Abstract
Peroxiredoxins (Prdxs), a family of antioxidant and redox-signaling proteins, are plentiful within the heart; however, their cardiac functions are poorly understood. These studies were designed to characterize the complex changes in Prdxs induced by oxidant stress in rat myocardium. Hydrogen peroxide, a Prdx substrate, was used as the model oxidant pertinent to redox signaling during health and to injury at higher concentrations. Rat hearts were aerobically perfused with a broad concentration range of hydrogen peroxide by the Langendorff method, homogenized, and analyzed by immunoblotting. Heart extracts were also analyzed by size-exclusion chromatography under nondenaturing conditions. Hydrogen peroxide-induced changes in disulfide bond formation, nonreversible oxidation of cysteine (hyperoxidation), and subcellular localization were determined. Hydrogen peroxide induced an array of changes in the myocardium, including formation of disulfide bonds that were intermolecular for Prdx1, Prdx2, and Prdx3 but intramolecular within Prdx5. For Prdx1, Prdx2, and Prdx5, disulfide bond formation can be approximated to an EC50 of 10–100, 1–10, and 100–1,000 μM peroxide, respectively. Hydrogen peroxide induced hyperoxidation, not just within monomeric Prdx (by SDS-PAGE), but also within Prdx disulfide dimers, and reflects a flexibility within the dimeric unit. Prdx oxidation was also associated with movement from the cytosolic to the membrane and myofilament-enriched fractions. In summary, Prdxs undergo a complex series of redox-dependent structural changes in the heart in response to oxidant challenge with its substrate hydrogen peroxide.
Keywords: hydrogen peroxide, redox signaling, cysteine
the peroxiredoxins (Prdxs), a diverse family of antioxidant proteins, exhibit peroxidase and chaperone activities and participate in redox signaling (18, 43). These proteins are highly abundant in the heart, yet relatively little is known about their response during cardiac oxidative stress. Most work has indicated that Prdxs within the heart and vasculature are cytoprotective. Within erythrocytes, Prdx1 and Prdx2 protect hemoglobin from irreparable damage inflicted by reactive oxygen species, which otherwise leads to anemia (20, 30) and disrupts the vasodilatory role of hemoglobin (33). Prdx1 and Prdx2 suppress tyrosine kinase signaling, potentially regulating vascular remodeling (18). Prdx2 also prevents monocyte attachment to endothelial cells in response to treatment with oxidized LDL (40), an otherwise injurious inflammatory event. Prdx3, Prdx4, Prdx5, and Prdx6 are downregulated in the failing myocardium, at least in terms of mRNA expression (4). Prdx3 confers protection against oxidative stress within the mitochondria of aortic cells and within the ischemic rat myocardium (1) and also prevents left ventricular remodeling and failure after myocardial infarction (24). Prdx6-null mice are more susceptible to ischemia-reperfusion injury (29) but not to atherosclerosis (41). Prdx6, similar to Prdx1, Prdx2, Prdx3, and Prdx4, is prone to hyperoxidation, but not within murine hearts null for protein kinase Cδ (25). Prdxs are well known to suppress apoptosis mediated by hydrogen peroxide (18). Previous studies highlight the high abundance of cardiac Prdx (3, 9, 25).
Mammalian Prdx enzymes can be subdivided into six subtypes (Prdx1, Prdx2, Prdx3, Prdx4, Prdx5, and Prdx6) on the basis of protein sequence and catalytic mechanism (14, 36, 43). All subtypes share a common mechanism that involves oxidation of a conserved peroxidatic cysteine to a sulfenic acid group by a peroxide molecule. For Prdx1, Prdx2, Prdx3, and Prdx4 (typical 2-Cys Prdxs), the peroxidatic cysteine SOH is attacked by a second conserved cysteine (the resolving cysteine) from a second monomer, and the resultant disulfide bond is reduced by a disulfide reductase, such as thioredoxin. Prdx5 is referred to as an atypical 2-Cys Prdx, because the resolving cysteine resides within the same monomer. In the case of Prdx6 (a 1-Cys Prdx), the peroxidatic sulfenic acid group is reduced by glutathione in a reaction catalyzed by π-glutathione transferase (23) or ascorbate (27).
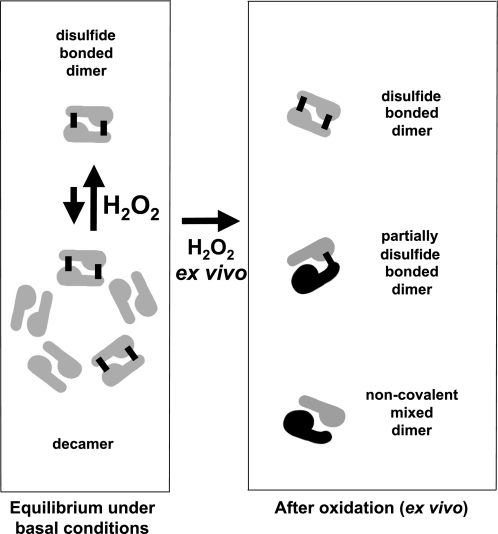
The dimer is the smallest structural subunit for all mammalian Prdxs. Catalytically, Prdx1, Prdx2, Prdx3, and Prdx4 function as dimers able to form disulfide bonds between both pairs of peroxidatic and resolving cysteine. Prdx5 is functional as a monomer and forms an intramolecular disulfide, whereas Prdx6 is functional as a dimer, although the catalytic cysteine is only known to form mixed disulfides with GSH within the reaction cycle. The peroxidatic and chaperone activities of Prdx are dependent on the oligomeric state of the protein (18, 32, 43). Prdxs most commonly exist as a dynamic equilibrium of dimers and decamers (5 dimers arranged as a decameric ring), although higher oligomers have been reported (43). Interconversion and stability of these various discrete structures are dependent on the oxidation state of the catalytic cysteines, that is, whether the peroxidatic cysteine is reduced (SH), sulfinated (SO2), or sulfonated (SO3; i.e., hyperoxidized), in a disulfide bond with the resolving cysteine, or in a disulfide bond formation involving dimers (in the case of Prdx1) (21).
