Abstract
Recent studies have suggested a genetic component to heart rate (HR) and HR variability (HRV). However, a systematic examination of the genetic contribution to the variation in HR and HRV has not been performed. This study investigated the genetic contribution to HR and HRV using a wide range of inbred and recombinant inbred (RI) mouse strains. Electrocardiogram data were recorded from 30 strains of inbred mice and 29 RI strains. Significant differences in mean HR and total power (TP) HRV were identified between inbred strains and RI strains. Multiple significant differences within the strain sets in mean low-frequency (LF) and high-frequency (HF) power were also found. No statistically significant concordance was found between strain distribution patterns for HR and HRV phenotypes. Genomewide interval mapping identified a significant quantitative trait locus (QTL) for HR [LOD (likelihood of the odds) score = 3.763] on chromosome 6 [peak at 53.69 megabases (Mb); designated HR 1 (Hr1)]. Suggestive QTLs for TP were found on chromosomes 2, 4, 5, 6, and 14. A suggestive QTL for LF was found on chromosome 16; for HF, we found one significant QTL on chromosome 5 (LOD score = 3.107) [peak at 53.56 Mb; designated HRV-high-frequency 1 (Hrvhf1)] and three suggestive QTLs on chromosomes 2, 11 and 15. In conclusion, the results demonstrate a strong genetic component in the regulation of resting HR and HRV evidenced by the significant differences between strains. A lack of correlation between HR and HRV phenotypes in some inbred strains suggests that different sets of genes control the phenotypes. Furthermore, QTLs were found that will provide important insight to the genetic regulation of HR and HRV at rest.
Keywords: electrocardiography, autonomic nervous system, ventilation
cardiovascular disorders are prominent public health concerns, and their complexity can be difficult to dissect. However, measurements of heart rate (HR) and heart rate variability (HRV) have become increasingly useful clinical tools for assessment of autonomic nervous system function and cardiovascular health (1). Reduced HRV and increased resting HR are well-established indicators of increased cardiovascular risk (5, 33). A number of factors have been identified as determinants of HRV (29) that do not fully explain the reported variance in HRV (24, 41). Importantly, genetic factors may significantly influence HR and HRV but have not been studied in sufficient detail.
HRV, in the frequency domain, is divided into a number of frequency ranges. In this study, the variation in HR in the low- (LF) and high-frequency (HF) ranges were of principal interest. LF HRV can be attributed to delays on baroreceptor feedback as a consequence of blood pressure fluctuations. HF HRV can be ascribed to respiratory sinus arrhythmia, where the HRV peak within the HF HRV range matches the breathing frequency (26).
Epidemiological studies have begun to address the genetic influence on HRV in human subjects, and heritability was estimated to account for 13–22% (31) and 28–34% (32) of the interindividual variance in HRV. Unfortunately, these studies can be difficult to interpret since the effect of respiration on HRV was not considered. Nevertheless, they do provide an excellent platform from which a more detailed study of the genetic contribution to HR and HRV can be performed while respiration is monitored.
Studies that have measured HR and HRV from conscious, unrestrained inbred rodents have used a limited number of strains, and the effect of respiration on HRV was not considered (21, 36). Initial evidence for a genetic contribution to HR was provided by Kreutz et al. (22) who reported an association between Hr-sp1 locus on rat chromosome 3 and HR regulation. Furthermore, differences in the HR of inbred mouse strains have been reported (35). However, Campen et al. (7) found no significant between-strain differences in HRV. Interestingly, the ECG was recorded during periods of sleep, and therefore the physical activity of these mice was presumably similar and between-strain differences in respiration may have been minimized, making the ECG record more appropriate for HRV analysis. Hoit et al. (19) investigated cardiovascular variation and reported a 91 and a 79 beats/min difference in HR between A/J and C57BL/6J mice using echocardiography and tail-cuff measurements, respectively. Tankersley et al. (36, 39) have reported an ∼80 beats/min difference between C3H/HeJ and C57BL/6J mice using radiotelemetry.
The current ambiguity in the literature regarding the genetic regulation of HR and HRV in humans and rodents warrants further and more detailed investigation. In the present study, HR and HRV were calculated from ECG recorded from inbred strains of mice using telemetric methods while monitoring minute ventilation (V̇e). The purpose of this study was to assess the within- and between-strain variation in HR and HRV and to identify quantitative trait loci (QTLs) that account for significant genetic variance in HR and HRV phenotypes.
MATERIALS AND METHODS
Animals
Thirty male inbred strains of mice (20–30 g) were studied (Table 1). All mice, except for CAST (24 wk old), SPRET (37 wk old), and MOLF (49 wk old), were 8–12 wk of age (Table 1). CAST, SPRET, and MOLF mice were studied after additional aging because the body mass of these mice was not sufficient to meet surgery requirements when at an age similar to the other strains. Twenty-nine male recombinant inbred (RI) strains (20–30 g) were studied. All mice were studied at 8–21 wk of age (Table 2).
Table 1.
Vt, f, and nV̇e and age of all inbred strains
| Strain | n | Vt, ml | f, breaths/min | nV̇e, ml/g | Mass, g | Age, wk |
|---|---|---|---|---|---|---|
| 129X1/SvJ (129Sv) | 10 | 0.22±0.008* | 272.45±24.60 | 2.53±0.33*† | 24.20±0.33 | 8 |
