Abstract
Baroreceptor afferents project to the cardiovascular region of the nucleus tractus solitarius (cvNTS), and their cvNTS target neurons may play a role in governing the sensitivity and operating range of the arterial baroreceptor reflex (baroreflexes). Recent studies have shown differential gene and protein expression in the cvNTS in response to changed arterial pressure. However, the extent of these responses is unknown. Therefore, we collected differential global gene expression data in a time series following acute hypertension in awake, freely moving rats. To acquire statistically significant results and place them in functional context, we overcame several quality control requirements and developed novel analytical approaches. The physiologically new findings from the study are that acute hypertension causes very extensive, time-varying gene regulatory changes, many involving neuronal function-specific genes and systems of genes. We use standard genomic analysis methods to manage the large data sets and to develop results such as heat maps to examine patterns and clusters in the gene regulation. We used the Gene Ontology categories to provide functional context. To place our findings in the context of the relevant literature, we developed two graphical representations of the networks implicated, linking receptors and channels to signaling pathways. The results point to the multivariate complexity of the response and implicate a group of receptors as candidates for mediating nucleus tractus solitarius baroreflex function in hypertension by identifying concurrent upregulation of receptor genes. We were able to make transcription factor binding predictions and record dysregulation of heart rate correlated with the transcriptional response.
Keywords: gene expression, central cardiovascular regulation, ANG II AT1 receptor, phenylephrine, adaptive molecular process, baroreflex resetting
the nucleus tractus solitarius (NTS) receives afferent input from high-pressure arterial baroreceptors involved in the baroreceptor reflex (baroreflex), a powerful regulatory mechanism (60) in blood pressure control. Within this negative feedback controller, an increase in blood pressure results in increased afferent drive to the NTS and elicits compensatory changes to cardiac output and decreases in peripheral vascular resistance due to adjustments in vagal cardiac and sympathetic nerve outputs. The NTS is essential for baroreflex function; lesions of the NTS eliminate the baroreflex and lead to fulminating hypertension. Previous investigations of NTS function have found neurons receiving baroreceptor inputs concentrated at intermediate levels of the NTS coextensive with the area postrema, primarily in the commissural and dorsomedial subnuclei, defining a cardiovascular region of the NTS (cvNTS) (55). In recent years, studies using molecular approaches have shown induction of Fos in this same region subsequent to activation of baroreceptor inputs by acute blood pressure increases, including by intravenous injection of phenylephrine (PE) (9, 18, 19, 33, 34, 41, 58).
The increase in Fos protein levels is of interest as an indicator of neuronal activity. But it is also an indicator of the induction of gene regulation because it is a component in the activator protein-1 (AP-1) protein complex, an important transcription factor (TF) with a regulatory role in the expression of many neuron-specific genes. Thus, the previous Fos studies imply that elevated baroreceptor afferent drive to the NTS may lead to gene regulatory responses of NTS neurons in the cvNTS. This suggestion is bolstered by additional studies showing that increased baroreceptor inputs regulate the gene expression in NTS for tyrosine hydroxylase (TH) and neuropeptide Y (NPY) neuropeptide genes among others (10, 28). Interestingly, the dynamics were different for these two gene regulatory responses, with the TH gene showing a faster time course than the NPY gene response. These findings suggest the hypothesis that cvNTS neurons respond to increased baroreceptor drive by transcriptional regulation of genes affecting neuronal function and that these responses may have complex temporal coordination. The present study tests this hypothesis, examining the initial global differential transcriptomic response patterns to hypertension in the cvNTS.
We expected to need to resolve 20–100% changes in regulation of neural function genes that are physiologically significant (68, 72). There were major technical challenges to surmount to acquire these data. Our approach requires carefully refined application of transcriptional profiling technology, involving stringent quality control and quantitative analysis, thereby permitting resolution of the expected range of differential activity changes. In addition, differential gene expression study of the cvNTS required precise microdissection and subsequent amplification of the RNA from this specific, small cvNTS cell group.
Our analysis of the transcriptional results uses standard genomic analysis methods to manage the large data sets and results, including heat maps to examine patterns and clusters in the gene regulation, literature analyses to connect the findings to prior studies, and Gene Ontology (GO) categorization to provide functional context for the extensive transcriptional response. This effort culminated in two graphical representations of the networks implicated by the present results in NTS baroreflex modulation, linking receptors and channels to pathways. Our analysis approach explored the multivariate complexity of the response by identifying concurrent upregulation of receptor genes. We analyzed the differential transcriptional results in terms of their gene regulatory network, yielding predicted TF binding. Finally, we analyzed the heart rate and blood pressure records for correlated changes in cardiovascular dynamics with the transcriptional regulation.
METHODS
Animals
Male Sprague-Dawley rats (Charles River Laboratories, Wilmington, MA) weighing ∼250 g were used. They were housed in pairs under 12:12 light-dark cycles (lights on at 0600). Food and water were available ad libitum. All animal protocols were approved by the Thomas Jefferson University Institutional Animal Care and Use Committee.
Acute Hypertensive Model and Experimental Design
Animals were anesthetized with isoflurane vaporized in O2 (5% induction; 1% maintenance), and one femoral artery and vein were cannulated (PE-50 tubing) via a small medial incision for measurement of arterial pressure and infusion of drugs, respectively. The cannulas were run subcutaneously to an exit incision between the scapulas. The leg wound was sutured and topical anesthetic (lidocaine) was applied to both skin incisions.