Molecular chaperone activity is also clearly associated with increased oligomer formation induced by oxidative or heat stress (18, 21). In contrast, comprehensive structure-peroxidase activity studies have only been conducted on the distantly related bacterial Prdx in vitro (32), and our understanding of the influence of oligomer formation on the in vivo peroxidatic activity of mammalian Prdx is poor. A more detailed understanding of the in vivo oligomeric status of cardiac Prdx is therefore of direct importance to understanding the activity of Prdx within the heart during redox signaling or oxidant stress. Therefore, the aim of the present study was to characterize changes in the redox-dependent structural status of Prdxs within cardiac tissue in response to hydrogen peroxide, a model oxidant important for cardiac redox signaling, as well as a perpetrator of injury at higher concentrations. The perfusion of isolated hearts with hydrogen peroxide in an ex vivo setting is a widely used and well-established procedure in cardiovascular research. Application of 0.01–1 mM hydrogen peroxide to isolated cells and intact organ tissues is a commonly used procedure that mimics the release of endogenous peroxide through growth factors and other effectors. This approach is considered relevant to physiology and, at higher concentrations, to pathology. Although endogenous peroxide levels are unlikely to ever exceed 0.1 mM (except perhaps very locally), application of exogenous peroxide results in a final intracellular concentration that is likely to be ≤1–15% of the applied concentration (37).
METHODS
Experimental procedures.
This investigation was performed in accordance with the Home Office Guidance on the Operation of the Animals (Scientific Procedures) Act 1986, published by Her Majesty's Stationery Office (London, UK). Animals were maintained humanely in compliance with the “Principles of Laboratory Animal Care” formulated by the National Society for Medical Research and the Guide for Care and Use of Laboratory Animals prepared by the National Academy of Sciences and published by the National Institutes of Health (NIH Pub. No. 85-23, revised 1985). All animal protocols were approved both by the local King's College Ethical Review Process Committee and by the UK Government Home Office (Animals Scientific Procedures Group).
Chemicals.
Chemicals were obtained from Sigma Chemical (Poole, UK) or VWR (Lutterworth, UK) unless otherwise stated and were of analytical research grade or above.
Preparation of isolated rat hearts perfused with hydrogen peroxide.
Male Wistar rats (4–6 wk old, 250–300 g body wt; B & K Universal, Hull, UK) were anesthetized with pentobarbitone sodium (40 mg ip) and injected with sodium heparin (200 IU) via the femoral vein. Hearts were rapidly excised, placed in cold (4°C) bicarbonate buffer (10), cannulated, and perfused at a constant flow of 12 ml·min−1·g tissue−1 (10) as follows: 30 min of aerobic perfusion with bicarbonate buffer followed by 5 min of perfusion with 0–10,000 μM hydrogen peroxide in bicarbonate buffer. At the end of the perfusion protocol, hearts were frozen and stored in liquid nitrogen until further analysis. Hearts were prepared as 10% homogenates (10 ml buffer/g cardiac tissue) in cold 100 mM Tris·HCl (pH 7.2), 100 mM maleimide, and protease inhibitors (Complete C, Roche). Hearts were disrupted by mechanical tissue disruption using a tissue grinder (Polytron). An unfractionated aliquot was reconstituted in nonreducing maleimide-SDS sample buffer. For subcellular fractions, the 10% ventricle homogenate was centrifuged at 24,000 g for 5 min, and the supernatant was designated the cytosol. The pellet was resuspended in homogenization buffer containing 1% Triton X-100 and centrifuged as described above, with the new supernatant enriched in membrane-associated proteins and the pellet enriched in integral membrane proteins, myofilament, and nuclear proteins. The pellet was resuspended in homogenization buffer supplemented with 1% Triton X-100 and centrifuged as described above. The new supernatant is known to be enriched in integral membrane marker proteins such as the Na-K-ATPase and sarco(endoplasmic) reticulum Ca-ATPase, whereas the pellet is enriched with myofilament and nuclear proteins.
Prdx subtype detection by immunoblotting and immunodetection.
Protein samples prepared from hearts were analyzed by SDS-PAGE using the Mini Protean 3 system (Bio-Rad, Hemel Hempstead, UK). Aliquots of each sample were treated with an equal volume of 2× SDS nonreduced sample buffer [100 mM Tris·HCl (pH 6.8), 4% SDS, 0.02% bromphenol blue, and 20% glycerol] before SDS-PAGE. Reduced samples were prepared with 10% (vol/vol) mercaptoethanol. After electrophoresis, samples were transferred onto polyvinylidene difluoride membranes (GE Healthcare, Little Chalfont, UK) using a semidry Transblot Transfer Cell (Bio-Rad). Blots were incubated with primary antibodies (diluted 1:1,000–10,000 in 5% milk + PBS-Tween) for 3 h at room temperature or overnight at 4°C. Horseradish peroxidase-coupled anti-mouse IgG secondary antibody (diluted 1:1,000 in 5% milk + PBS-Tween and applied for 1 h at room temperature) was used to detect primary antibodies bound to the blot, together with enhanced chemiluminescence reagent (GE Healthcare). Nonreduced and reduced immunoblots were probed with polyclonal rabbit antibodies against Prdx1, Prdx2, Prdx3, Prdx5, Prdx6, and Prdx SO2/SO3. Primary antibodies were purchased from Lab Frontier (Seoul, Korea), with the exception of purified rabbit polyclonal antibody raised against full-length human Prdx2, which was generously provided by Dr. Leslie Poole (Wake Forest University School of Medicine, Winston-Salem, NC). Nonreduced blots derived for Prdx1, Prdx2, and hyperoxidized Prdx were digitized and quantitatively analyzed using NIH Image software (Freeware, National Institutes of Health, Bethesda, MD). Values are means ± SE. Differences between groups were assessed using ANOVA followed by a t-test and were considered significant at the 95% confidence level.
RESULTS
Prdx subtype detection by immunoblotting.