| 129S1/SvlmJ (129Svlm) | 8 | 0.20±0.10 | 180.59±15.27 | 1.45±0.09 | 25.64±0.78 | 13 |
| A/J (A) | 10 | 0.19±0.007 | 186.25±13.80* | 1.46±0.07† | 24.69±0.50 | 8 |
| AKR/NCr (AKR) | 10 | 0.27±0.012* | 171.90±14.20* | 1.43±0.21 | 26.33±0.96 | 9 |
| BALB/cByJ (BALB) | 10 | 0.18±0.006 | 355.01±33.59† | 2.78±0.27* | 25.45±0.28 | 7 |
| BPH/2J (BPH) | 8 | 0.13±0.004† | 376.51±22.39 | 2.54±0.13* | 20.02±0.32 | 12 |
| BPL/1J (BPL) | 8 | 0.15±0.005† | 292.3±28.31† | 2.18±0.21 | 22.69±1.16 | 15 |
| BPN/3J (BPN) | 8 | 0.16±0.005† | 352.93±15.13 | 2.32±0.09* | 25.28±0.47 | 15 |
| BTBR T+tfl J (BTBR T) | 8 | 0.20±0.010 | 252.89±18.29 | 1.70±0.08 | 28.19±0.66 | 9 |
| C3H/HeJ (HeJ) | 10 | 0.26±0.013* | 167.94±15.60* | 1.75±0.12 | 24.18±0.71 | 9 |
| C3H/HeOuJ (OuJ) | 10 | 0.21±0.008* | 191.80±11.10* | 1.43±0.08† | 30.30±0.56 | 10 |
| C57BL/10J (B10) | 6 | 0.16±0.004† | 332.50±17.30† | 2.28±0.21* | 22.94±0.65 | 11 |
| C57BL/6J (B6) | 10 | 0.21±0.007 | 239.54±18.96 | 2.27±0.19* | 23.70±0.49 | 9 |
| C57 BLKS/J (C57Blks) | 7 | 0.17±0.009 | 266.27±25.33 | 1.87±0.10 | 23.7±0.40 | 13 |
| C57L/J (C57L) | 8 | 0.22±0.008* | 327.65±14.10† | 3.25±0.15* | 22.94±0.65 | 10 |
| CAST/EiJ (CAST) | 7 | 0.22±0.006* | 311.19±9.78† | 3.31±0.19* | 19.14±0.62 | 24 |
| CBA/J (CBA) | 8 | 0.18±0.008 | 183.10±8.64* | 1.30±0.06† | 26.37±0.53 | 10 |
| DBA1/J (DBA1) | 8 | 0.71±0.01 | 322.13±15.67 | 2.25±0.13* | 24.62±0.58 | 10 |
| DBA2/J (DBA2) | 10 | 0.23±0.014* | 279.93±20.35 | 2.69±0.11* | 23.88±0.92 | 11 |
| FVB/NJ (FVB) | 8 | 0.22±0.030 | 320.60±32.60† | 2.55±0.29* | 24.48±0.28 | 10 |
| ICRTac:ICR (ICR) | 8 | 0.26±0.004 | 125.33±9.85 | 1.02±0.09† | 31.96±0.30 | 9 |
| LP/J | 8 | 0.17±0.007 | 298.29±22.10 | 1.61±0.13† | 27.16±0.73 | 13 |
| MOLF/EiJ (MOLF) | 4 | 0.14±0.003 | 315.88±11.85 | 1.80±0.05 | 21.7±0.55 | 49 |
| NOD/LtJ (NOD) | 8 | 0.16±0.005† | 298.10±21.40† | 2.03±0.17 | 23.90±1.00 | 10 |
| NZB/BinJ (NZB) | 10 | 0.30±0.007* | 129.50±5.47* | 1.19±0.06† | 31.12±0.67 | 9 |
| NZW/LacJ (NZW) | 10 | 0.27±0.010* | 222.53±20.30 | 1.63±0.15† | 36.88±1.02 | 11 |
| PL/J (PL) | 8 | 0.18±0.004 | 232.40±5.43 | 2.08±0.07 | 21.83±0.48 | 10 |
| SJL/J (SJL) | 8 | 0.16±0.010† | 263.50±21.50 | 1.56±0.13† | 25.01±0.37 | 11 |
| SM/J (SM) | 8 | 0.19±0.0086 | 262.25±35.98 | 1.85±0.20 | 23.85±0.51 | 14 |
| SPRET/EiJ (SPRET) | 8 | 0.32±0.060* | 196.90±9.90* | 2.11±0.11 | 22.69±1.22 | 37 |
Values are means ± SE; n, number of mice. Ranges for tidal volume (Vt), breathing frequency (f), and minute ventilation normalized to body mass (nV̇e) were 0.13 (BPH)–0.32 ml (SPRET), 129.5 (NZB)–376.51 breaths/min (BPH), and 1.02 (ICR)–3.31 ml/g (CAST), respectively. *P < 0.05, significantly different compared to SPRET mice; †P < 0.05, significantly different from BPH mice for Vt. *P < 0.05, significantly different compared to BPH mice; †P < 0.05, significantly different compared to NZB mice for f. *P < 0.05, significantly different compared with ICR mice; †P < 0.05, significantly different compared with CAST mice for nV̇e.
Table 2.
Vt, f, body mass and age of the RI strains
| Strain | n | Vt, ml | f, breaths/min | nV̇e, ml/g | Mass, g | Age, wk |
|---|---|---|---|---|---|---|
| AXB1/PgnJ (AXB1) | 4 | 0.17±0.02 | 360.70±6.15 | 2.07±0.38 | 29.0±2.8 | 13 |
| AXB2/PgnJ (AXB2) | 4 | 0.17±0.01 | 295.32±59.18 | 1.87±0.30 | 25.3±2.6 | 12 |
| AXB4/PgnJ (AXB4) | 4 | 0.13±0.01 | 337.68±47.91 | 0.99±0.58 | 19.5±0.2 | 11 |
| AXB5/PgnJ (AXB5) | 4 | 0.16±0.01 | 237.56±25.92 | 1.64±0.13 | 23.2±0.7 | 9 |
| AXB6/PgnJ (AXB6) | 4 | 0.18±0.02 | 313.22±37.21 | 2.31±0.55 | 25.5±0.6 | 9 |
| AXB8/PgnJ (AXB8) | 2 | 0.13±0.01 | 328.58±66.76 | 1.31±0.76 | 19.6±0.5 | 8 |
| AXB10/PgnJ (AXB10) | 4 | 0.16±0.01 | 351.59±20.53 | 2.39±0.09 | 23.6±0.9 | 10 |
| AXB12/PgnJ (AXB12) | 4 | 0.17±0.01 | 247.33±49.97 | 2.19±0.48 | 24.4±0.1 | 12 |
| AXB13/PgnJ (AXB13) | 2 | 0.13±0.02 | 406.28±71.59 | 1.67±0.30 | 20.3±0.5 | 8 |
| AXB15/PgnJ (AXB15) | 4 | 0.18±0.02 | 294.61±12.74 | 2.05±0.23 | 27.1±0.7 | 13 |
| AXB17/PgnJ (AXB17) | 4 | 0.14±0.02 | 434.14±28.72 | 2.49±0.54 | 24.8±0.9 | 13 |
| AXB18/PgnJ (AXB18) | 4 | 0.16±0.02 | 346.29±10.91 | 1.94±0.20 | 21.2±0.7 | 9 |
| AXB19/PgnJ (AXB19) | 4 | 0.14±0.01 | 335.24±14.72 | 2.18±0.08 | 21.1±0.5 | 11 |
| AXB20/PgnJ (AXB20) | 3 | 0.14±0.01 | 388.47±48.69 | 1.79±0.68 | 21.9±1.3 | 12 |
| AXB23/PgnJ (AXB23) | 4 | 0.21±0.02 | 232.94±59.20 | 1.67±0.28 | 24.5±0.1 | 9 |
| AXB24/PgnJ (AXB24) | 4 | 0.15±0.01 | 257.96±61.92 | 1.76±0.44 | 22.4±0.4 | 11 |
| BXA1/PgnJ (BXA1) | 4 | 0.17±0.01 | 252.59±36.73 | 1.63±0.20 | 29.4±0.4 | 11 |
| BXA2/PgnJ (BXA2) | 4 | 0.21±0.02 | 269.91±34.76 | 1.41±0.15 | 38.4±1.4 | 21 |
| BXA4/PgnJ (BXA4) | 3 | 0.15±0.02 | 345.23±68.11 | 1.80±0.21 | 27.5±0.5 | 21 |
| BXA7/PgnJ (BXA7) | 4 | 0.17±0.03 | 261.57±16.41 | 1.53±0.29 | 29.7±0.4 | 14 |
| BXA8/PgnJ (BXA8) | 4 | 0.15±0.01 | 279.05±27.47 | 1.67±0.14 | 25.0±0.8 | 13 |
| BXA11/PgnJ (BXA11) | 4 | 0.16±0.03 | 335.98±48.35 | 1.47±0.24 | 35.6±2.6 | 16 |
| BXA12/PgnJ (BXA12) | 2 | 0.15±0.00 | 248.43±36.52 | 1.61±0.07 | 22.7±1.9 | 13 |
| BXA13/PgnJ (BXA13) | 4 | 0.18±0.03 | 329.04±36.69 | 2.30±0.17 | 24.8±1.1 | 13 |
| BXA14/PgnJ (BXA14) | 4 | 0.18±0.02 | 298.20±51.83 | 1.87±0.29 | 28.5±0.7 | 17 |
| BXA16/PgnJ (BXA16) | 4 | 0.15±0.01 | 351.16±17.75 | 2.01±0.09 | 25.2±0.5 | 15 |
| BXA24/PgnJ (BXA24) | 4 | 0.17±0.02 | 419.48±45.15 | 2.16±0.37 | 29.7±0.6 | 13 |
| BXA25/PgnJ (BXA25) | 4 | 0.18±0.02 | 277.86±15.08 | 2.01±0.14 | 25.8±0.2 | 13 |
| BXA26/PgnJ (BXA26) | 4 | 0.12±0.01 | 409.17±42.14 | 2.26±0.23 | 22.3±0.4 | 13 |
Values are means ± SE; n, number of mice. Ranges for Vt, f, and nV̇e were 0.12 (BXA26)–0.21 ml (AXB23), 232.9 (AXB23)–434.1 breaths/min (AXB17), and 0.99 (AXB4)–2.49 ml/g (AXB17), respectively. RI, recombinant inbred. No significant differences were found.