Following the surgery and 1-h recovery, characterized by stable, normal resting blood pressure and heart rate, either 1 ml/h saline (control), or ∼200 μg/ml phenylephrine (∼1 ml/h), was infused. The latter was manually titered to maintain an increase of 25 mmHg diastolic arterial pressure.
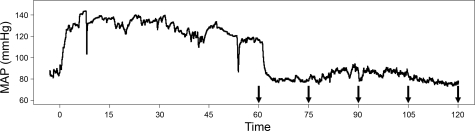
Our experimental design used a standard 60-min acute hypertension experimental treatment, taking the first samples after 60 min of PE or saline infusion, and we then took additional samples at 15-min intervals thereafter. We took samples to acquire time series data at 60, 75, 90, 105 and 120 min (see Fig. 1).
Fig. 1.
A representative mean arterial blood pressure (MAP) trace during and following a 1 h acute hypertensive stimulus. Arrows indicate the five time points when gene expression was measured. MAP, diastolic + 1/3* (systolic-diastolic).
Gas anesthesia has previously been reported to induce p-ERK1/2 phosphorylation in some central autonomic structures (59). To control for these effects, the present study reports only differential data that compares to controls that experience the same surgery and fluid (saline) infusion volume.
Cardiovascular NTS Microdissection
At the appropriate time points, rats were rapidly decapitated, the brains were quickly removed, and the medullas were sectioned transversely at 250 μm using a tissue chopper (McIlwain; Gamshall, England). The cvNTS region was microdissected from the tissue sections containing it using size-matched micropunches (<1 mm; Stoelting, Wood Dale, IL). The cvNTS and appropriate tissue sections were identified by neuroanatomical landmarks as the NTS region containing the great majority of c-fos induction following acute hypertension, encompassing the rostral pole of the dorsal commissural subnucleus and the dorsal, dorsolateral, and medial subnuclei at levels coextensive with the area postrema in the rat. This neuronal group contains neurons with extensive local circuit interactions and intermixing dendritic trees (47, 48, 54), and many cells in this region respond to baroreceptor afferent nerve stimulation, both as recipients of this input and with complex longer-latency responses indicative of higher-order processing (47, 55).
We used quantitative standards to minimize potential variability in the tissue obtained and ensure accuracy. Microdissections were guided (within the conservative boundaries defined by the prior c-fos positive baroreceptor afferent studies) using clearly demarcated neuroanatomical landmarks. All samples were collected, and the mRNA was protected within a stringent 30-min postmortem time window. Then data from each sample were analyzed individually to maximize the statistical power of the experimental design and to capture any biological variability. We did not pool mRNA across animals but used one array per animal.
Gene Expression Data Generation and Validation
The data requirements of the present study necessitated development of novel microarray methods providing increased sensitivity to detect changes in the physiological range of neuronal gene regulation: 20–80% (11, 28, 30). Further, the genes on the microarray are the full set of all genes for which functional annotation (LocusLink ID) was available at the time of array manufacture plus additional genes annotated as “similar to” genes in other organisms with neuronal specific functions. These, and the methods for NTS mRNA isolation, amplification, labeling, microarray hybridization, microarray manufacture, and quantitative real-time polymerase chain reaction (qRT-PCR) validation were as previously described (17) and also described in detail in the supplement. In brief, we used quantitative quality control measures to guide protocols that minimized technical noise. We employed a universal reference design (30) that contained a reference with large expression for all genes to eliminate dye bias, which we found to be the largest contributor to technical noise. We further minimized more subtle effects such as day effects, molecular biology reagent batch effects, hybridization condition effects, and scanning effects by processing each control animal together with its paired hypertensive animal throughout all steps. For example, we used arrays from a single batch and performed wet lab procedures as a batch on single time point samples. We also analyzed the microarrays using quality control criteria such as weak signal (median absolute signal less than 2,000), deviations in signal distribution from the general signal distribution of other array data, and abnormal MA plots compared with other microarrays, suggesting a systematic bias. We discarded and reprocessed the RNA samples from the microarrays that failed these quality control criteria. To maximize the statistical power of the experimental design and capture biological variability, we did not pool mRNA across animals but used one array per animal. This quality assurance methodology, although tedious, produced data with sufficient signal-to-noise ratio using a reasonable number of replicates (discussed below) to reliably discern differential expression changes as small as 20%.
In our previous studies (17, 30), we found that four replicates in each condition were necessary to accurately and reproducibly measure small, physiologically relevant changes as low as 1.2-fold (20%). Therefore, four control and four treated animals were taken for each time point for a total of 40 animals (5 time points, 2 conditions, 4 biological replicates). Samples corresponding to microarrays failing our quality control criteria (see Supplemental Methods, which can be found at http://www.dbi.tju.edu/ajp-hypertension and on the American Journal of Physiology—Regulatory, Integrative and Comparative Physiology Web site) were reprocessed along with their paired control, and new microarrays were generated. On average, 40% of the samples processed produced microarray data not meeting our stringent criteria and were reprocessed. In all, 64 arrays were generated for 40 samples (only the best 40 arrays that passed quality control criteria were used). Our target was a noise level (including technical and biological variance) of 15% standard deviation of log2 fold changes of genes across samples and arrays, as this level would permit resolution of 20% changes with eight samples at a 10% false discovery rate (FDR), which was adequate for our purposes (31, 61).