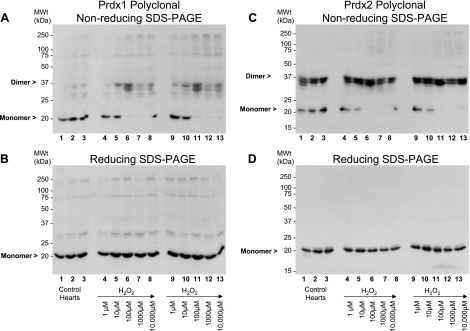
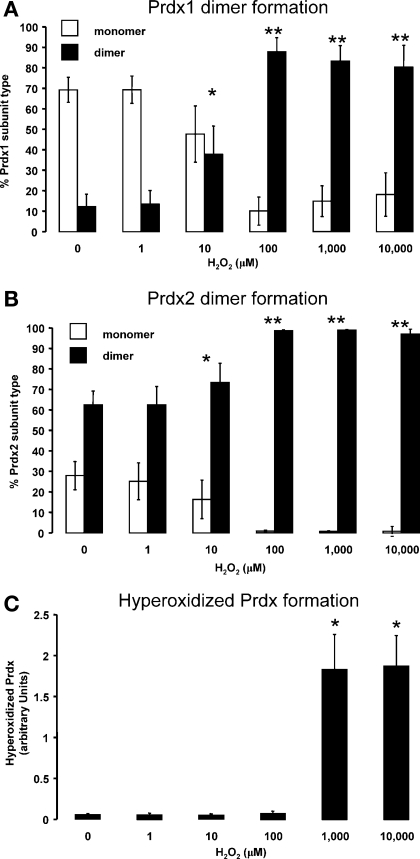
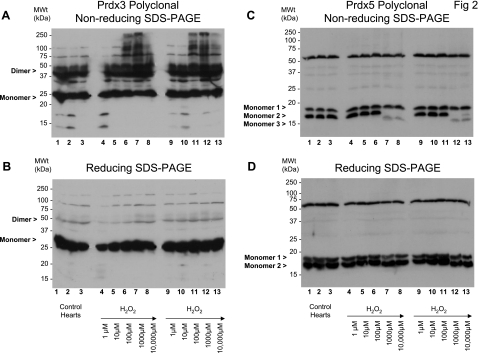
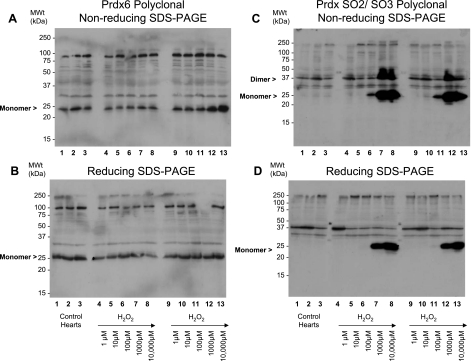
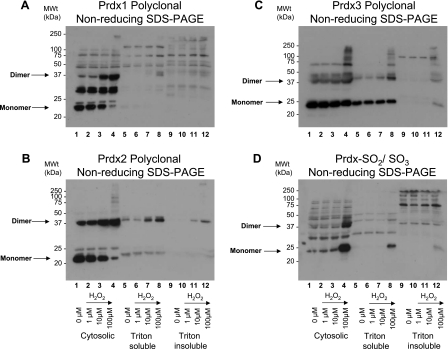
Rat hearts were perfused with 0–10,000 μM hydrogen peroxide and subsequently prepared in the presence of maleimide, which should prevent any further thiol modifications by alkylating free cysteinyl thiols. Under basal (control) conditions, Prdx1 migrates predominantly as a monomer (Fig. 1A, lanes 1–3). Treatment of a series of hearts with 1–10,000 μM hydrogen peroxide induced the formation of disulfide dimers (Fig. 1A, lanes 4–8 and 9–13, and Fig. 4A). These Prdx1 disulfide dimers were absent under reducing conditions (Fig. 1B). Hydrogen peroxide at 10–100 μM was adequate to completely disulfide dimerize Prdx1. In comparison, approximately two-thirds of Prdx2 already existed as disulfide-dependent dimers under basal conditions (Fig. 1C, lanes 1–3). Treatment of a series of hearts with 1–10,000 μM hydrogen peroxide further induced the formation of disulfide dimers (Fig. 1C, lanes 4–8 and 9–13, and Fig. 4B). Hydrogen peroxide at 10–100 μM was sufficient to completely dimerize Prdx2. Multiple dimeric species are clearly present and may differ in terms of minor degradation or phosphorylation state. A small fraction of the total Prdx1 and Prdx2 appears to form large (>250-kDa) oligomers in response to high (>100 μM) levels of hydrogen peroxide (Fig. 1, A and C, lanes 6–8 and 11–13). For Prdx2 at least, these species break down after reduction (Fig. 1D). Approximately half of all the Prdx3 already existed as disulfide-dependent dimers under basal conditions, and hydrogen peroxide did not appear to induce the formation of additional dimers. Instead, treatment of a series of hearts with 1–10,000 μM hydrogen peroxide induces the formation of a succession of disulfide-dependent species (∼75, 100, and >250 kDa) that are larger than the disulfide dimer (Fig. 2, A lanes 6–8 and 11–13). However, these species are readily reducible (Fig. 2B, lanes 6–8 and 11–13). Prdx5 forms two monomer species in the 15- to 20-kDa range under basal conditions (referred to as monomer 1 and monomer 2), although an increase in the hydrogen peroxide concentration from 100 to 1,000 μM causes disappearance of monomer 2 and emergence of a smaller species, monomer 3. However, these are only observed under nonreducing conditions (cf. Fig. 2C, lanes 7 and 8 and lanes 12 and 13, with Fig. 2B, lanes 7 and 8 and lanes 12 and 13). This transition is consistent with Prdx5 forming an intramolecular (rather than intermolecular) disulfide bond that displays a faster mobility by SDS-PAGE, as reported by others (11), although half of the total monomeric Prdx5 appears not to do so (monomer 1). Finally, Prdx6 runs as a ∼25-kDa species under basal conditions that correlates with the cDNA-derived mass under reduced (Fig. 3B) or nonreduced (Fig. 3A) conditions. Treatment with hydrogen peroxide did not alter Prdx6 migration, which is a notable contrast to the other isoforms. The bands that migrate at >25 kDa are unlikely to be Prdx6, inasmuch as they are not lost under reducing conditions and their banding pattern is not modulated by any of the peroxide interventions. Consequently, these bands, including the strong one at 75–100 kDa, are likely nonspecific.
