0.12 | 4.74 ± 0.26 | 0.23 ± 0.01 | 0.63 ± 0.08 | 2.60 ± 0.47 |
| 4-OHE1(E2)-2-NAcCys | 0.37 ± 0.08 | 0.87 ± 0.12 | 2.14 ± 0.13 | 0.50 ± 0.13 | 1.42 ± 0.11 | 2.73 ± 0.11 | 0.23 ± 0.01 | 0.64 ± 0.07 | 1.56 ± 0.19 |
| 4-OHE1(E2)-1-N3Ade | 0.07 ± 0.01 | 0.34 ± 0.09 | 0.94 ± 0.15 | 0.02 ± 0.01 | 0.20 ± 0.01 | 0.59 ± 0.05 | nd3 | nd | nd |
| 4-OHE1(E2)-1-N7Gua | 0.06 ± 0.01 | 0.26 ± 0.04 | 0.93 ± 0.14 | 0.02 ± 0.01 | 0.16 ± 0.03 | 0.57 ± 0.09 | nd | nd | nd |
4-OHE2 was incubated with MCF-10F cells at 37 °C for 24 h in the presence or absence of Resv.
The compounds were identified and quantified by UPLC-MS/MS, and values are an average of three replicates.
Not detected.
Figure 5.
Profile of 4-OCH3E1(E2) metabolites and depurinating DNA adducts in MCF-10F cells treated with Resv and/or 4-OHE2. (A) Levels of 4-OCH3E1(E2) in culture medium of cells treated with 4-OHE2 with or without Resv. (B) Levels of depurinating DNA adducts in culture medium from cells treated with 4-OHE2 with or without Resv. The levels of DNA adducts in Resv-treated cells were significantly lower than those in the cells not treated with Resv, p< 0.05 as determined by ANOVA.
When the MCF-10F cells were preincubated with Resv, a significant increase in 4-OCH3E1(E2) (~ 1.5-fold) was observed (Table 1, Figure 5A). When preincubation with Resv was followed by incubation with both 4-OHE2 and Resv, the level of 4-OCH3E1(E2) was further increased (Table 1, Figure 5A). The level obtained with Resv preincubation is compatible with oxidation of 4-OHE2 to quinone and subsequent reduction of the quinone back to catechol, catalyzed by Resv-induced NQO1. The enhancement in 4-OCH3E1(E2) when Resv is included with 4-OHE2 is interpreted as the reduction of E1(E2)-3,4-semiquinone back to catechol, with subsequent COMT-catalyzed methylation (Figures 1 and 5A).
The levels of GSH, Cys and NAcCys conjugates remained low under all three sets of experimental conditions (Table 1). The levels of the two depurinating DNA adducts were decreased when the cells were preincubated with Resv and became undetectable when the cells were preincubated with Resv and incubated with 4-OHE2 plus Resv (Table 1, Figure 5B).
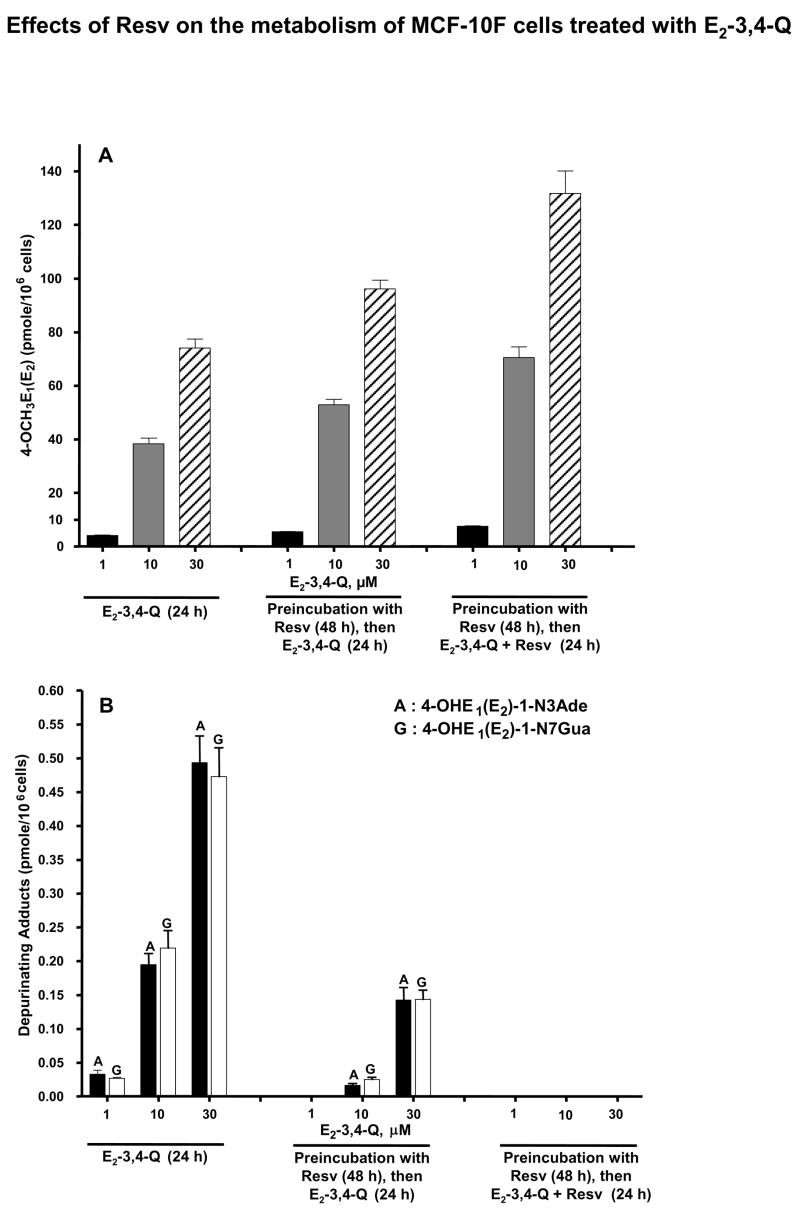