Statistical Analysis
Images were scanned at 10 μm/pixel with a ScanArray 5000XL (PerkinElmer, Wellesley, MA) scanner, and image analysis was performed on the microarray images using ScanArray Express v2.2 software. Spot intensities were log transformed and normalized to the on-chip universal reference (30) using median-scale correction. The microarray data are available at the supplemental site, as well as in the Gene Expression Omnibus with accession no. GSE8585.
Testing for significant gene expression was performed using an ANOVA design (49): normalized signal intensity = treatment + timepoint + treatment timepoint + animal pair random effect + residual error.
The analysis methods, implemented in R, can be replicated using the programs in the supplement. P values were calculated for the treatment effect at each time point using the appropriate contrast tests. Significant genes determined at a 10% FDR threshold (61) were used for subsequent analyses.
Gene ontology analysis.
Enrichment tests of functional annotation were calculated using a Fisher-exact test and were corrected for multiple testing where noted. The complex nature of the correlations in the annotation data makes meaningful multiple-test correction very difficult and thus a naïve Bonferroni or FDR correction was too conservative in some cases. The justification and specifics regarding our statistical approach are provided in the supplement.
Transcription factor analysis.
The differential expression of genes potentially regulated by AP-1 and cAMP-responsive element-binding protein (CREB), individually and together, was tested using the SAM-GS algorithm (21). The 5′ flanking promoter sequences (2,000 base pairs) corresponding to the genes on the microarray were analyzed using the MATCH tool in the TRANSFAC database (39) to predict AP-1 and CREB binding sites (MATCH options used: vertebrate database, minimize false positives). These results were filtered further to consider only the hits with 100% similarity to the core binding site descriptions in the database. A gene was considered a potential target of AP-1 or CREB if any corresponding binding site was found in its promoter sequence. Out of 8,832 genes represented in the microarray, a total of 2,498 genes were predicted to be AP-1 targets, 2,220 genes as CREB targets, and 806 genes as potentially regulated by both transcription factors (TFs).
Given the expression data of TF target genes, the SAM-GS method estimates a T-like statistic for each gene. A SAM-GS score was computed as a sum of squared T-like statistics for all genes potentially regulated by AP-1, CREB, or both TFs. The statistical significance of these scores was estimated by generating null hypothesis distributions of SAM-GS scores based on permuting the control and hypertension samples. The P value was computed as the fraction of randomized data scores higher than the original score. In this study, the T-like statistic employed in the original SAM-GS formulation was modified to consider paired samples. We considered gene expression data at all of the time points together, as well as individually to test whether these TFs are playing a role at any time point and at individual time points, respectively.
Cardiovascular function analysis.
Pulse interval between successive diastolic blood pressure times was calculated from blood pressure traces. Heart rate at each beat was calculated as the reciprocal of the pulse interval (beats/s). Heart rate variability was taken as the standard deviation of heart rate in a window of 20 beats. The median variability was calculated for beats 150 s before the onset of infusion and 250 s at the end of the experiment before death of the animal. Differences between median heart rate variability from the end of the experiment and the baseline variability before infusion were calculated for all animals. Heart rate variability differences in hypertensive animals were compared with that of control animals using a two-sided two-sample Student's t-test.
Pathway Conceptual/Graphical Representation
To visualize and to help interpret the systems of differentially expressed genes associated with the various receptors whose genes were differentially expressed, the signaling pathways associated with these receptors were placed in an in silico prototypical neuron model using CellDesigner v3.1 (32). A set of 25 baroreflex implicated receptor-associated pathways were obtained from the Panther Pathways Database (40), http://www.patherdb.org, and two pathways (AT1R receptor and GABA channel families) were constructed by assembling information from the literature. The in silico model is available as an XML file in the systems biology markup language (SBML) format (see supplement). Gene identifiers were mapped to pathway components through the Panther Pathways Database and by manual curation. The in silico graphical representation was colored according to the gene expression data at each time point (as described in the legend for Fig. 2). Details of the mapping between genes and pathway components for color annotation are available in the Supplemental Methods.
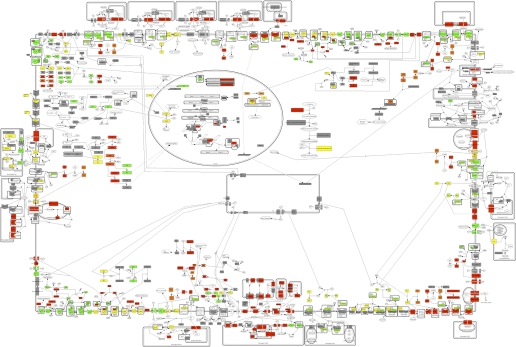
Fig. 2.
Pathway analysis of signaling networks relevant to nucleus tractus solitarius (NTS) control of baroreflex in response to acute hypertension. An in silico prototypical neuron model was constructed from 27 curated signaling pathways that have been implicated in the baroreflex response of NTS neurons. Figure depicts the physiological transcriptional response 60 min after the onset of hypertension. Proteins corresponding to regulated genes are colored red (upregulated) and green (downregulated). Proteins corresponding to both upregulated and downregulated genes are colored with a weighted mixture of red and green. Pathway components whose genes were not significantly regulated are colored gray. Unmeasured pathway components are not colored. The enlarged versions of the in silico model, colored based on transcriptional response at each of the five time points, are available (see Supplemental Figs. S1 through S5).