Fig. 1.
Effect of hydrogen peroxide on cardiac peroxiredoxin (Prdx) 1 (Prdx1) and Prdx2. Nonreduced (A and C) and reduced (B and D) immunoblots of whole rat cardiac extracts were probed with polyclonal antibodies against Prdx1 and Prdx2. Treatment of a series of hearts with 1–10,000 μM hydrogen peroxide induced formation of disulfide dimers for Prdx1 (A lanes 4–8 and 9–13) and Prdx2 (C, lanes 4–8 and 9–13). Treatments were carried out in duplicate hearts. Dimers are absent under reducing conditions (B and D).
Fig. 4.
Quantification of formation of disulfide dimers of Prdx1 (A) and Prdx2 (B) in rat hearts after treatment with 0–10,000 μM hydrogen peroxide. Data from ≥3 independent experiments are shown as percentage of Prdx1 or Prdx2 that exists as monomer or dimer after peroxide treatment. Measurements were made from nonreducing blots, examples of which are shown in Fig. 1. C: quantitation of Prdx hyperoxidation, which is predominantly derived from Prdx1, Prdx2, Prdx3, and Prdx4, after hydrogen peroxide treatment. Values are means ± SE. Significantly different from nonperoxide (control): *P < 0.05; **P < 0.01.
Fig. 2.
Effect of hydrogen peroxide on cardiac Prdx3 and Prdx5. Nonreduced (A and C) and reduced (B and D) immunoblots of whole rat cardiac extracts were probed with polyclonal antibodies that detect Prdx3 and Prdx5 from hearts treated in duplicate. Hydrogen peroxide induces formation of a succession (∼75, 100, and >250 kDa) of disulfide-dependent species (A, lanes 6–8 and 11–13). These species are readily reducible (B, lanes 6–8 and 11–13). Under basal conditions, Prdx5 forms 2 monomer species, although an increase in hydrogen peroxide concentration causes disappearance of monomer 2 and emergence of a smaller species, monomer 3, but only under nonreducing conditions (cf. C, lanes 7 and 8 and lanes 12 and 13, with D, lanes 7 and 8 and lanes 12 and 13).
Fig. 3.
Effect of hydrogen peroxide on cardiac Prdx6 and Prdx hyperoxidized to cysteine sulfinic/sulfonic acid (SO2/SO3) at peroxidatic cysteine. Nonreduced (A) and reduced (B) immunoblots of cardiac extracts were probed with a polyclonal antibody against Prdx6 from hearts treated in duplicate. Prdx6 runs as a ∼25-kDa species under all conditions, regardless of hydrogen peroxide concentration. Nonreduced (C) and reduced (B and D) immunoblots of cardiac extracts were probed with polyclonal antibodies against hyperoxidized Prdx (Prdx1, Prdx2, Prdx3, and Prdx4). Significant amounts of hyperoxidized Prdx are only detected after treatment with ≥100 μM hydrogen peroxide (C and D, lanes 7 and 8 and lanes 12 and 13). Hyperoxidized Prdx can associate with nonhyperoxidized Prdx and form a hybrid dimer that is part disulfide, part hyperoxidized (C, lanes 7 and 8 and lanes 12 and 13) and is reducible (D, lanes 7 and 8 and lanes 12 and 13).
Hyperoxidation of cardiac Prdx in response to hydrogen peroxide.
Hyperoxidized Prdx was detected with an antibody raised against an active site peptide, the sequence of which is common to four Prdx isozymes (Prdx1, Prdx2, Prdx3, and Prdx4) and is hyperoxidized to a mixture of sulfinic (SO2) and sulfonic (SO3) acids at the peroxidatic cysteine (42). Significant amounts of hyperoxidized Prdx were only detected after treatment with >100 μM hydrogen peroxide (Fig. 3, C and D, lanes 7 and 8 and lanes 12 and 13). Hyperoxidation of Prdx1, Prdx2, Prdx3, and Prdx4 was quantitated (Fig. 4C) using data from three separate experiments. Unfortunately, no pan-specific Prdx1, Prdx2, Prdx3, and Prdx4 “total” antibody is available to serve as a loading control. However, probing the same samples under reducing conditions with antibodies to Prdx1, Prdx2, Prdx3, Prdx5, and Prdx6 indicates that the expression levels of these subtypes do not change with the interventions tested. The nonreduced profile suggests that hyperoxidized Prdx can associate with nonhyperoxidized Prdx and form a hybrid dimer that contains a single disulfide bond and a single hyperoxidized peroxidatic cysteine residue (Fig. 3C, lanes 7 and 8 and lanes 12 and 13), as represented in schematic form in Fig. 6. This dimer is readily reducible (Fig. 3D, lanes 7 and 8 and lanes 12 and 13).
Fig. 6.
Oligomeric behavior of cardiac Prdx2 during ex vivo treatment with hydrogen peroxide. Under basal conditions, Prdx2 exists as a mixture of disulfide-bonded dimers and partially reduced decamers. Disulfide bonds are indicated by black bars. Ex vivo treatment with 100 μM hydrogen peroxide drives oxidation of decamers and reduced dimers into disulfide-bound dimers, coincident with breakdown of remaining decamers and hyperoxidation of some monomers.
Effect of hydrogen peroxide on subcellular distribution of Prdx1, Prdx2, and Prdx3.
Fractionated heart (control or hydrogen peroxide treated) samples prepared for Western blot analysis were probed with polyclonal antibodies against Prdx1, Prdx2, Prdx3, or hyperoxidized Prdx. Prdx1 and Prdx2 are mostly confined to the soluble fraction (Figs. 5A and 4B), although a proportion of Prdx2 disulfide dimer translocated from the cytosol to the membrane-enriched fraction in response to treatment with 10–100 μM hydrogen peroxide. Prdx3 also translocated to the membrane fraction, as well as the Triton-insoluble fraction enriched in myofilament proteins in response to oxidant (Fig. 5C). Treatment with elevated levels of hydrogen peroxide (100 μM) induced hyperoxidation of Prdx, and this also appears in the membrane and, to a lesser extent, in the myofilament compartment (Fig. 5D). Some of the increased amounts of dimer at the membrane may have resulted from dimerization events at the membrane, rather than solely from translocation of Prdx from the cytosol.