All mice were purchased from Jackson Laboratory (Bar Harbor, ME) with the exception of the AKR mice, which were purchased from the National Cancer Institute (Bethesda, MD), and the ICR mice, which were purchased from Taconic Farms (Raleigh, NC). Information regarding the known characteristic features of the different strains is publicly available through The Jackson Laboratory Mouse Phenome Project (http://phenome.jax.org/pub-cgi/phenome/mpdcgi?rtn=docs/home). All mice were housed individually with a 12-h:12-h (7:00 am to 7:00 pm) light-dark cycle. Food (NIH-31) and water were provided ad libitum. Animals were handled in accordance with The National Institutes of Health Humane Care and Use of Laboratory Animals guidelines and the American Physiological Society's “Guiding Principles in the Care and Use of Animals.” The study protocol was reviewed and approved by the National Institute of Environmental Health Science Animal Care and Use Committee.
Surgical Implantation of Radiotelemetry Transmitter
Mice were anesthetized using inhaled isoflurane, and buprenorphine (0.1 mg/kg) was given for analgesia. A 3-cm midline dorsal incision was made in the skin, and a subcutaneous tissue pocket was made using a blunt instrument. An ETA-F20 transmitter (DSI; Arden Hills, MN) was placed inside the tissue pocket and sutured to the left latissimus dorsi muscle. The anodal and cathodal leads were tunneled subcutaneously and sutured over the left superficial gluteus and right trapezius muscles, respectively. All incisions were closed using wound clips, and animals were allowed 5 days to recover.
Measurements
All data were recorded from conscious, unrestrained mice at the same time of day (9:00–11:00 am) to control for circadian variation. Animals were placed individually in whole body plethysmographs (Buxco Electronics, Wilmington, NC) and allowed at least 30 min to acclimate. To monitor the possible confounding effects of breathing on HRV (6), we simultaneously recorded 20 min of radiotelemetry ECG and pulmonary function data from each mouse during periods of quiet rest or sleep when breathing rate and depth were consistent.
With 20 min of ECG from individual mice, R-waves were marked and R-R intervals were extracted using specialist software (Dataquest A.R.T. v. 3.1, DSI). The resultant R-R series was then uploaded to custom software (HRV Webstart edition development) created in collaboration with Drs. Todd Jenkins (East Carolina University, NC) and Matthew J. Campen (The Lovelace Foundation, NM). Any R-R interval that was 150% greater or less than the median R-R intervals for a given series was considered an arrhythmia and removed; HRs were then calculated. HRV was calculated using a Lomb periodogram, which was developed specifically for spectral analysis of unevenly spaced data.
Currently, a standard LF cutoff has not been agreed on for mice. However, it is important to consider statistical as well as physiological rationales when choosing the LF cutoff. We analyzed 120 s of ECG data at a time, and it is well known that the LF cutoff should be limited to 1/6 of the sampling length (i.e., 0.05 Hz). In our case and in that of Campen et al. (7), we used a LF cutoff where spectral power commonly reduced almost to zero (∼0.2 Hz). Since 0.2 Hz is greater than the minimum cutoff of 0.05 Hz, we feel confident that our LF range is appropriate.
The LF range was set at 0.2–1.5 Hz, and HF was 1.5–5.0 Hz. Total power (TP) was determined by the summation of the LF and HF range values.
Breathing frequency (f) tidal volume (Vt), and minute ventilation normalized to body mass (nV̇e) were used to indicate changes in pulmonary function, which are thought to affect HRV assessments. Values for f, Vt, and V̇e were calculated using specialist software (Buxco Electronics).
Linkage Analysis
Genomewide scans for QTLs were done with the entire RI data set and parental phenotypes. We used Web QTL (www.genenetwork.org) (43) for interval analyses by fitting a regression equation for the effect of a hypothetical QTL at the position of each marker and at intervals between the markers. Over 8,500 informative markers and 2,400 unique strain distribution patterns (SDPs) exist for the AXB/BXA RI set. Likelihood of the odds (LOD) score curves on the interval map are provided for each chromosome and graphically represent the approximate position of the QTL(s). The dominance and additive properties of each putative QTL were also evaluated. The significance of each association (the base-10 LOD score) was determined and plotted against the linear position on the chromosome. Permutation tests were used to establish empirically the significance thresholds for the genomewide QTL mapping results following Churchill and Doerge (10) and Doerge and Churchill (12). For each genome scan, 1,000 permutations were done to establish significant and suggestive linkage threshold values as indicated in each figure. These values correspond to the genomewide probabilities proposed by Lander and Kruglyak (23) and are as have been published previously for mouse and rat RI data sets (9, 11, 16, 42). The significant threshold level approximately corresponds to a LOD score of 3.0 (i.e., P = 10−4) or higher, whereas the suggestive threshold level approximately corresponds to a LOD score of 2.1 (i.e., P = 10−3).
Statistical Analysis
Data from individual mice were used to calculate the strain average (mean). Strain averaged data were then used to make comparisons between strains. Means ± SE are presented. Differences between strains in nV̇e, f, Vt, HR, and HRV were assessed independently using a one-way ANOVA with Tukey's post hoc test. However, the LF and HF HRV phenotypes failed the normality test, and therefore between strains differences were assessed using a Kruskal-Wallis ANOVA on ranks with Dunn's post hoc test. The relationships between HR and TP, body mass or age, and nV̇e were assessed using a linear regression analysis. Statistical significance was accepted at <0.05.
RESULTS
Inbred Strain Comparison
Pulmonary function.