RESULTS
NTS Transcriptional Regulatory Response to Acute Hypertension: Character and Extent of the Response
Differential transcriptional response.
High-throughput system-wide data sets of the kind in the present study offer the opportunity for unbiased discovery of genes functionally associated with the acute hypertensive stimulus. All the microarray data are available in the supplement to this paper at http://www.dbi.tju.edu/ajp-hypertension, as well as in the Gene Expression Omnibus with accession no. GSE8585.
On the basis of earlier c-fos results in the literature showing the earliest responses at 40 min, we hypothesized that our initial 60-min time point would capture an early and modestly extensive differential gene expression response. However, the results at the 60-min time point show extensive, rather than modest, differential regulation of genes. This differential response includes many neuronal function-related genes encoding 236 receptors, 108 ion channels, 266 signal transduction proteins, and 185 proteins involved in other neurological processes, of which 98 are involved in synaptic transmission. There were also 337 differentially expressed genes that encode proteins located at the plasma membrane, including 49 at the synapse and 28 in dendrites. Tables 1 and 2 are a partial listing of brain-related receptors and signal transduction proteins, showing the broad effects of the hypertensive stimulus in these classes of genes. The complete list of differentially expressed genes and the five-time point time series for regulated receptors and channels are presented in Supplemental Tables S1 and S2 (found at http://www.dbi.tju.edu/ajp-hypertension and on the American Journal of Physiology—Regulatory, Integrative and Comparative Physiology Web site).
Table 1.
Time series expression profile for receptors implicated in baroreflex modulation in NTS
| 60 | 75 | 90 | 105 | 120 | Gene Name |
|---|---|---|---|---|---|
| 0.30 | 0.22 | Purinergic receptor P2Y, G-protein coupled 2 | |||
| 0.65 | 0.54 | Natriuretic peptide receptor 1 | |||
| 0.24 | 0.43 | 0.45 | 0.50 | Glutamate receptor, ionotropic, AMPA3 α3 | |
| 0.61 | 0.23 | 0.37 | Glutamate receptor, metabotropic 4 | ||
| 0.18 | −0.37 | Glutamate receptor, ionotropic, NMDA2C | |||
| 0.27 | −0.29 | Glutamate receptor, ionotropic, kainate 1 | |||
| 0.52 | −0.66 | Glutamate receptor, ionotropic, NMDA 1 | |||
| 0.43 | −0.26 | Glutamate receptor, ionotropic, NMDA 2A | |||
| 0.36 | −0.23 | Glutamate receptor, ionotropic, AMPA1 α1 | |||
| 0.30 | −0.21 | Glutamate receptor, ionotropic, kainate 2 | |||
| 0.36 | −0.67 | Adrenergic receptor, beta 2 | |||
| 0.29 | −0.32 | Adrenergic receptor, alpha 2c | |||
| 0.36 | −0.45 | ANG II receptor, type 1 (AT1A) | |||
| 0.30 | −0.29 | −0.29 | Dopamine receptor 1A | ||
|
0.52 |
−0.31 | Tachykinin receptor 1 | |||
| −0.19 | −0.23 | 0.28 | Purinergic receptor P2X 1 | ||
| −0.41 | 0.25 | Purinergic receptor P2X 2 | |||
| −0.17 | 0.14 | Glutamate receptor, ionotropic, kainate 5 | |||
| −0.34 | 0.41 | 5-hydroxytryptamine (serotonin) receptor 1D | |||
| −0.31 | −0.41 | 0.33 | Gamma-aminobutyric acid A receptor, α5 | ||
| −0.24 | 0.27 | Cholinergic receptor, nicotinic, β polypeptide 1 | |||
| −0.23 | −0.17 | 0.17 | 0.18 | Cholinergic receptor, nicotinic, δ polypeptide | |
| −0.33 | −0.49 | −0.50 | Cholinergic receptor, muscarinic 3 | ||
| −0.28 | −0.29 | Arginine vasopressin receptor 1A | |||
| −0.35 | −0.18 | −0.22 | Natriuretic peptide receptor 3 | ||
| −0.21 | −0.21 | Adenosine A2B receptor | |||
| −0.24 | −0.38 | −0.32 | Hypocretin (orexin) receptor 1 | ||
| −0.17 | −0.24 | 5-hydroxytryptamine (serotonin) receptor 1A | |||
| −0.44 | −0.42 | Glutamate receptor, metabotropic 2 | |||
| −0.18 | −0.28 | Tachykinin receptor 3 | |||
| −0.15 | −0.22 | TNF receptor superfamily, member 4 |
Bold values denote downregulation; italic values denote upregulation. 60, 75, 90, 105, and 120 in the header row correspond to minutes after hypertensive stimulus began (see Fig. 1). AMPA, dl-α-amino-3-hydroxy-5-methylisoxazole-propionic acid; NMDA, N-methyl-d-aspartate. The numbers are log fold change in expression.
Table 2.