Fig. 5.
Effect of hydrogen peroxide on compartmental distribution of Prdx1, Prdx2, and Prdx3 within whole heart extracts. Rat hearts were exposed to increasing concentrations of hydrogen peroxide and fractionated into cytosolic, membrane-associated (Triton-soluble), and integral membrane (Triton-insoluble) compartments. Extracts were treated without reducing agent, and fractions were probed with polyclonal antibodies to Prdx1, Prdx2, Prdx3, and Prdx4 and to hyperoxidized Prdx (hyperoxidized Prdx1, Prdx2, Prdx3, and Prdx4). Prdx1 (A), Prdx2, and Prdx3 are almost entirely confined to the cytosol, although translocation is apparent for some Prdx2 (B) and Prdx3 (C) from the cytosol to the membrane in response to hydrogen peroxide. Hyperoxidation causes Prdx translocation to membrane-associated and integral membrane compartments (D).
DISCUSSION
Treatment of rat hearts with an increasing concentration of exogenously applied hydrogen peroxide clearly alters the oligomeric structure of a number of Prdx proteins, which reflects changes in the redox status of one or both of the catalytic cysteine residues. Prdx1 and Prdx2 react in a similar fashion, in that they readily form disulfide bonds in response to 10–100 μM hydrogen peroxide; under basal conditions, however, Prdx1 is more reduced than Prdx2. This is consistent with the observation that hydrogen peroxide elicits less reaction with Prdx1 than Prdx2 (21). Multiple dimeric bands are present for Prdx1 and Prdx2 in the samples illustrated in Fig. 1, although the extent of the dimeric bands is less for the equivalent samples shown in Fig. 5. This discrepancy is most probably due to differences in the extent of degradation between samples. The monomer-to-(covalent) dimer transitions on gels reported here compare well with oligomeric changes characterized for Prdx within eukaryotes (5, 22, 39) and prokaryotes (32), although Low et al. (22) detected complete dimerization of erythrocyte Prdx2 with only 5 μM hydrogen peroxide. Our approach of extracting cardiac tissues in the presence of the alkylating reagent maleimide is intended to trap the redox state of the cysteine residues within all proteins, by preventing further modification of Cys-SH, and is a commonly applied and accepted approach (3). However, alkylation may only be partially effective in preventing artifactual oxidation during sample preparation, especially under native conditions in the absence of SDS. As a result, the extent of dimer formation as indicated in Figs. 1–3 (and quantitated in Fig. 4) may be overstated. Nevertheless, because all samples were prepared equally, our data still present a valid analysis of the changes in the extent of Prdx dimerization in response to oxidation within cardiac tissues.
Prdx3 differs from Prdx1 and Prdx2, in that it appears unaffected by exposure to exogenous hydrogen peroxide in terms of increased dimer formation. Instead, Prdx3 appears to be more prone to oligomer formation than Prdx1 and Prdx2 and, in response to hydrogen peroxide, forms species that represent multiples of monomers (75 kDa = 3 × 25 kDa, 100 kDa = 4 × 25 kDa, ≥250 kDa ≥ 10 × 25 kDa). Prdx3 oligomer formation has been observed by crystallographic and electron-microscopic studies (6, 12, 13) and by gel and circular dichroism analysis of recombinant Prdx3 (12).
As an atypical 2-Cys Prdx, Prdx5 differs from Prdx1, Prdx2, Prdx3, and Prdx4 in having a pair of catalytic cysteines (consisting of 1 peroxidatic and 1 resolving cysteine) within one polypeptide (as opposed to 2 pairs of cysteines split between 2 polypeptides within each dimer), such that the monomer is a single functional unit capable of forming an intramolecular disulfide bond between these two residues, although Prdx5 generally always exists as a noncovalent dimer (11). We observed Prdx5 intramolecular disulfide bond formation after 100 μM hydrogen peroxide treatment, manifesting itself as a small, but reversible, increase in mobility by SDS-PAGE (11). Recently, Prdx5 was found to be translated from two alternate initiation sites, generating a smaller (17.0-kDa) protein in addition to the previously reported 21.5-kDa protein, although both variants contain the pair of reactive cysteines required for intramolecular disulfide bond formation (31); therefore, monomer 1 may relate to this species and monomer 2 to the 21.5-kDa species, in which case only the 21.5-kDa species appears to be redox active. The nature of the ∼50-kDa species is unknown and probably does not relate to Prdx5.
Prdx6 appears to show no gross structural change in response to exogenous peroxide, as evidenced by migration changes on nonreducing immunoblots. Prdx6 can form aggregated protein that is dependent on the noncatalytic Cys91 (8) and, therefore, possibly in a disulfide-dependent manner. However, we did not observe its oxidant-induced oligomerization by SDS-PAGE. In summary, disulfide bond formation within Prdx1, Prdx2, and Prdx5 can be approximated to an EC50 of 10–100, 1–10, and 100–1,000 μM, respectively, for hydrogen peroxide.
Prdxs are prone to hyperoxidation in the presence of >100 μM hydrogen peroxide in a reducing environment (e.g., DTT in vitro or thioredoxin in vivo), such that the protein is in a continual cycle of oxidation and reduction, and within each cycle a small fraction of the sulfenic acid form becomes hyperoxidized to the sulfinate or sulfonate form (44). Hyperoxidation has been reported for purified Prdx in vitro (38, 44) and for Prdx in cells and tissues treated with exogenous hydrogen peroxide (7, 26, 44). Importantly, hyperoxidation has also been demonstrated in response to peroxides generated endogenously in response to an effector molecule, such as glucose oxidase and TNFα (35), underlying the notion that Prdx hyperoxidation is, in all likelihood, a physiological event. Sulfination of the peroxidatic cysteine has been characterized in terms of the exogenous concentration of hydrogen peroxide that is required (>100 μM), in terms of its lowered isoelectric point (∼5.2 for Prdx2-SO2 compared with ∼5.5 for Prdx2-SH) (26, 42), its enzymatic repair by sulfiredoxin (2), and its influence on the protein environment at the molecular structural level (38). Within cardiac cells, the isoelectric acidification of Prdx1, Prdx2, Prdx3, Prdx4, and Prdx6 has been reported as a result of hyperoxidation (9, 25). Therefore, hyperoxidation has been observed for typical 2-Cys and for 1-Cys Prdx, but not (so far) for atypical 2-Cys Prdx, possibly because the intramolecular disulfide formation is more rapid than for typical 2-Cys Prdx, which require intermolecular disulfide bond formation. In our studies, hyperoxidation of Prdx1, Prdx2, Prdx3, and Prdx4 is apparent after treatment with >100 μM hydrogen peroxide. After treatment with 1,000–10,000 μM peroxide, most of the immunoreactivity is attributable to monomeric Prdx (Fig. 3C), although virtually all Prdx1 and Prdx2 are now in the dimeric form (Fig. 1). This would suggest that this antibody is either extremely sensitive to trace amounts of hyperoxidized Prdx or the hyperoxidized monomeric Prdx observed in Fig. 3C is predominantly Prdx3 (or Prdx4, not studied here), rather than Prdx1 or Prdx2. Because the hyperoxidized antibody used is specific for typical 2-Cys Prdx only (42), we cannot make any judgments on the hyperoxidation of Prdx5 or Prdx6 from our immunoblots.