Multiple statistically significant differences in nV̇e (in ml·min−1·g−1) and f (in ml·min−1·g−1) were found between inbred strains (Table 1). Interestingly, the SDPs for nV̇e, Vt (in ml), and f were discordant (Table 1). No statistically significant relationship was found between age (in wk) or body mass and nV̇e, Vt, or f (data not shown), although all strains (except the wild-derived CAST, SPRET, and MOLF strains) were age matched (Table 1).
HR and HRV.
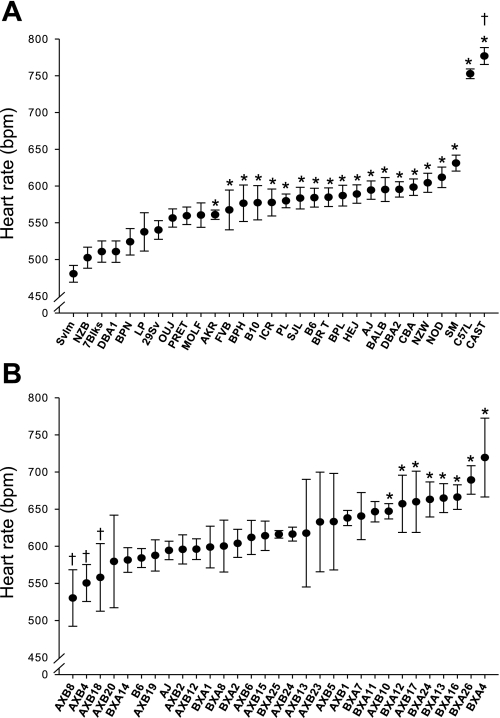
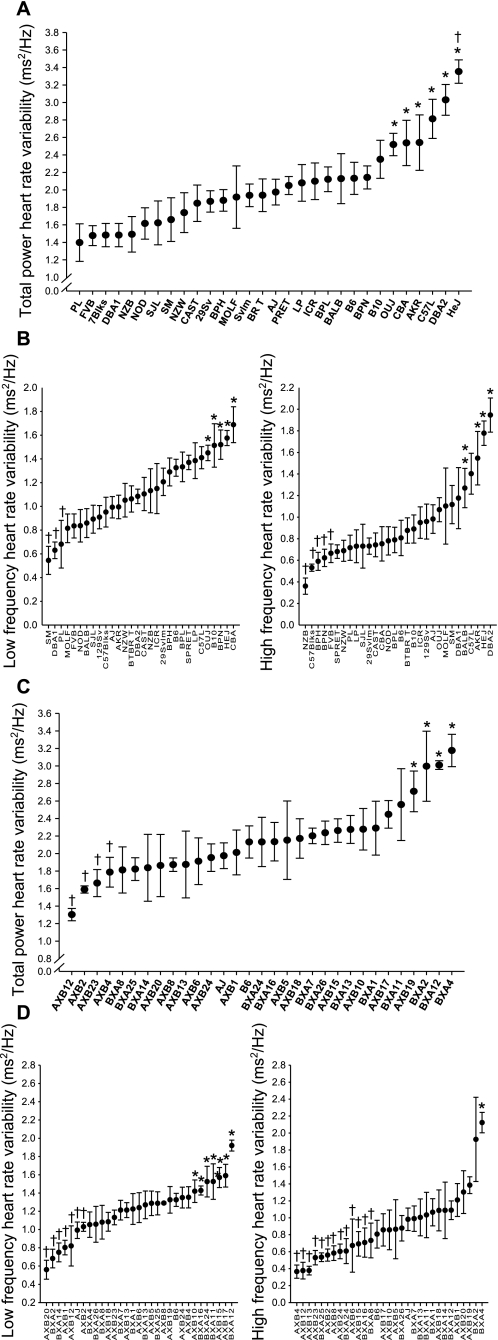
Statistically significant differences were found in baseline HR and HRV between the inbred strains (Figs. 1A and 2A). Mean (±SE) HR (in beats/min) ranged from 480.5 ± 11.4 (129Svlm) to 776.9 ± 11.5 (CAST). Whereas mean HR for C57L mice was significantly lower than CAST HR, the mean HRs of both strains were significantly higher than all other strains (P < 0.05; Fig. 1A). The range in mean (±SE) TP (in ms2/Hz) was 1.39 ± 0.2 (PL) to 3.35 ± 0.1 (HeJ; Fig. 2A) for the inbred strains. Significant between-strain differences in LF and HF power, the two components of TP, were also found (Fig. 2B), and the SDPs were not the same as TP. Indeed, the relationship between mean LF and HF was different and in some cases reversed (e.g., NZB vs. DBA2; Fig. 2). Furthermore, no correlation between baseline HRV and nV̇e, Vt, or f was found between these inbred strains.
Fig. 1.
A: baseline heart rate [HR, in beats/min (bpm)] values recorded from 30 inbred strains of mice. Values are means ± SE from 20 min of ECG recording. *P < 0.05, significantly different compared with 129Svlm mice; †P < 0.05, significantly different compared with all other strains. B: baseline HR (in beats/min) values recorded from 29 AXB/BXA recombinant-inbred (RI) inbred strains of mice. Values are means ± SE from 20 min of ECG recording. *P < 0.05, significantly different compared with AXB8 mice; †P < 0.05, significantly different compared with BXA4 mice.
Fig. 2.
A: baseline total power (TP) HR variability (HRV; in ms2/Hz) in 30 inbred strains of mice. TP HRV is the summation of values from the low-frequency (LF) and high-frequency (HF) HRV ranges. Values are means ± SE from 20 min of ECG recording. *P < 0.05, significantly different compared with PL mice; †P < 0.05, significantly different compared with all strains except DBA/2 and C57L mice. B, left: baseline LF HRV (in ms2/Hz) in 30 inbred strains of mice. Values are means ± SE from 20 min of ECG recording. *P < 0.05, significantly different compared with SM mice; †P < 0.05, significantly different compared with CBA mice. B, right: baseline HF HRV (in ms2/Hz) in 30 inbred strains of mice. Values are means ± SE from 20 min of ECG recording. *P < 0.05, significantly different compared with NZB mice; †P < 0.05, significantly different compared with DBA2 mice. C: baseline TP HRV (in ms2/Hz) in 29 AXB/BXA RI strains of mice. Values are means ± SE from 20 min of ECG recording. *P < 0.05, significantly different compared with AXB12 mice; †P < 0.05, significantly different compared with BXA4 mice. D, left: baseline LF HRV (in ms2/Hz) in 29 AXB/BXA RI strains of mice. Values are means ± SE from 20 min of ECG recording. *P < 0.05, significantly different compared with AXB20 mice; †P < 0.05, significantly different compared with BXA12 mice. D, right: baseline HF HRV (in ms2/Hz) in 29 AXB/BXA RI strains of mice. Values are means ± SE from 20 min of ECG recording. *P < 0.05, significantly different compared with AXB4 mice; †P < 0.05, significantly different compared with BXA4 mice.