Time series expression profile for upstream components of PKC, MAPK, PI3K, PKA, and SNARE pathways
| 60 | 75 | 90 | 105 | 120 | Gene Name | |
|---|---|---|---|---|---|---|
| −0.19 | −0.24 | AC | Adenylate cyclase 3 | |||
| −0.20 | −0.17 | AC | Adenylate cyclase 4 | |||
| −0.27 | −0.32 | AC | Adenylyl cyclase 8 | |||
| 0.18 | AC | Adenylate cyclase 5 | ||||
| 0.36 | −0.45 | AC | Adenylate cyclase 9 (predicted) | |||
| 0.41 | −0.40 | −0.26 | −0.38 | PKA | Protein kinase, cAMP dependent regulatory, type I, alpha | |
| 0.28 | −0.27 | −0.27 | PKA | Protein kinase, cAMP-dependent, regulatory, type 2, alpha | ||
| −0.53 | PKA | Protein kinase, cAMP dependent regulatory, type I, beta | ||||
| −0.52 | PKA | Protein kinase, cAMP dependent regulatory, type II beta | ||||
| −0.33 | PKA | Sperm autoantigenic protein 17 | ||||
| −0.28 | −0.30 | PI3K | Phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 | |||
| 0.43 | PI3K | Phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 2 | ||||
| 0.16 | PI3K | Phosphoinositide-3-kinase, class 2, beta polypeptide (predicted) | ||||
| 0.29 | PI3K | Phosphoinositide-3-kinase, class 3 | ||||
| 0.93 | PI3K | Phosphatidylinositol 3-kinase catalytic delta polypeptide (predicted) | ||||
| 0.27 | −0.15 | PI3K | Phosphatidylinositol 4-kinase, catalytic, beta polypeptide | |||
| 0.44 | PI3K | Ataxia telangiectasia mutated homolog (human) (mapped) | ||||
| 0.27 | PI3K | Similar to Expressed sequence BB220380 | ||||
| −0.24 | 0.24 | 0.25 | Gqa | Guanine nucleotide binding protein, alpha 11 | ||
| 0.41 | Gqa | Guanine nucleotide binding protein, alpha q polypeptide | ||||
| −0.15 | PLC | Phospholipase C, beta 3 | ||||
| −0.20 | PLC | Phospholipase C, beta 4 | ||||
| −0.31 | −0.55 | PLC | Phospholipase C, epsilon 1 | |||
| −0.45 | PLC | Phospholipase C, delta 1 | ||||
| 0.44 | 0.15 | PLC | Phospholipase C, delta 4 | |||
| 0.18 | PLC | Phospholipase C-like 2 (predicted) | ||||
| −0.18 | −0.29 | PKC | Protein kinase C, beta 1 | |||
| 0.26 | −0.29 | PKC | Protein kinase C, delta | |||
| −0.32 | PKC | Protein kinase C, epsilon | ||||
| −0.48 | PKC | Protein kinase C, zeta | ||||
| −0.32 | −0.27 | PKC | Protein kinase C, eta | |||
| 1.03 | 0.66 | RAS | RAB1, member RAS oncogene family | |||
| 0.38 | 0.15 | 0.37 | RAS | RAB8B, member RAS oncogene family | ||
| 0.34 | RAS | RAB10, member RAS oncogene family | ||||
| 0.27 | RAS | RAB13, member RAS oncogene family | ||||
| 0.59 | RAS | RAB28, member RAS oncogene family | ||||
| 0.42 | RAS | Kirsten rat sarcoma viral oncogene homolog 2 (active) | ||||
| 0.42 | RAS | Related RAS viral (r-ras) oncogene homolog 2 | ||||
| 0.24 | 0.34 | −0.14 | −0.23 | RAS | RAB25, member RAS oncogene family (predicted) | |
| 0.22 | RAS | RAB11a, member RAS oncogene family | ||||
| 0.95 | RAS | RAB11B, member RAS oncogene family | ||||
| −0.33 | −0.38 | SNARE | Vesicle-associated membrane protein 1 | |||
| −0.51 | SNARE | Vesicle-associated membrane protein 2 | ||||
| −0.44 | SNARE | Vesicle-associated membrane protein 8 | ||||
| −0.21 | SNARE | Vesicle transport through interaction with t-SNAREs | ||||
| −0.28 | SNAP29 | Synaptosomal-associated protein 29 | ||||
| −0.20 | −0.40 | STX | Epimorphin | |||
| 0.29 | 0.24 | STX | Syntaxin 3 | |||
| 0.26 | 0.30 | STX | Syntaxin 4A (placental) | |||
| 0.60 | STX | Syntaxin 12 |
Bold values denote downregulation; italic values denote upregulation. The upstream components of PKC, MAPK, phosphatidylinositol 3-kinase (PI3K), PKA, and soluble N-ethylmalemide sensitive fusion protein attachment receptor (SNARE) pathways are coordinately regulated in response to a 1-h acute hypertensive pulse. 60, 75, 90, 105, and 120 in the header row correspond to minutes after hypertensive stimulus began (see Fig. 1). RAS, renin angiotensin system. The numbers are log-fold change in expression.
Dynamics of the response.
The common approach in the field has been to use a hypertensive stimulus (in the present case 60 min) and then to examine the molecular responses at a series of time points following the end of the stimulus (9, 12–14, 23, 33, 51, 70). Consistent with this practice, we sampled the NTS in a time series initiated at the end of the hypertensive stimulus. Across the full series of five time points, distinct patterns of differential gene expression were observed involving 579 receptors, 332 channels, and 803 signal transduction proteins. Tables 1 and 2 show the dynamics in the gene responses across a sampling of genes. These short-term dynamics within the 60-min time series, distinct across different genes, are consistent with those previously reported for the dynamics of tyrosine hydroxylase and neuropeptide Y mRNA expression following hypotension from hemorrhage (12). In the present study, both the hypertensive stimulus and the subsequent return to baseline blood pressure might have contributed to the gene expression dynamics, and future studies will compare the present data to the dynamics during sustained hypertensive stimuli beyond 1 h.