We were surprised to find that hyperoxidation does not preclude the formation of disulfide-dependent dimers, presumably because only a single (of a possible 2) disulfide bond is required to resist SDS-dependent breakdown, such that the other peroxidatic cysteine is now hyperoxidized, inactive, and unable to form disulfide bonds. This is only the second time that this phenomenon has been reported (22). A crystallographic analysis that compared the structure for the reduced dimer of Prdx1 with that for the hyperoxidized dimer of Prdx2 suggested subtle but significant conformational differences between the two forms (38). Nevertheless, these two alternate monomer structures can clearly still accommodate one another within one dimer that contains only a single disulfide bond. This suggests that the stringency of interaction between different monomeric forms is quite low, and, in support of this, the dimerization of different, but closely related, subtypes has been reported (17). Partial disulfide dimers have also been observed for Prdx2 from erythrocytes (22).
The large complexes formed by Prdx3 (and, to a much lesser extent, Prdx1 and Prdx2) are resistant to SDS and, therefore, covalent but are readily reducible and, therefore, disulfide dependent. As such, they are presumed to involve the participation of other cysteines (apart from the peroxidatic and resolving cysteine residues) and are distinct from the well-characterized decamer composed of five dimers (which may or may not be disulfide bonded) packed noncovalently into a toroidal ring (43). Prdx1 also contains Cys71 and Cys83, and Prdx2 also contains Cys70. Unique to Prdx1, Cys83 can form disulfide bonds between adjacent dimers of the decameric toroid (21), although not apparently at the same time as the well-established disulfide bond within each dimer (between the peroxidatic and resolving cysteines); hence, these decamers are still SDS labile. High-molecular-mass Prdx oligomers that have lost their peroxidase activity but gained chaperone activity, which enables them to protect other proteins from unfolding during conditions of oxidant stress and heat shock, have been reported (15, 16, 21, 28). For Prdx1 and Prdx2, formation of these “chaperone oligomers” can be induced not just by oxidation (21, 28), but also by phosphorylation at Thr90, at least for Prdx1 (15). However, all these species appear to be SDS labile, suggesting that the disulfide-dependent complexes that we have observed are related to, but distinct from, these chaperone oligomers. The only comparable SDS-resistant species that has been reported is a Prdx2 complex isolated from erythrocyte membranes (34). Cellular compartmental analysis of hearts indicates that a proportion of Prdx2 translocates from the cytosol- to the membrane-associated fraction in response to treatment with 10–100 μM hydrogen peroxide, mostly in the form of SDS-resistant oligomers that are enriched in hyperoxidized Prdx.
Overall, these results demonstrate that Prdxs readily form intermolecular (or intramolecular) bonds in response to treatment of rat hearts ex vivo with 10–100 μM exogenous hydrogen peroxide. These results are consistent with those observed for Prdx in other tissues and purified Prdx in vitro. However, in contrast to previously published in vitro data, hyperoxidized Prdx is still able to form a covalent dimer (containing a single disulfide bond) with nonhyperoxidized Prdx, which reflects a previously unsuspected flexibility of interaction within the dimer. In terms of functional consequences, increased Prdx disulfide dimer formation is probably associated with lessened peroxidase activity once the concentration of reduced thioredoxin becomes limiting, if the Prdx reaction cycle is considered (43, 44). Hyperoxidation does inhibit Prdx activity, at least temporarily, before enzymatic repair (2). No attempt was made to measure the peroxidatic activities of the various Prdx enzymes, because there is no peroxidatic assay that is specific to Prdx activity within crude extracts. Furthermore, the alkylation of hearts with maleimide during sample preparation was a requirement if the Prdx redox status were to be preserved, but this inadvertently inhibits Prdx peroxidatic activity. A summary and model of these redox-modulated changes in Prdxs are outlined in Fig. 6. Within this scheme, oxidation of the catalytic cysteines to disulfide bonds promotes the breakdown of decamers to dimers, as previously reported (32, 42). Prdx enzymes play cardioprotective and signaling roles within the heart by regulating peroxide concentrations under basal conditions and during oxidative stress and also by acting as chaperones under conditions of heat and oxidative stress.