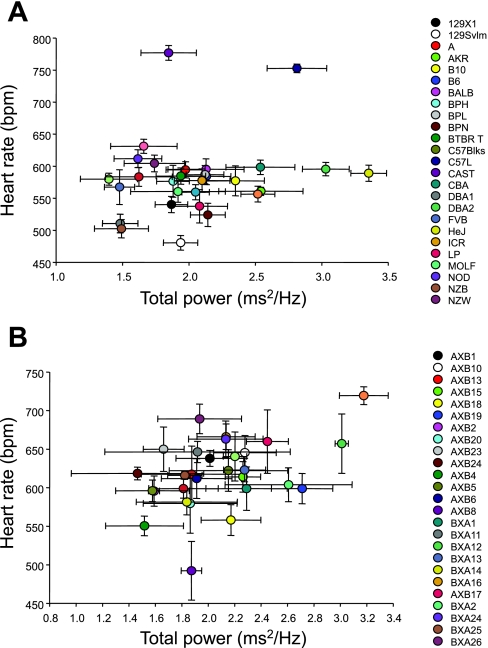
Mean HR and TP did not cosegregate (R = 0.18) among the strains investigated (Fig. 3A); that is, HR did not predict TP, and in the majority of strains, a wide range of values for TP was observed at similar HRs. The most notable exceptions were the CAST and C57L strains. Despite the higher HRs, TP remained within the range of the other strains, suggesting that HR and TP are independent from one another in these strains.
Fig. 3.
A: cosegregation plot of HR and TP HRV from 30 inbred strains of mice. No correlation between these variables was found (R = 0.18). Values are means ± SE from 20 min of ECG recording. B: cosegregation plot of HR and TP HRV from 29 AXB/BXA RI strains of mice. No correlation between these variables was found (R = 0.42). Values are means ± SE from 20 min of ECG recording.
RI Strain Comparison
Pulmonary function.
nV̇e, f, and Vt were not significantly different between the RI strains (Table 2). The SDPs for each phenotype were different among these strains. It was not possible to predict nV̇e based on f or Vt alone. The strains used different Vt to f ratios to achieve the required nV̇e.
HR and HRV.
Mean (±SE) HR (in beats/min) ranged from 530.3 ± 38.1 (AXB8) to 719.5 ± 53.2 (BXA4; Fig. 1B). This range in RI strain HRs was observed despite no significant difference between the A and B6 parental strains in the present study. The range in mean (±SE) TP (in ms2/Hz) was 1.47 ± 0.5 (AXB24) to 3.18 ± 0.2 (BXA4) for the RI strains (Fig. 2C). As observed for the inbred strains, the SDPs for the LF and HF were different from each other (e.g., AXB13 vs. BXA4; Fig. 3B). No overall concordance was found (R = 0.42; Fig. 3B) between HR and TP, although a wide range of HRs was observed at similar TP values.
QTL mapping with AXB/BXA RI strains.
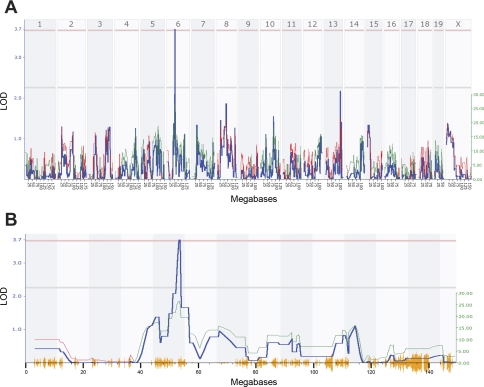
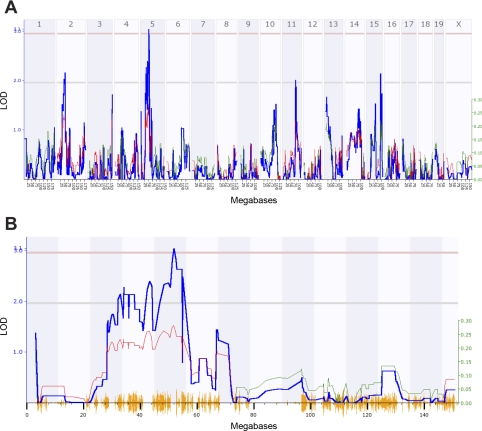
A genomewide search for QTLs for each of the HR and HRV phenotypes was performed with the entire RI data set. Interval mapping identified a significant QTL for HR (Fig. 4), five suggestive QTLs for TP, a suggestive QTL for LF (Table 3), and one significant and three suggestive QTLs for HF (Fig. 5 and Table 3). The significant HR QTL [designated HR 1 (Hr1)] was found on chromosome 6 between 52 and 56 megabases (Mb), with the peak linkage at marker rs6263715 (53.564327 Mb), and the LOD score of 3.763 exceeded the significance threshold as determined empirically by the permutation test (Fig. 4). The positive additive effect at this and neighboring markers indicates that A/J alleles accounted for the difference in heart rate. A significant HF QTL [designated HR variability-HF 1 (Hrvhf1)] was identified on chromosome 5 between 46 and 56 Mb, with the peak linkage at marker rs6263715 (53.564327 Mb) and a LOD score of 3.107 (Fig. 5). The negative additive effect for this QTL indicates that B6 alleles are responsible for the difference in HF (Table 3). Composite interval mapping was done to estimate the influence of the HF suggestive QTLs on Hrvhf1, but controlling for each did not affect Hrvhf1.
Fig. 4.
A: genomewide linkage map for HR in 29 AXB/BXA RI strains of mice. The x-axis represents the length of the each chromosome, the left y-axis shows the likelihood of the odds statistic (LOD; blue line), and the right y-axis shows the degree to which either A/J (green line) or B6 (red line) alleles increase phenotypic values. The numbers along the top of the graph indicate chromosome number. The bottom horizontal line indicates suggestive linkage, and the top horizontal line indicates significant (P < 0.05) linkage. A significant quantitative trait locus (QTL) was found on chromosome 6 with a LOD score of 3.763 at ∼54 megabases (Mb). B: a detailed linkage map of chromosome 6 indicates the significant QTL for HR in 29 AXB/BXA RI strains of mice. The lines in this figure are the same as those described in A.
Table 3.
QTL for basal heart rate and heart rate variability phenotypes identified by interval mapping in AXB and BXA RI strains
| Phenotype/QTL Type | Chromosome | Marker | Location, Mb | LRS | LOD | P Value | Additive Effect* |
|---|---|---|---|---|---|---|---|
| Heart Rate | |||||||
| Significant | 6 | rs3653600 | 53.694055 | 17.350 | 3.763 | 0.00003 | 26.552 |
| Total Power | |||||||
| Suggestive | 2 | rs13476480 | 48.027981 | 10.087 | 2.188 | 0.0015 | −0.247 |
| Suggestive | 4 | rs3676423 | 51.811463 | 10.087 | 2.188 | 0.0015 | 0.238 |
| Suggestive | 5 | rs3682333 | 32.226277 | 13.830 | 3.000 | 0.0002 | −0.260 |
| Suggestive | 6 | rs13478972 | 111.946504 | 10.087 | 2.188 | 0.0015 | 0.237 |
| Suggestive | 14 | rs13482327 | 96.084261 | 11.239 | 2.438 | 0.0008 | −0.243 |
| Low Frequency | |||||||
| Suggestive | 16 | rs4202837 | 73.172021 | 10.737 | 2.329 | 0.0011 | 0.170 |
| High Frequency | |||||||
| Suggestive | 2 | rs3720957 | 51.218796 | 9.981 | 2.165 | 0.0008 | −0.235 |
| Significant | 5 | rs6263715 | 53.564327 | 14.321 | 3.107 | 0.0002 | −0.242 |
| Suggestive | 11 | rs13481126 | 83.099703 | 9.299 | 2.017 | 0.0023 | −0.221 |
| Suggestive | 15 | rs13482701 | 89.703046 | 9.897 | 2.147 | 0.0017 | 0.219 |
Additive effect indicates which strain alleles increase the trait value: positive additive effect indicates A/J alleles increase trait value; negative additive effect indicates C57BL/6J alleles increase trait value. QTL, quantitative trait locus; Mb, megabases; LRS, likelihood ratio statistic; LOD, likelihood of the odds.