Functional Analysis Using Gene Ontology Enrichment
We have functionally annotated the upregulated and downregulated genes at each time point using Gene Ontology (GO) terms, which provide a controlled vocabulary to describe gene/gene product attributes and tools to evaluate their significance in gene expression results (www.geneontology.org). GO results of particular interest to neuronal function in baroreflexes are presented below, while the full list of statistically significant GO terms (Bonferroni adjusted P < 0.05) and GO term enrichment tests are provided in the supplement (see Supplemental Tables S3–S5, which may be found at http://www.dbi.tju.edu/ajp-hypertension and on the American Journal of Physiology—Regulatory, Comparative and Integrative Physiology Web site).
Transcription factors.
Transcription factors are classified in GO under the term “transcription factor activity.” The large number of genes regulated after the 1-h pulse suggests that many transcription factors were activated by the hypertension. Consistent with this hypothesis, at the 60-min time point, 83 of the 488 transcription factors measured are upregulated and are overrepresented in the set of upregulated genes (P < 0.05). However, 30 min later, at the 90-min time point, 135 of the 488 transcription factors are downregulated and are overrepresented in the set of downregulated genes (P < 0.01). At later points in the time series, transcription factors are underrepresented in the upregulated genes (P < 0.001). The upregulated, then downregulated pattern of these data tracks the upregulation, then downregulation pattern of the blood pressure stimulus used and suggests the hypothesis that transcription factors are responsive correlatively to the stimulus pattern.
Metabolism.
At the 90-min time point, most of the significant GO terms focus on metabolism, specifically oxidative phosphorylation. There are 13 biological process GO terms related to aerobic respiration through acetyl-CoA enzyme metabolic and catabolic processes using the tricarboxylic acid cycle (see Supplemental Table S3). Oxidoreductase activity, mitochondria function-related, and ribosomal structural components are enriched GO molecular function terms. These results suggest an initial increase in NTS metabolic activity in response to the stimulus. This is consistent with the findings in NTS of Chan et al. (9) for ultrastructural changes in response to PE-induced hypertension and are consistent with adaptation to facilitate increased gene expression.
RNA processing.
At the 105-min time point, most of the significant GO terms are related to translation and mRNA processing. Enriched GO biological processes include translation, ribosome biogenesis, and assembly (see Supplemental Table S3). The enriched molecular activities of upregulated genes are structural constituents of ribosomes, RNA binding, and single-stranded DNA binding (see Supplemental Table S4), and all significant GO cellular components are related to the ribosome (see Supplemental Table S5). These results are consistent with the hypothesis that the observed transcriptional response initiates downstream translational activities.
Gene Ontology Terms for Neuronal Function-Specific Responses
GO analysis of the initial response to the hypertensive stimulus showed overrepresentation of genes related to both ionotropic and metabotropic glutamate receptor activity (see Supplemental Table S4). Also, synapse and dendrite GO terms were enriched for upregulated genes. Downregulated genes were overrepresented in Ras protein signal transduction and small GTPase regulator activity, suggesting a downregulation of MAPK pathways (see Supplemental Table S4 and S5).
Transcriptional regulation of ion channels.
We analyzed transcriptional regulation of channel types with a primary focus on calcium (L, N, and P/Q types) and potassium [G protein-coupled K inwardly rectifying (GIRK), K inwardly rectifying (KIR), Kv, KCa, and leak current types] channels because they are known to mediate immediate (e.g., electric potential changes), intermediate (e.g., calcium signaling), and long term (e.g., gene expression) changes in neurons (25). The alpha subunits of L and P/Q type channels are upregulated after 1 h of hypertension at the 60 and 75 min time points. This suggests or predicts new, additional involvement of channels by which NTS may produce functional adaptation to the stimulus (see Supplemental Table S2).
PKC is downregulated at the 60-min time point (Table 2) and PKC can inhibit or activate L, N, and P/Q-type calcium channels (4, 22, 29), whereas PKA (which shows an upregulation, then downregulation pattern) activates l-type calcium channels (29). l-type channels are involved in CREB activation and gene expression changes (3). N and P/Q type calcium channels are involved in neurotransmitter release (53). How the cell responds to these various calcium-signaling events in unique ways is still an open question, but studies suggest that somatic or dendritic localization of calcium currents and/or proteins associated with calcium channels can differentiate the different sources of calcium signaling and respond appropriately (2). Additional channel results are in Supplemental Table S2.
Transcriptional regulation of receptors.
A diverse set of receptors are differentially regulated by the 1-h hypertensive pulse and the subsequent 1-h time series (see Supplemental Table S1). As noted above, ionotropic glutamate receptors are upregulated at the 60-min time point and downregulated subsequently. Receptors with transmembrane activity (as annotated by GO) show similar patterns [enrichment of upregulated genes at the 60-min time point (P < 0.1) and downregulation subsequently (P < 0.001)] with over half being G protein-coupled receptors. Fibroblast growth factor receptors, glial cell line-derived neurotropic factor receptors, and glucagon receptors are downregulated over all time points studied. Interleukin receptors are generally downregulated at the 90-min time point but have mixed regulation at earlier time points. The time series gene expression dynamics of other receptors were also mixed and difficult to interpret using qualitative logic.