Effect of Resv on E2-3,4-Q metabolism in MCF-10F cells
Much lower levels of 4-OCH3E1(E2) were observed when the MCF-10F cells were incubated with E2-3,4-Q (Table 2) compared to 4-OHE2 (Table 1). When the cells were preincubated with Resv, the level of 4-OCH3E1(E2) increased about 30%, and inclusion of Resv with E2-3,4-Q led to a doubling of the levels of 4-OCH3E1(E2) (Table 2, Figure 6A). The levels of thiol conjugates showed a small dose-response, but were unaffected by preincubation or incubation with Resv (Table 2). The levels of the depurinating DNA adducts were lower with E2-3,4-Q than with 4-OHE2 (Table 1), and they decreased dramatically with Resv preincubation, once again becoming undetectable when the cells were preincubated with Resv and incubated with E2-3,4-Q + Resv (Table 2, Figure 6B).
Table 2.
Incubation of MCF-10F cells with E2-3,4-Q with or without 25 μM Resv1
| Detected Compounds2 | E2-3,4-Q treatment | 48 h Resv Pre-incubation E2-3,4-Q (24 h) | 48 h Resv Pre-incubation E2-3,4-Q + Resv (24 h) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 10 | 30 | 1 | 10 | 30 | 1 | 10 | 30 | |
| pmol/106 cells | |||||||||
| 4-OHE1(E2) | 0.30 ± 0.04 | 0.75 ± 0.09 | 1.34 ± 0.16 | 0.22 ± 0.07 | 0.70 ± 0.09 | 1.17 ± 0.10 | 0.18 ± 0.09 | 1.48 ± 0.16 | 1.02 ± 0.13 |
| 4-OCH3E1(E2) | 4.08 ± 0.42 | 38.4 ± 3.5 | 70.0 ± 5.7 | 5.51 ± 0.22 | 52.8 ± 3.4 | 96.0 ± 5.7 | 7.57 ± 0.28 | 70.5 ± 6.8 | 132 ± 14 |
| 4-OHE1(E2)-2-SG | 0.05 ± 0.01 | 0.28 ± 0.05 | 0.57 ± 0.05 | 0.07 ± 0.01 | 0.34 ± 0.06 | 0.73 ± 0.07 | 0.06 ± 0.01 | 0.29 ± 0.02 | 0.61 ± 0.03 |
| 4-OHE1(E2)-2-Cys | 0.36 ± 0.06 | 0.84 ± 0.17 | 2.60 ± 0.18 | 0.46 ± 0.04 | 1.25 ± 0.09 | 3.41 ± 0.23 | 0.39 ± 0.06 | 0.95 ± 0.07 | 2.92 ± 0.06 |
| 4-OHE1(E2)-2-NAcCys | 0.10 ± 0.01 | 0.57 ± 0.06 | 1.26 ± 0.07 | 0.16 ± 0.01 | 1.09 ± 0.06 | 1.61 ± 0.06 | 0.12 ± 0.01 | 0.86 ± 0.05 | 1.39 ± 0.05 |
| 4-OHE1(E2)-1-N3Ade | 0.03 ± 0.01 | 0.19 ± 0.03 | 0.49 ± 0.07 | nd3 | 0.02 ± 0.01 | 0.14 ± 0.03 | nd | nd | nd |
| 4-OHE1(E2)-1-N7Gua | 0.03 ± 0.002 | 0.22 ± 0.04 | 0.47 ± 0.07 | nd | 0.03 ± 0.01 | 0.14 ± 0.02 | nd | nd | nd |
E2-3,4-Q was incubated with MCF-10F cells at 37 °C for 24 h in the presence or absence of Resv.