GRANTS
This research was supported by grants from the Biotechnology and Biological Sciences Research Council and the Wellcome Trust.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Araki M, Nanri H, Ejima K, Murasato Y, Fujiwara T, Nakashima Y, Ikeda M. Antioxidant function of the mitochondrial protein SP-22 in the cardiovascular system. J Biol Chem 274: 2271–2278, 1999. [DOI] [PubMed] [Google Scholar]
- 2.Biteau B, Labarre J, Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 425: 980–984, 2003. [DOI] [PubMed] [Google Scholar]
- 3.Brennan JP, Wait R, Begum S, Bell JR, Dunn MJ, Eaton P. Detection and mapping of widespread intermolecular protein disulfide formation during cardiac oxidative stress using proteomics with diagonal electrophoresis. J Biol Chem 279: 41352–41360, 2004. [DOI] [PubMed] [Google Scholar]
- 4.Brixius K, Schwinger RH, Hoyer F, Napp A, Renner R, Bolck B, Kumin A, Fischer U, Mehlhorn U, Werner S, Bloch W. Isoform-specific downregulation of peroxiredoxin in human failing myocardium. Life Sci 81: 823–831, 2007. [DOI] [PubMed] [Google Scholar]
- 5.Budde H, Flohe L, Hecht HJ, Hofmann B, Stehr M, Wissing J, Lunsdorf H. Kinetics and redox-sensitive oligomerisation reveal negative subunit cooperativity in tryparedoxin peroxidase of Trypanosoma brucei brucei. Biol Chem 384: 619–633, 2003. [DOI] [PubMed] [Google Scholar]
- 6.Cao Z, Roszak AW, Gourlay LJ, Lindsay JG, Isaacs NW. Bovine mitochondrial peroxiredoxin III forms a two-ring catenane. Structure 13: 1661–1664, 2005. [DOI] [PubMed] [Google Scholar]
- 7.Chevallet M, Wagner E, Luche S, van Dorsselaer A, Leize-Wagner E, Rabilloud T. Regeneration of peroxiredoxins during recovery after oxidative stress: only some overoxidized peroxiredoxins can be reduced during recovery after oxidative stress. J Biol Chem 278: 37146–37153, 2003. [DOI] [PubMed] [Google Scholar]
- 8.Choi HJ, Kang SW, Yang CH, Rhee SG, Ryu SE. Crystallization and preliminary X-ray studies of hORF6, a novel human antioxidant enzyme. Acta Crystallogr D 54: 436–437, 1998. [DOI] [PubMed] [Google Scholar]
- 9.Cullingford TE, Wait R, Clerk A, Sugden PH. Effects of oxidative stress on the cardiac myocyte proteome: modifications to peroxiredoxins and small heat shock proteins. J Mol Cell Cardiol 40: 157–172, 2006. [DOI] [PubMed] [Google Scholar]
- 10.Eaton P, Bell RM, Cave AC, Shattock MJ. Ischemic preconditioning: a potential role for protein S-thiolation? Antioxid Redox Signal 7: 882–888, 2005. [DOI] [PubMed] [Google Scholar]
- 11.Evrard C, Capron A, Marchand C, Clippe A, Wattiez R, Soumillion P, Knoops B, Declercq JP. Crystal structure of a dimeric oxidized form of human peroxiredoxin 5. J Mol Biol 337: 1079–1090, 2004. [DOI] [PubMed] [Google Scholar]
- 12.Gourlay LJ, Bhella D, Kelly SM, Price NC, Lindsay JG. Structure-function analysis of recombinant substrate protein 22 kDa (SP-22). A mitochondrial 2-Cys peroxiredoxin organized as a decameric toroid. J Biol Chem 278: 32631–32637, 2003. [DOI] [PubMed] [Google Scholar]
- 13.Harris JR, Schroder E, Isupov MN, Scheffler D, Kristensen P, Littlechild JA, Vagin AA, Meissner U. Comparison of the decameric structure of peroxiredoxin-II by transmission electron microscopy and X-ray crystallography. Biochim Biophys Acta 1547: 221–234, 2001. [DOI] [PubMed] [Google Scholar]
- 14.Hofmann B, Hecht HJ, Flohe L. Peroxiredoxins. Biol Chem 383: 347–364, 2002. [DOI] [PubMed] [Google Scholar]
- 15.Jang HH, Kim SY, Park SK, Jeon HS, Lee YM, Jung JH, Lee SY, Chae HB, Jung YJ, Lee KO, Lim CO, Chung WS, Bahk JD, Yun DJ, Cho MJ, Lee SY. Phosphorylation and concomitant structural changes in human 2-Cys peroxiredoxin isotype I differentially regulate its peroxidase and molecular chaperone functions. FEBS Lett 580: 351–355, 2006. [DOI] [PubMed] [Google Scholar]
- 16.Jang HH, Lee KO, Chi YH, Jung BG, Park SK, Park JH, Lee JR, Lee SS, Moon JC, Yun JW, Choi YO, Kim WY, Kang JS, Cheong GW, Yun DJ, Rhee SG, Cho MJ, Lee SY. Two enzymes in one: two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 117: 625–635, 2004. [DOI] [PubMed] [Google Scholar]
- 17.Jin DY, Chae HZ, Rhee SG, Jeang KT. Regulatory role for a novel human thioredoxin peroxidase in NF-κB activation. J Biol Chem 272: 30952–30961, 1997. [DOI] [PubMed] [Google Scholar]
- 18.Kang SW, Rhee SG, Chang TS, Jeong W, Choi MH. 2-Cys peroxiredoxin function in intracellular signal transduction: therapeutic implications. Trends Mol Med 11: 571–578, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kristensen P, Rasmussen DE, Kristensen BI. Properties of thiol-specific anti-oxidant protein or calpromotin in solution. Biochem Biophys Res Commun 262: 127–131, 1999. [DOI] [PubMed] [Google Scholar]
- 20.Lee TH, Kim SU, Yu SL, Kim SH, Park DS, Moon HB, Dho SH, Kwon KS, Kwon HJ, Han YH, Jeong S, Kang SW, Shin HS, Lee KK, Rhee SG, Yu DY. Peroxiredoxin II is essential for sustaining life span of erythrocytes in mice. Blood 101: 5033–5038, 2003. [DOI] [PubMed] [Google Scholar]
- 21.Lee W, Choi KS, Riddell J, Ip C, Ghosh D, Park JH, Park YM. Human peroxiredoxin 1 and 2 are not duplicative proteins: the unique presence of Cys83 in PRX1 underscores the structural and functional differences between PRX1 and PRX2. J Biol Chem 282: 22011–22022, 2007. [DOI] [PubMed] [Google Scholar]
- 22.Low FM, Hampton MB, Peskin AV, Winterbourn CC. Peroxiredoxin 2 functions as a noncatalytic scavenger of low-level hydrogen peroxide in the erythrocyte. Blood 109: 2611–2617, 2007. [DOI] [PubMed] [Google Scholar]