Coordinate Relationships in Gene Regulation
Table 1 contains selected time series of differential expression for receptors that show patterned expression of either upregulated at all time points, upregulated initially and then downregulated, downregulated initially and then upregulated, or downregulated at all time points. Several of these receptors were ionotropic glutamate alpha 2c adrenergic, tachykinin 1, and P2X receptors, which have been previously implicated by pharmacological studies in the bradycardic component of the baroreflex (44, 62, 71) and ANG II receptor type 1A and GABAA, which have been previously implicated in the tachycardic component of the baroreflex (1, 15, 16, 50, 63, 66). However, many receptors that have not been previously linked to the baroreflex were also found to be coordinately regulated. In particular, purinergic P2Y, dopamine 1A, serotonin 1, and nicotinic cholinergic receptors may be plausible candidates for a role in blood pressure regulation.
Generally speaking, the NTS response shows either consistent regulation of genes in one direction, or upregulation at 60- and 75-min time points and downregulation at the other time points (or vice versa). The latter could be a 30-min delayed response to the blood pressure's return to baseline.
Correlated Transcription Factor Predictions and Cardiovascular Measures
Our observations imply extensive and dynamic regulation of gene expression in the NTS following hypertension and increased baroreceptor afferent drive into the NTS. These observations have led us to hypothesize that these changes reflect altered transcription factor (TF) binding to the coregulated gene promoters. It has been shown that PE-induced acute hypertension leads to immediate early gene c-fos induction and Fos protein accumulation in the NTS (13, 34, 70), and phosphorylation of CREB has been suggested to mediate this response (8, 14). Fos is a component in the AP-1 protein complex, a key TF with a regulatory role in the expression of many neuronal function-specific genes. We tested whether AP-1 and CREB are playing a role in the system-wide gene regulation by analyzing the differential expression of genes potentially regulated by AP-1 and CREB, individually and together, using the SAM-GS algorithm (21). We considered gene expression data at all of the time points together, as well as individually, to test whether these TFs are playing a role at any time point and at individual time points, respectively. Our results indicate that AP-1 and CREB target genes are significantly differentially expressed at all of the time points, together and individually, following PE-induced acute hypertension. The statistical significance P values from SAM-GS are shown in Table 3.
Table 3.
Statistical significance of AP-1 and CREB transcription factor binding site enrichment for regulated genes at all time points and each time point individually using the SAM-GS algorithm
| TFs | All Time Points | 60 min | 75 min | 90 min | 105 min | 120 min |
|---|---|---|---|---|---|---|
| AP-1 | 0.019 | 0.082 | 0.041 | 0.003 | 0.003 | 0.036 |
| CREB | 0.028 | 0.086 | 0.036 | 0.003 | 0.011 | 0.031 |
| AP-1 and CREB | 0.028 | 0.068 | 0.038 | 0.003 | 0.004 | 0.051 |
AP-1, activator protein-1; CREB, cAMP-responsive element-binding protein.
Finally, we analyzed the heart rate and blood pressure records and found that, correlated with the transcriptional regulation, there was a statistically significant increase in heart rate variability after the 1 h hypertensive episode compared with baseline values and controls (15% increase; P < 0.05, data not shown).
DISCUSSION
We have built on the extensive literature that has studied molecular changes in the NTS consequent to PE-induced acute hypertension (9, 11–14, 18, 19, 23, 28, 33, 34, 41, 43, 58, 70) that suggest the hypothesis that cvNTS neurons respond to increased baroreceptor drive by transcriptional regulation of genes affecting neuronal function and that these responses may have complex temporal coordination. We have tested this hypothesis, examining the differential transcriptomic response patterns in the cvNTS and evaluating and characterizing the molecular response to baroreceptor afferent modulation. Our analyses identify a broad response of neuronal function-related genes, including receptors, and these results point to the multivariate complexity of the response.
The gene expression changes that we report implicate a wide range of receptors in the response to a hypertensive stimulus. Although some of these have been implicated by previous findings, many are novel (such as purinergic P2Y, dopamine 1A, serotonin 1, and nicotinic cholinergic receptors). Thus, the presented findings are consistent with previous data on NMDA and angiotensin AT1 receptors. Activation of NMDA receptors is known to have bradycardic effects (Table 4), and our results suggest that NMDA receptors may be upregulated when the system is challenged by a hypertensive stimulus. This is also consistent with previous studies that pharmacologically associated ionotropic glutamate receptors in baroreflex function, including NMDA, APMA, and kainite (44, 62, 71). Our gene expression data add complementary physiological evidence to these previous results, as well as those showing that angiotensin receptor 1A can modulate baroreflex when activated in the NTS (1, 50, 63, 66). Furthermore, our pathway results have shown angiotensin-related pathway enrichment with upregulation of both MAPK and phosphatidylinositol 3-kinase (PI3K) pathways are consistent with the important role of AT1R in NTS baroreflex function.
Table 4.