The compounds were identified and quantified by UPLC-MS/MS, and values are an average of three replicates.
Not detected.
Figure 6.
Profile of 4-OCH3E1(E2)-3,4-Q metabolites and depurinating DNA adducts in MCF-10F cells co-treated with Resv and/or E2-3,4-Q. (A) Levels of 4-OCH3E1(E2) in culture medium of cells treated with E2–3,4-Q with or without Resv. (B) Levels of depurinating DNA adducts in culture medium of cells treated with E2–3,4-Q with or without Resv. The levels of DNA adducts in Resv-treated cells were significantly lower than those in the cells not treated with Resv, p< 0.05 as determined by ANOVA.
Dose-Response of Resv
MCF-10F cells were treated with 10 μM 4-OHE2 and varying concentrations (0, 1, 10, 30 μM) of Resv, and the metabolic profile was analyzed by UPLC-MS/MS. There was a dose-dependent increase in the level of 4-methoxycatechol estrogens, with a small increase at equimolar 4-OHE2 and Resv and a 3-fold increase when the concentration of Resv was 3-times that of 4-OHE2 (Table 3). There was a marginal, dose-dependent decrease in the corresponding levels of the 4-OHE1(E2)-2-SG, 4-OHE1(E2)-2-Cys and 4-OHE1(E2)-2-NAcCys conjugates compared with control (Table 3). Significant inhibition of 4-OHE1(E2)-1-N3Ade and 4-OHE1(E2)-1-N7Gua adduct formation was observed with increased Resv concentrations and the adducts were not detected with 30 μM Resv (Table 3).
Table 3.
Incubation of MCF-10F cells with 10 μM 4-OHE2 with or without Resv1
| Detected Compounds2 | 4-OHE2/Resv | 4-OHE2/Resv | 4-OHE2/Resv | 4-OHE2/Resv |
|---|---|---|---|---|
| 10:0 | 10:1 | 10:10 | 10:30 | |
| pmol/106 cells | ||||
| 4-OHE1(E2) | 0.86 ± 0.15 | 1.21 ± 0.03 | 1.06 ± 0.08 | 1.73 ± 0.16 |
| 4-OCH3E1(E2) | 326 ± 36 | 373 ± 30 | 430 ± 25 | 990 ± 87 |
| 4-OHE1(E2)-2-SG | 0.68 ± 0.08 | 0.64 ± 0.05 | 0.60 ± 0.03 | 0.49 ± 0.04 |
| 4-OHE1(E2)-2-Cys | 1.07 ± 0.07 | 0.97 ± 0.07 | 0.88 ± 0.11 | 0.69 ± 0.11 |
| 4-OHE1(E2)-2-NAcCys | 0.97 ± 0.05 | 0.89 ± 0.07 | 0.85 ± 0.06 | 0.75 ± 0.04 |
| 4-OHE1(E2)-1-N3Ade | 0.31 ± 0.05 | 0.29 ± 0.05 | 0.17 ± 0.01 | nd3 |
| 4-OHE1(E2)-1-N7Gua | 0.24 ± 0.03 | 0.22 ± 0.03 | 0.13 ± 0.02 | nd |
10μM 4-OHE2 was incubated with MCF-10F cells at 37 °C for 24 h in the presence or absence of Resv at different ratios.