Fig. 5.
A: genomewide linkage map for HF HRV in 29 AXB/BXA RI strains of mice. The x-axis represents the length of the each chromosome, the left y-axis shows the LOD statistic (blue line), and the right y-axis shows the degree to which either A/J (green line) or B6 (red line) alleles increase phenotypic values. The numbers along the top of the graph indicate chromosome number. The bottom horizontal line indicates suggestive linkage, and the top horizontal line indicates significant (P < 0.05) linkage. A significant QTL was found on chromosome 5 with a LOD score of 3.107 at ∼54 Mb. B: a detailed linkage map of chromosome 5 indicates the significant QTL for HF in 29 AXB/BXA RI strains of mice. The lines in this figure are the same as those described in A.
DISCUSSION
We studied 30 inbred strains and 29 RI strains of mice to determine the genetic contribution to the variation in HR and HRV. Significant between-strain differences were found in HR, HRV, and pulmonary function between the two strain sets. Furthermore, the comparison of HR and HRV SDPs indicates that the traits do not cosegregate, suggesting that different sets of genes may determine the two phenotypes (Fig. 3). To dissect specific chromosomal regions responsible for the variation in these phenotypes, a genomewide QTL analysis was performed for the HR and HRV phenotypes. Significant and suggestive QTLs were found for HR, LF, HF, and TP (Figs. 4 and 5; and Table 3).
Pulmonary Function: nV̇e, Vt, and f
Significantly greater interstrain differences in nV̇e, Vt, and f were found compared with within-strain variance, suggesting a genetic contribution to pulmonary function (Table 1). This observation was not in agreement with previous reports, which found no significant differences in V̇e among five of the inbred strains used in this study (37, 38). Furthermore, after the values are normalized for body mass, the nV̇e values in this study were notably higher than those reported by Tankersley et al. (37). The principal cause of the differences in nV̇e between these studies was breathing frequency since Vt values were similar. A number of factors may account for the differences in nV̇e between studies, including seasonal variation, differential behavioral characteristics between strains, and equipment sensitivity. Nonetheless, the new data suggest that genetic background accounts for a significant portion of the interstrain variation in nV̇e (Table 1).
HR and HRV
The strain most commonly used to investigate murine HR and HRV has been C57BL/6J, and reported HRs have not been consistent. Values for mean (±SE) HR recorded in this strain include 662 ± 12 (21) and 564 ± 24 beats/min (7). This lack of consistency in the published data makes estimates of normal murine HR values difficult and may be in part due to differences in animal arousal levels. Mean resting HR values for C57BL/6J mice (584 ± 12 beats/min) in the present study were similar to those reported by Campen et al. (7), who provided an excellent standard because their HR values were calculated over a 24-h period. These HR values are ∼10 times higher than those in the normal human HR range.
The multiple inbred strain SDP for HR revealed at least two distinct phenotypes (Fig. 1A). C57L and CAST HRs were significantly higher when compared with the remaining strains despite all ECG records being made during periods of rest. The mechanisms related to this difference are currently unclear but may become evident when candidate genes are identified and investigated. However, continuous distribution of HR values was found for the majority of the strains studied, which suggests that HR regulation is a complex trait and influenced by multiple genes. A continuous distribution for HR in the RI strain SDP supports this notion. Interestingly, we did not find a significant difference in HR between the AXB and BXA parental strains A/J and B6, where only a 10 beats/min difference was observed (Fig. 1A); this differs from previous reports of much larger differences between A/J and B6 mice (19). Hoit et al. (19) reported higher HR values for both strains during tail-cuff measurements compared with echocardiography. These conflicting values may be attributable to the differing conditions under which the measures were taken. It is assumed, although not reported, that a form of anesthesia was used in their study during echocardiography which could have depressed HR. Conversely, the tail-cuff procedure presents the potential for increased stress to the mouse, thus stimulating HR.
Differences in HR between BALB/cJ and CBA/CaJ mice have been associated with QTLs on chromosomes 2 (Hrq1) and 15 (Hrq2) (35). Sugiyama et al. (35) also demonstrated a significant gene-gene interaction for HR between Hrq1 and D1Mit10, referred to as Hrq3. However, no HR differences were found between BALB/cByJ and CBA/J in the present study. Again, these contrasting results could be due to differences in the methods used for data collection since Sugiyama et al. (35) used the tail-cuff method.
It is important to note that QTL analysis using RI strain sets does not require a significant trait difference between the parental strains but rather a broad range of trait values between the RI strains (Figs. 1 and 3). The recombination of alleles that occurs during derivation of the RI strain set enables different gene × gene interactions that may give rise to trait values that are beyond the range observed in the parental strains. This wide range of trait values can then be analyzed for QTLs that associate with the phenotype of interest. Linkage analysis with the AXB/BXA RI set identified Hr1 on chromosome 6, which contains a number of potentially important genes in association with HR regulation. These candidate genes include corticotropin-releasing factor receptor 2 (Crhr2) and neuropeptide Y (Npy), both of which have been associated with heart rate regulation (34, 40). Although speculative, it is important to note that the effects of these genes may be strain dependent and associate with the higher or lower inbred and RI strain HRs in the present study, warranting further investigation.
In this study, higher LF relative to the HF HRV in 15 inbred strains and in 21 RI strains was found. A number of studies have indicated that the ratio between LF and HF is an index of the relative sympathetic and vagus nerve tone in regulating HR (2, 28, 39a). Previous studies using C57BL/6J mice also reported a higher level of LF relative to HF power (7, 21). This relationship between LF and HF power is the reverse of that expected in resting humans (27) at HRs lower than the anticipated human intrinsic heart rate of 120 beats/min. However, the ratio between LF and HF as an index of sympathovagal balance has been criticized in recent years. The interpretation of the LF and HF components of HRV is a subject of current debate (13, 25, 26, 39a). The LF and HF values most likely indicate only the degree of variability within the HR and not nerve activity.