- 23.Manevich Y, Feinstein SI, Fisher AB. Activation of the antioxidant enzyme 1-Cys peroxiredoxin requires glutathionylation mediated by heterodimerization with π-GST. Proc Natl Acad Sci USA 101: 3780–3785, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsushima S, Ide T, Yamato M, Matsusaka H, Hattori F, Ikeuchi M, Kubota T, Sunagawa K, Hasegawa Y, Kurihara T, Oikawa S, Kinugawa S, Tsutsui H. Overexpression of mitochondrial peroxiredoxin-3 prevents left ventricular remodeling and failure after myocardial infarction in mice. Circulation 113: 1779–1786, 2006. [DOI] [PubMed] [Google Scholar]
- 25.Mayr M, Chung YL, Mayr U, McGregor E, Troy H, Baier G, Leitges M, Dunn MJ, Griffiths JR, Xu Q. Loss of PKC-δ alters cardiac metabolism. Am J Physiol Heart Circ Physiol 287: H937–H945, 2004. [DOI] [PubMed] [Google Scholar]
- 26.Mitsumoto A, Takanezawa Y, Okawa K, Iwamatsu A, Nakagawa Y. Variants of peroxiredoxins expression in response to hydroperoxide stress. Free Radic Biol Med 30: 625–635, 2001. [DOI] [PubMed] [Google Scholar]
- 27.Monteiro G, Horta BB, Pimenta DC, Augusto O, Netto LE. Reduction of 1-Cys peroxiredoxins by ascorbate changes the thiol-specific antioxidant paradigm, revealing another function of vitamin C. Proc Natl Acad Sci USA 104: 4886–4891, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moon JC, Hah YS, Kim WY, Jung BG, Jang HH, Lee JR, Kim SY, Lee YM, Jeon MG, Kim CW, Cho MJ, Lee SY. Oxidative stress-dependent structural and functional switching of a human 2-Cys peroxiredoxin isotype II that enhances HeLa cell resistance to H2O2-induced cell death. J Biol Chem 280: 28775–28784, 2005. [DOI] [PubMed] [Google Scholar]
- 29.Nagy N, Malik G, Fisher AB, Das DK. Targeted disruption of peroxiredoxin 6 gene renders the heart vulnerable to ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol 291: H2636–H2640, 2006. [DOI] [PubMed] [Google Scholar]
- 30.Neumann CA, Krause DS, Carman CV, Das S, Dubey DP, Abraham JL, Bronson RT, Fujiwara Y, Orkin SH, Van Etten RA. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature 424: 561–565, 2003. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen-Nhu NT, Berck J, Clippe A, Duconseille E, Cherif H, Boone C, Van der Eecken V, Bernard A, Banmeyer I, Knoops B. Human peroxiredoxin 5 gene organization, initial characterization of its promoter and identification of alternative forms of mRNA. Biochim Biophys Acta 1769: 472–483, 2007. [DOI] [PubMed] [Google Scholar]
- 32.Parsonage D, Youngblood DS, Sarma GN, Wood ZA, Karplus PA, Poole LB. Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry 44: 10583–10592, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pawloski JR, Hess DT, Stamler JS. Export by red blood cells of nitric oxide bioactivity. Nature 409: 622–626, 2001. [DOI] [PubMed] [Google Scholar]
- 34.Plishker GA, Chevalier D, Seinsoth L, Moore RB. Calcium-activated potassium transport and high molecular weight forms of calpromotin. J Biol Chem 267: 21839–21843, 1992. [PubMed] [Google Scholar]
- 35.Rabilloud T, Heller M, Gasnier F, Luche S, Rey C, Aebersold R, Benahmed M, Louisot P, Lunardi J. Proteomics analysis of cellular response to oxidative stress. Evidence for in vivo overoxidation of peroxiredoxins at their active site. J Biol Chem 277: 19396–19401, 2002. [DOI] [PubMed] [Google Scholar]
- 36.Rhee SG, Kang SW, Chang TS, Jeong W, Kim K. Peroxiredoxin, a novel family of peroxidases. IUBMB Life 52: 35–41, 2001. [DOI] [PubMed] [Google Scholar]
- 37.Schroder E, Eaton P. Hydrogen peroxide as an endogenous mediator and exogenous tool in cardiovascular research: issues and considerations. Curr Opin Pharmacol 8: 153–159, 2008. [DOI] [PubMed] [Google Scholar]
- 38.Schroder E, Littlechild JA, Lebedev AA, Errington N, Vagin AA, Isupov MN. Crystal structure of decameric 2-Cys peroxiredoxin from human erythrocytes at 1.7 A resolution. Structure 8: 605–615, 2000. [DOI] [PubMed] [Google Scholar]
- 39.Schroder E, Willis AC, Ponting CP. Porcine natural-killer-enhancing factor-B: oligomerisation and identification as a calpain substrate in vitro. Biochim Biophys Acta 1383: 279–291, 1998. [DOI] [PubMed] [Google Scholar]
- 40.Shau H, Kim AT, Hedrick CC, Lusis AJ, Tompkins C, Finney R, Leung DW, Paglia DE. Endogenous natural killer enhancing factor-B increases cellular resistance to oxidative stresses. Free Radic Biol Med 22: 497–507, 1997. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, Phelan SA, Petros C, Taylor EF, Ledinski G, Jurgens G, Forsman-Semb K, Paigen B. Peroxiredoxin 6 deficiency and atherosclerosis susceptibility in mice: significance of genetic background for assessing atherosclerosis. Atherosclerosis 177: 61–70, 2004. [DOI] [PubMed] [Google Scholar]
- 42.Woo HA, Kang SW, Kim HK, Yang KS, Chae HZ, Rhee SG. Reversible oxidation of the active site cysteine of peroxiredoxins to cysteine sulfinic acid. Immunoblot detection with antibodies specific for the hyperoxidized cysteine-containing sequence. J Biol Chem 278: 47361–47364, 2003. [DOI] [PubMed] [Google Scholar]
- 43.Wood ZA, Schroder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci 28: 32–40, 2003. [DOI] [PubMed] [Google Scholar]
- 44.Yang KS, Kang SW, Woo HA, Hwang SC, Chae HZ, Kim K, Rhee SG. Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. J Biol Chem 277: 38029–38036, 2002. [DOI] [PubMed] [Google Scholar]