The compounds were identified and quantified by UPLC-MS/MS, and values are an average of three replicates.
Not detected.
Discussion
Resv induced NQO1 protein and activity in MCF-10F cells in a dose- and time- dependent manner, with maximal induction after 48 h (Figure 3). The induction of NQO1 by Resv occurs through transcriptional events [24,32,33] and is accompanied by localization of NQO1 in the nucleus [24]. When the cells were pre-incubated with Resv for 48 h, the effects of NQO1 induction on the metabolism of 4-OHE2 can be observed from the increasing formation of 4-OCH3E1(E2) and decreasing amounts of 4-OHE1(E2)-1-N3Ade and 4-OHE1(E2)-1-N7Gua adducts (Table 1, Figure 5). The level of methoxycatechol estrogens increased about 50% and the level of adducts decreased about 2-fold at all three doses of 4-OHE2. A similar effect of Resv was seen when the cells were incubated with E2-3,4-Q, following pre-incubation with Resv for 48 h (Table 2, Figure 6). In this case, the levels of 4-OCH3E1(E2) were much lower than with 4-OHE2 as the estrogen, but the increase in methoxycatechol estrogens across doses of E2-3,4-Q was about 30%, while the decrease in depurinating estrogen-DNA adducts was greater than 50% and they were no longer detectable at the lowest dose of the estrogen. Marginal changes were seen in the levels of thiol conjugates (Table 1).
Pre-incubation with Resv had no effect on the expression of COMT as detected by its level of protein or catalytic activity (Figure 4). The level of COMT in the MCF-10F cells was sufficiently high to methylate virtually all of the 4-OHE1(E2) present in the cells incubated with either 4-OHE2 or E2-3,4-Q (Tables 1 and 2).
To investigate the antioxidant effect of Resv, the MCF-10F cells were pre-incubated with Resv for 48 h, then the cells were incubated with 4-OHE2 or E2-3,4-Q plus a second dose of Resv (Tables 1 and 2, Figures 5 and 6). Little further induction of NQO1 during this 24 h period would be expected, based on the results shown in Figure 3. Thus, the effects of Resv observed correspond to its antioxidant properties.
With 4-OHE2 as the estrogen, the levels of 4-OCH3E1(E2) were 2–2.5 times those seen with 4-OHE2 alone, and no depurinating estrogen-DNA adducts could be detected (Table 1, Figure 5). The ratio of 4-OHE2 to Resv affected the estrogen metabolism (Table 3). Little effect was observed when the concentration of Resv was 10% that of 4-OHE2; at equimolar doses, the methoxycatechol estrogens increased about 30% and a similar decrease in adducts was seen. When the level of Resv was 3-times that of the catechol estrogen, however, the level of 4-OCH3E1(E2) approximately tripled, and the adducts were not detected (Table 3).
The antioxidant effect of Resv is due to the high energy of resonance of its one-electron oxidized form, namely, the 4′-oxyradical [34]. This chemical property can give Resv the capacity to reduce the E2-3,4-semiquinone to 4-OHE2, as already observed in vitro when enzyme-activated 4-OHE2 was reacted with DNA [22]. This reduction has also been observed for cysteine and N-acetylcysteine [21,22]. With E2-3,4-Q as the estrogen, the levels of methoxycatechol estrogens were almost twice those seen with the quinone alone, and the adducts were not detected (Table 2, Figure 6). Inclusion of Resv with E2-3,4-Q had little effect on the formation of thiol conjugates (Table 2).
The ability of Resv to modulate estrogen metabolism may be important in the prevention of breast, prostate and other estrogen-initiated cancers. Men with prostate cancer [12] and women with breast cancer or at high risk for the disease [11] form significantly higher amounts of the depurinating estrogen-DNA adducts than healthy men or women. Thus, formation of these adducts appears to play a critical role in the initiation of these cancers [9,11,12]. The results reported here clearly show that Resv can induce NQO1, which serves as a protective enzyme, by reducing the amount of catechol estrogen quinones available to react with DNA and form depurinating adducts. In addition, Resv inhibits CYP1B1, which catalyzes the formation of 4-OHE1(E2) [24,25].