The continuous distribution of HRV among both strain sets (Fig. 2) suggests that HRV is a complex physiological trait. A significant QTL on chromosome 5 (Hrvhf1) and 3 suggestive QTLs were identified for HF HRV in the RI strains. Candidate genes in Hrvhf1 include Drd5, which encodes the D5 dopamine receptor, and mice deficient in Drd5 develop hypertension caused by a CNS defect increasing sympathetic outflow (20). Peroxisome proliferative-activated receptor-γ coactivator-1α (Pcg1α) has a critical role in cardiac mitochondrial function, and hearts from Pcg1α−/− mice had a reduced capacity for increasing cardiac output in response to stimulation (3), and the overexpression of PCG1α led to cardiomyopathy (30). Endothelial nitric oxide synthase (eNOS) has also been shown to be important of cardiac function and pathology (4, 8). With the consideration of the effect of these genes on cardiovascular function, it is reasonable to suggest an effect on HF. However, given the size of Hrvhf1 and other QTLs identified in this study, it is possible that other genes important in HR and HRV regulation may be identified after fine mapping of these regions.
Genetic analysis of factors regulating HF HRV did not produce evidence of genes involved in respiratory sinus arrhythmia, the principal determinant of HRV at these higher frequencies (18). However, no differences in baseline breathing frequency were observed between the RI strains (Table 2). Therefore, respiratory sinus arrhythmia did not account for the differences in HRV in these data, and other factors were perhaps more important in these resting mice. QTLs containing genes associated with respiratory sinus arrhythmia may only be found if variability in breathing frequency is used a phenotype.
Figure 2 highlights the potential differences between murine and human cardiac regulation. It is well known that in resting humans higher HF HRV is observed relative to LF HRV (39a). However, differences in the HRV characteristics were found between these strains. Fifteen of the inbred strains had higher LF compared with HF HRV than the others (Fig. 2B). Moreover, in PL and BALB mice, higher HF HRV levels were observed, but these strains also had relatively low HR and TP (Fig. 3A). Conversely, in the C57L and CAST strains, high HRs but different levels of HRV were found. The present data demonstrate the complexity of between-strain differences in the autonomic nervous system regulation of the murine heart at rest, but its relationship between murine cardiac responses to acute cardiovascular perturbations remains unclear.
This study reports a clear genetic influence on HR, HRV, and nV̇e using a broad range of inbred mouse strains. However, the influence of genotype on HR and, in particular, HRV is highly complex. This is supported by the continuous distribution of HR and HRV among the strain sets and the multiple QTLs for each phenotype. The complexity of cardiovascular regulation is well known (17) and should be kept in mind when discussing the importance of candidate genes that may play a role in cardiac regulation. The candidate genes discussed were selected based on previously reported associations with cardiovascular function. However, this list of genes was not intended to be definitive. The QTLs identified may contain many other genes that are involved in HR and HRV regulation. This study provides the basis to investigate the precise interaction between genotype and the underlying mechanisms associated with murine heart regulation.
GRANTS
This research was funded by the Intramural Research program of the National Institute of Environmental Health Sciences, National Institutes of Health, Department of Health and Human Services.
Acknowledgments
We thank Drs. Donald Cook and Abraham Nyska for reviewing the manuscript.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Abildstrom SZ, Jensen BT, Agner E, Torp-Pedersen C, Nyvad O, Wachtell K, Ottesen MM, Kanters JK. Heart rate versus heart rate variability in risk prediction after myocardial infarction. J Cardiovasc Electrophysiol 14: 168–173, 2003. [DOI] [PubMed] [Google Scholar]
- 2.Acharya U, Rajendra NK, Sing O, Ping L, Chua T. Heart rate analysis in normal subjects of various age groups. BioMedical Engineering Online 3: 24, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arany Z, He H, Lin J, Hoyer K, Handschin C, Toka O, Ahmad F, Matsui T, Chin S, Wu PH, Rybkin II, Shelton JM, Manieri M, Cinti S, Schoen FJ, Bassel-Duby R, Rosenzweig A, Ingwall JS, Spiegelman BM. Transcriptional coactivator PGC-1 alpha controls the energy state and contractile function of cardiac muscle. Cell Metab 1: 259–271, 2005. [DOI] [PubMed] [Google Scholar]
- 4.Barouch LA, Cappola TP, Harrison RW, Crone JK, Rodriguez ER, Burnett AL, Hare JM. Combined loss of neuronal and endothelial nitric oxide synthase causes premature mortality and age-related hypertrophic cardiac remodeling in mice. J Mol Cell Cardiol 35: 637–644, 2003. [DOI] [PubMed] [Google Scholar]
- 5.Bigger JT Jr, Fleiss JL, Steinman RC, Rolnitzky LM, Schneider WJ, Stein PK. RR variability in healthy, middle-aged persons compared with patients with chronic coronary heart disease or recent acute myocardial infarction. Circulation 91: 1936–1943, 1995. [DOI] [PubMed] [Google Scholar]
- 6.Brown TE, Beightol LA, Koh J, Eckberg DL. Important influence of respiration on human R-R interval power spectra is largely ignored. J Appl Physiol 75: 2310–2317, 1993. [DOI] [PubMed] [Google Scholar]
- 7.Campen MJ, Tagaito Y, Jenkins TP, Smith PL, Schwartz AR, O'Donnell CP. Phenotypic differences in the hemodynamic response during REM sleep in six strains of inbred mice. Physiol Genomics 11: 227–234, 2002. [DOI] [PubMed] [Google Scholar]
- 8.Cappola TP, Cope L, Cernetich A, Barouch LA, Minhas K, Irizarry RA, Parmigiani G, Durrani S, Lavoie T, Hoffman EP, Ye SQ, Garcia JGN, Hare JM. Deficiency of different nitric oxide synthase isoforms activates divergent transcriptional programs in cardiac hypertrophy. Physiol Genomics 14: 25–34, 2003. [DOI] [PubMed] [Google Scholar]
- 9.Cho HY, Jedlicka AE, Reddy SP, Zhang LY, Kensler TW, Kleeberger SR. Linkage analysis of susceptibility to hyperoxia. Nrf2 is a candidate gene. Am J Respir Cell Mol Biol 26: 42–51, 2002. [DOI] [PubMed] [Google Scholar]
- 10.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conti LH, Jirout M, Breen L, Vanella JJ, Schork NJ, Printz MP. Identification of quantitative trait Loci for anxiety and locomotion phenotypes in rat recombinant inbred strains. Behav Genet 34: 93–103, 2004. [DOI] [PubMed] [Google Scholar]