In summary, the inhibitory effects of Resv are the sum of three factors. One is the induction of NQO1 that provides a decrease in estrogen quinones and an increase in catechol estrogen [24]. The second factor is the inhibition of CYP1B1 expression, which decreases the formation of 4-catechol estrogens [24,25]. The third factor is the antioxidant properties of Resv that reduce the estrogen semiquinones to catechol estrogens, as indirectly determined in vitro [22]. Thus, Resv may be very effective in preventing cancer initiation by estrogens because it induces the protective enzyme NQO1, as reported here and in reference 24, it inhibits overexpression of CYP1B1 [24,25], and it acts as an antioxidant, presumably reducing catechol estrogen semiquinones to catechol estrogens [21,22,34].
Acknowledgments
Grant Support: This research was supported by U.S. Army Breast Cancer Research Program grant DAMD-17-03-0229 and by Prevention LLC. Core support of the Eppley Institute was provided by grant P30 CA36727 from the National Cancer Institute.
Abbreviations
- Resv
resveratrol
- CE
catechol estrogen
- E2-3
4-Q, estradiol-3,4-quinone
- 4-OHE1(E2)
4-ydroxyestrone(estradiol)
- 4-OCH3E1(E2)
4-methoxyestrone(estradiol)
- NQO1
NAD(P)H quinone xidoreductase 1
- COMT
Catechol-O-methyltransferase
- UPLC-MS/MS
Ultraperformance liquid chromatography/tandem mass spectrometry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Agarwal BB, Bhardwaj A, Agarwal RS, Seeram NP, Shishodia S, Takada Y. Role of resveratrol in the prevention and therapy of cancer: preclinical and clinical studies. Anticancer Res. 2004;24:3–53. [PubMed] [Google Scholar]
- 2.Bhat KP, Pezzuto JM. Cancer chemopreventive activity of resveratrol. Ann NY Acad Sci. 2002;957:210–229. doi: 10.1111/j.1749-6632.2002.tb02918.x. [DOI] [PubMed] [Google Scholar]
- 3.Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF, Beecher CW. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275:218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- 4.Tessitone L, Davit A, Sarotto I, Caderni G. Resveratrol depresses the growth of colorectal aberrant crypt foci by affecting bax and p21(CIP) expression. Carcinogensis. 2000;21:1619–1622. [PubMed] [Google Scholar]
- 5.Aziz MH, Kumar R, Ahmed N. Cancer chemoprevention by resveratrol: in vitro and in vivo studies and the underlying mechanisms (Review) Int J Oncol. 2003;23:17–28. [PubMed] [Google Scholar]
- 6.Kim Y-A, Choi BT, Lee YT, Park DI, Rhee S-H, Park K-Y, Choi YH. Resveratrol inhibits cell proliferation and induces apoptosis of human breast carcinoma MCF-7 cells. Oncol Rep. 2004;11:441–446. [PubMed] [Google Scholar]
- 7.Pozo-Guisado E, Merino JM, Mulero-Novarro S, Lorenzo-Benayas MJ, Centeno F, Alvarez-Barrientos A, Salguero PMF. Resveratrol induced apoptosis in MCF-7 human breast cancer cells involves caspase-independent mechanism with down regulation of Bcl-2 and NF-kB. Int J Caner. 2005;115:74–84. doi: 10.1002/ijc.20856. [DOI] [PubMed] [Google Scholar]
- 8.Mgbonyebi OP, Russo J, Russo IH. Antiproliferative effect of synthetic resveratrol on human breast epithelial cells. Int J Oncol. 1998;12:865–869. [PubMed] [Google Scholar]
- 9.Cavalieri E, Chakravarti D, Guttenplan J, Hart E, Ingle J, Jankowiak R, Muti P, Rogan E, Russo J, Santen R, Sutter T. Catechol estrogen quinones as initiators of breast and other human cancers: implications for biomarkers of susceptibility and cancer prevention. BBA Reviews on Cancer. 2006;1766:63–78. doi: 10.1016/j.bbcan.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Zahid M, Kohli E, Saeed M, Rogan E, Cavalieri E. The greater reactivity of estradiol-3,4-quinone versus estradiol-2,3-quinone with DNA in the formation of depurinating DNA adducts. Implications for tumor–initiating activity. Chem Res Toxicol. 2006;19:164–172. doi: 10.1021/tx050229y. [DOI] [PubMed] [Google Scholar]
- 11.Gaikwad NW, Yang L, Muti P, Meza J, Pruthi S, Ingle J, Rogan EG, Cavalieri EL. The molecular etiology of breast cancer: evidence from biomarkers of risk. Int J Cancer. 2008;122:1949–1953. doi: 10.1002/ijc.23329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Markushin Y, Gaikwad N, Zhang H, Kapke P, Rogan EG, Cavalieri EL, Trock BJ, Pavlovich C, Jankowiak R. Potential biomarker for early risk assessment of prostate cancer. Prostate. 2006;66:1565–1571. doi: 10.1002/pros.20484. [DOI] [PubMed] [Google Scholar]
- 13.Markushin Y, Zhong W, Cavalieri EL, Rogan EG, Small GJ, Yeung ES, Jankowiak R. Spectral characterization of catechol estrogen quinone (CEQ)-derived DNA adducts and their identification in human breast tissue extract. Chem Res Toxicol. 2003;16:1107–1117. doi: 10.1021/tx0340854. [DOI] [PubMed] [Google Scholar]
- 14.Chakravarti D, Mailander P, Li K-M, Higginbotham S, Zhang HL, Gross ML, Meza JL, Cavalieri EL, Rogan EG. Evidence that a burst of DNA depurination in SENCAR mouse skin induces error-prone repair and form mutations in the H-ras gene. Oncogene. 2001;20:7945–7953. doi: 10.1038/sj.onc.1204969. [DOI] [PubMed] [Google Scholar]
- 15.Mailander PC, Meza JL, Higginbotham S, Chakravarti D. Induction of A.T to G.C mutations by erroneous repair of depurinated DNA following estrogen treatment of the mammary gland of ACI rats. J Steroid Biochem Mol Biol. 2006;101:204–215. doi: 10.1016/j.jsbmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 16.Zhao Z, Kosinska W, Khmelnitsky M, Cavalieri EL, Rogan EG, Chakravarti D, Sacks PG, Guttenplan JB. Mutagenic activity of 4-hydroxyestradiol, but not 2-hydroxyestradiol, in BB rat2 embryonic cells, and the mutational spectrum of 4-hydroxyestradiol. Chem Res Toxicol. 2006;19:475–479. doi: 10.1021/tx0502645. [DOI] [PubMed] [Google Scholar]
- 17.Lu F, Zahid M, Saeed M, Cavalieri EL, Rogan EG. Estrogen metabolism and formation of estrogen DNA-adducts in estradiol-treated MCF-10F cells. The effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin induction and catechol-O-methyltransferase inhibition. J Steroid Biochem Mol Biol. 2007;105:150–158. doi: 10.1016/j.jsbmb.2006.12.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zahid M, Saeed M, Lu F, Gaikwad N, Cavalieri EL, Rogan EG. Inhibition of catechol-O-methyltransferase increases estrogen-DNA adduct formation. Free Radic Biol Med. 2007;43:1534–1540. doi: 10.1016/j.freeradbiomed.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaikwad NW, Rogan EG, Cavalieri EL. Evidence by ESI-MS for NQO1-catalyzed reduction of estrogen ortho-quinones. Free Radic Biol Med. 2007;43:1289–1298. doi: 10.1016/j.freeradbiomed.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaikwad NW, Yang L, Rogan EG, Cavalieri EL. Evidence for NQO2-mediated reduction of the carcinogenic estrogen ortho-quinones. Free Radic Biol Med. 2008 doi: 10.1016/j.freeradbiomed.2008.10.029. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samuni AM, Chuang EY, Krishna MC, Stein W, DeGraff W, Russo A, Mitchell JB. Semiquinone radical intermediate in catecholic estrogen-mediated cytotoxicity and mutagenesis: chemoprevention strategies with antioxidants. Proc Natl Acad Sci USA. 2003;100:5390–5395. doi: 10.1073/pnas.0930078100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zahid M, Gaikwad N, Cavalieri EL, Rogan EG. Inhibition of depurinating estrogen-DNA adducts by natural compounds. Chem Res Toxicol. 2007;20:1947–1953. doi: 10.1021/tx700269s. [DOI] [PubMed] [Google Scholar]