- 12.Doerge RW, Churchill GA. Permutation tests for multiple loci affecting a quantitative character. Genetics 142: 285–294, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckberg DL Sympathovagal balance: a critical appraisal. Circulation 96: 3224–3232, 1997. [DOI] [PubMed] [Google Scholar]
- 16.Gill KJ, Boyle AE. Genetic basis for the psychostimulant effects of nicotine: a quantitative trait locus analysis in AcB/BcA recombinant congenic mice. Genes Brain Behav 4: 401–411, 2005. [DOI] [PubMed] [Google Scholar]
- 17.Guyton AC, Coleman TG, Granger HJ. Circulation: overall regulation. Annu Rev Physiol 34: 13–46, 1972. [DOI] [PubMed] [Google Scholar]
- 18.Hayano J, Mukai S, Sakakibara M, Okada A, Takata K, Fujinami T. Effects of respiratory interval on vagal modulation of heart rate. Am J Physiol Heart Circ Physiol 267: H33–H40, 1994. [DOI] [PubMed] [Google Scholar]
- 19.Hoit BD, Kiatchoosakun S, Restivo J, Kirkpatrick D, Olszens K, Shao H, Pao YH, Nadeau JH. Naturally occurring variation in cardiovascular traits among inbred mouse strains. Genomics 79: 679–685, 2002. [DOI] [PubMed] [Google Scholar]
- 20.Hollon TR, Bek MJ, Lachowicz JE, Ariano MA, Mezey E, Ramachandran R, Wersinger SR, Soares-da-Silva P, Liu ZF, Grinberg A, Drago J, Young WS 3rd, Westphal H, Jose PA, Sibley DR. Mice lacking D5 dopamine receptors have increased sympathetic tone and are hypertensive. J Neurosci 22: 10801–10810, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Just A, Faulhaber J, Ehmke H. Autonomic cardiovascular control in conscious mice. Am J Physiol Regul Integr Comp Physiol 279: R2214–R2221, 2000. [DOI] [PubMed] [Google Scholar]
- 22.Kreutz R, Struk B, Stock P, Hubner N, Ganten D, Lindpaintner K. Evidence for primary genetic determination of heart rate regulation: chromosomal mapping of a genetic locus in the rat. Circulation 96: 1078–1081, 1997. [DOI] [PubMed] [Google Scholar]
- 23.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11: 241–247, 1995. [DOI] [PubMed] [Google Scholar]
- 24.Liao D, Myers R, Hunt S, Shahar E, Paton C, Burke G, Province M, Heiss G. Familial history of stroke and stroke risk: The Family Heart Study. Stroke 28: 1908–1912, 1997. [DOI] [PubMed] [Google Scholar]
- 25.Malliani A The pattern of sympathovagal balance explored in the frequency domain. News Physiol Sci 14: 111–117, 1999. [DOI] [PubMed] [Google Scholar]
- 26.Malpas SC Neural influences on cardiovascular variability: possibilities and pitfalls. Am J Physiol Heart Circ Physiol 282: H6–H20, 2002. [DOI] [PubMed] [Google Scholar]
- 27.Marks BL, Lightfoot JT. Reproducibility of resting heart rate variability with short sampling periods. Can J Appl Physiol 24: 337–348, 1999. [DOI] [PubMed] [Google Scholar]
- 28.Pagani M, Lombardi F, Guzzetti S, Rimoldi O, Furlan R, Pizzinelli P, Sandrone G, Malfatto G, Dell'Orto S, Piccaluga E. Power spectral analysis of heart rate and arterial pressure variabilities as a marker of sympathovagal interaction in man and conscious dog. Circ Res 59: 178–193, 1986. [DOI] [PubMed] [Google Scholar]
- 29.Parati G, Di Rienzo M. Determinants of heart rate and heart rate variability. J Hypertens 21: 477–480, 2003. [DOI] [PubMed] [Google Scholar]
- 30.Russell LK, Mansfield CM, Lehman JJ, Kovacs A, Courtois M, Saffitz JE, Medeiros DM, Valencik ML, McDonald JA, Kelly DP. Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor γ coactivator-1α promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ Res 94: 525–533, 2004. [DOI] [PubMed] [Google Scholar]
- 31.Singh JP, Larson MG, O'Donnell CJ, Tsuji H, Evans JC, Levy D. Heritability of heart rate variability: The Framingham Heart Study. Circulation 99: 2251–2254, 1999. [DOI] [PubMed] [Google Scholar]
- 32.Sinnreich R, Friedlander Y, Luria MH, Sapoznikov D, Kark JD. Inheritance of heart rate variability: the kibbutzim family study. Hum Genet 105: 654–661, 1999. [DOI] [PubMed] [Google Scholar]
- 33.Stein KM, Borer JS, Hochreiter C, Okin PM, Herrold EM, Devereux RB, Kligfield P. Prognostic value and physiological correlates of heart rate variability in chronic severe mitral regurgitation. Circulation 88: 127–135, 1993. [DOI] [PubMed] [Google Scholar]
- 34.Stiedl O, Meyer M. Cardiac dynamics in corticotropin-releasing factor receptor subtype-2 deficient mice. Neuropeptides 37: 3–16, 2003. [DOI] [PubMed] [Google Scholar]
- 35.Sugiyama F, Churchill GA, Li R, Libby LJ, Carver T, Yagami K, John SW, Paigen B. QTL associated with blood pressure, heart rate, and heart weight in CBA/CaJ and BALB/cJ mice. Physiol Genomics 10: 5–12, 2002. [DOI] [PubMed] [Google Scholar]
- 36.Tankersley CG, Bierman A, Rabold R. Variation in heart rate regulation and the effects of particle exposure in inbred mice. Inhal Toxicol 19: 621–629, 2007. [DOI] [PubMed] [Google Scholar]
- 37.Tankersley CG, Fitzgerald RS, Kleeberger SR. Differential control of ventilation among inbred strains of mice. Am J Physiol Regul Integr Comp Physiol 267: R1371–R1377, 1994. [DOI] [PubMed] [Google Scholar]
- 38.Tankersley CG, Fitzgerald RS, Levitt RC, Mitzner WA, Ewart SL, Kleeberger SR. Genetic control of differential baseline breathing pattern. J Appl Physiol 82: 874–881, 1997. [DOI] [PubMed] [Google Scholar]
- 39.Tankersley CG, Irizarry R, Flanders S, Rabold R. Circadian rhythm variation in activity, body temperature, and heart rate between C3H/HeJ and C57BL/6J inbred strains. J Appl Physiol 92: 870–877, 2002. [DOI] [PubMed] [Google Scholar]
- 39a.Task Force of the European Society of Cardiology and the North American Society of Pacing and Electrophysiology. Heart rate variability. Standards of measurement, physiological interpretation, and clinical use. Task Force of the European Society of Cardiology and the North American Society of Pacing and Electrophysiology. Circulation 93: 1043–1065, 1996. [PubMed] [Google Scholar]
- 40.Tovote P, Meyer M, Beck-Sickinger AG, von Horsten S, Ove Ogren S, Spiess J, Stiedl O. Central NPY receptor-mediated alteration of heart rate dynamics in mice during expression of fear conditioned to an auditory cue. Regul Pept 120: 205–214, 2004. [DOI] [PubMed] [Google Scholar]
- 41.Tsuji H, Larson MG, Venditti FJ Jr, Manders ES, Evans JC, Feldman CL, Levy D. Impact of reduced heart rate variability on risk for cardiac events. The Framingham Heart Study. Circulation 94: 2850–2855, 1996. [DOI] [PubMed] [Google Scholar]
- 42.Voigt B, Kuramoto T, Mashimo T, Tsurumi T, Sasaki Y, Hokao R, Serikawa T. Evaluation of LEXF/FXLE rat recombinant inbred strains for the genetic dissection of complex traits. Physiol Genomics 32: 335–342, 2007. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Williams RW, Manly KF. WebQTL: web-based complex trait analysis. Neuroinformatics 1: 299–308, 2003. [DOI] [PubMed] [Google Scholar]