- 23.Floreani M, Napoli E, Quintieri L, Palatini P. Oral administration of trans-resveratrol to guinea pigs increases cardiac DT-diaphorase and catalase activities, and protects isolated atria from menadione toxicity. Life Sci. 2003;24:2741–2750. doi: 10.1016/s0024-3205(03)00179-6. [DOI] [PubMed] [Google Scholar]
- 24.Lu F, Zahid M, Wang C, Saeed M, Cavalieri EL, Rogan EG. Resveratrol prevents estrogen-DNA adduct formation and neoplastic transformation in MCF-10F cells. Cancer Prevention Research. 2008 doi: 10.1158/1940-6207.CAPR-08-0037. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guengerich FP, Chun YJ, Kim D, Gillam EM, Shimada T. Cytochrome P450 1B1: a target for inhibition in anticarcinogenesis strategies. Mutat Res. 2003;523–524:173–182. doi: 10.1016/s0027-5107(02)00333-0. [DOI] [PubMed] [Google Scholar]
- 26.Li K-M, Todorovic R, Devanesan P, Higginbotham S, Kofeler H, Ramanathan R, Gross ML, Rogan EG, Cavalieri EL. Metabolism and DNA binding studies of 4-hydroxyestradiol and estradiol-3,4-quinone in vitro and in female ACI rat mammary gland in vivo. Carcinogenesis. 2004;25:289–297. doi: 10.1093/carcin/bgg191. [DOI] [PubMed] [Google Scholar]
- 27.Saeed M, Zahid M, Rogan E, Cavalieri E. Synthesis of the catechols of natural and synthetic estrogens by using 2′-iodoxybenzoic acid (IBX) as the oxidizing agent. Steroids. 2005;70:173–178. doi: 10.1016/j.steroids.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 28.Stack DE, Byun J, Gross ML, Rogan EG, Cavalieri EL. Molecular characteristics of catechol estrogen quinones in reactions with deoxyribonucleosides. Chem Res Toxicol. 1996;9:851–859. doi: 10.1021/tx960002q. [DOI] [PubMed] [Google Scholar]
- 29.Cao K, Stack DE, Ramanathan R, Gross ML, Rogan EG, Cavalieri EL. Synthesis and structure elucidation of estrogen quinones conjugated with cysteine, N-acetylcysteine, and glutathione. Chem Res Toxicol. 1998;11:909–916. doi: 10.1021/tx9702291. [DOI] [PubMed] [Google Scholar]
- 30.Denizot F, Lang R. Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J Immunol Methods. 1986;89:271–277. doi: 10.1016/0022-1759(86)90368-6. [DOI] [PubMed] [Google Scholar]
- 31.Dawling S, Roodi N, Mernaugh R, Wang X, Parl FF. Catechol-O-methyltransferase (COMT)-mediated metabolism of catechol estrogens: comparison of wild-type and variant COMT isoforms. Cancer Res. 2001;61:6716–6722. [PubMed] [Google Scholar]
- 32.Li Y, Cao Z, Zhu H. Upregulation of endogenous antioxidants and phase 2 enzymes by the red wine polyphenol, resveratrol in cultured aortic smooth muscle cells leads to cytoprotection against oxidative and electrophilic stress. Pharmacol Res. 2006;53:6–15. doi: 10.1016/j.phrs.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Hsieh TC, Lu X, Wang Z, Wu JM. Induction of quinone reductase NQO1 by resveratrol in human K562 cells involves the antioxidant response element ARE and is accompanied by nuclear translocation of transcription factor Nrf2. Med Chem. 2006;2:275–285. doi: 10.2174/157340606776930709. [DOI] [PubMed] [Google Scholar]
- 34.Stivala LA, Savio M, Carafoli F, Perucca P, Bianchi L, Maga G, Forti L, Pagnoni UM, Albini A, Prosperi E, Vannini V. Specific structural determinants are responsible for the antioxidant activity and the cell cycle effects of resveratrol. J Biol Chem. 2001;276:22586–22594. doi: 10.1074/jbc.M101846200. [DOI] [PubMed] [Google Scholar]