Tabular representation of connections between receptors and pathways for receptors implicated in baroreflex function by pharmacological studies
| Receptor Name | Bradycardic | PKC | MAPK | PI3K | PKA |
|---|---|---|---|---|---|
| CB1 | 1–3 | X 4 | X 5 | X 5 | 5 |
| 5HT2 | 6, 7 | 8, 9 | |||
| AMPA | 10–12 | 13–15 | 16 | 16 | |
| Estrogen | 17, 18 | 19 | 19, 20 | 19, 20 | 19 |
| NMDA | 12, 24, 25 | 26 | 26 | ||
| NK1 | 10, 27, 28 | 29 | X 30 | ||
| mGluR group 1 | 31 | 32 | 33 | X 32 | |
| P2X Purinoreceptor | 34, 35 | ||||
| α2-adrenergic | 36–38 | 39, 40 | 40 | 41 | |
| P1 purinoreceptor | 34 | ||||
| Atrial natriuretic peptide | 46 | ||||
| Delta opioid | 48 | 49 | 49 | 49 | X 50 |
| Insulin | 51 | 52 | 53 | 51, 53 | 53 |
| NPY Y1 | 55–58 | 59 | 59 | ||
| Endothelin-A | 62, 63 | 64 | 64 | 64 |
| Tachycardic | PKC | MAPK | PI3K | PKA | |
|---|---|---|---|---|---|
| 5HT3 | 6, 66–70 | ||||
| Nociceptin (ORL1) | 71, 72 | 73 | X 5 | X 5 | X 73 |
| GABAA | 62, 74, 75 | 76 | |||
| GABAB | 62, 78 | 79 | 80 | ||
| mu opioid | 85 | X 5 | X 5 | X 5 | X 5 |
| ANG II | 86 | 87 | 88 | 89, 90 | X 91 |
| Adrenomedullin | 92 | 93 | 94 | 95 | 92 |
| Vasopressin 1 | 96 | 97 | 97 | ||
| NPY Y2 | 100 |
Tachycardic and bradycardic refer to the physiological response associated with binding of the receptor in the nucleus tractus solitarius (NTS). Numbers cite which studies found each connection and correspond to the supplemental references. X denotes no interaction found. Figure 3 is a graphical representation of the connections shown in this table. NPY, neuropeptide Y.
Experimental design choices.
We used PE because previous studies have convincingly shown that the NTS response to PE-induced acute hypertension is mediated by the buffer nerves (9, 34, 51). PE does not cross the blood-brain barrier (69), and the central neuronal effects of PE-induced hypertension have been shown to depend on increased arterial baroreceptor afferent drive to the NTS by both pharmacological and sinoaortic denervation studies (9, 34, 51).
We examined the transcriptional response in a 1-h window following 1-h of hypertension. This design was chosen to be consistent with previous practice, in which the central neuronal effects of PE-induced hypertension utilized a defined period of hypertension and then measured effects on the NTS at time points following that standard period of blood pressure disturbance (13, 33, 34). These studies show that blood pressure elevations of 25–30 mmHg above baseline induce c-fos expression in the NTS after 30–40 min and that this induction is time dependent over a period of hours (34). On the basis of these findings, we hypothesized that transcriptional regulation in the NTS might be observed following 60 min of hypertension and also show time dependency. For this reason, we used a standard 60-min acute hypertension experimental treatment, taking the first samples after 60 min of PE or saline infusion, and we then took additional samples at 15-min intervals thereafter.
The use of 15-min intervals for the time series again emulates the prior literature (34), and these are the closest intervals that we were confident we could do reliably with our experimental methods. Our experimental design is for time series data out to 120 min (see Fig. 1). Although later time points are of interest, the present use of 5 time points already is a very large design, particularly in consideration of the need for four replicates (yielding eight samples, experimental plus control) at each time point and that the demands for high-quality data required several attempts to produce a useful sample/data point (see methods section on data generation). However, we recognize that, in future studies, the use of more sustained and even chronic hypertension will make an important addition that will be helpful to the analysis of the present time series data.
As explained in methods, we focused on the cardiovascular NTS as a meaningful biochemical/genomic system for these initial studies. This is consistent with previous literature performing molecular studies of microdissected NTS or its subnuclei, such as the cvNTS in the rat. The cvNTS contains neurons with extensive local circuit interactions and intermixing dendritic trees (47, 48, 54), and many cells in this region respond to baroreceptor afferent nerve stimulation, both as recipients of this input and with complex longer-latency responses indicative of higher-order processing. Thus the cvNTS region can be microdissected as a meaningful neuronal population for molecular biochemical/genomic study, and we elected to initially study it to acquire the full extent of the molecular response of the NTS to the acute hypertensive stimulus.
Interpretation of the broad molecular response.
While previous studies associate particular genes or proteins with baroreflexes and/or NTS, it is clear from the present results that increases in baroreceptor input activity engage and orchestrate a broad molecular response. This discovery of an unexpected, extensive differential gene regulatory response is consistent with previous transcript profiling studies of other functions that showed, for example, very broad molecular involvement in systems as diverse as yeast cell cycle (67) and the circadian rhythm in the neuronal suprachiasmatic nucleus (26, 30, 38, 46). In these systems, extensive additional experiments have gradually shown how these molecular processes operate in concert as a multivariate system to produce the observed response. For example, gene knockout/knockdown studies have been useful when done systematically and in combinations, together with a battery of complementary functional measures and computational analyses (27, 36, 56). In the present case, the complex molecular responses that we report in the NTS may point to functional processes that provide redundancy and robustness in the face of an array of environmental challenges. In this context, the functional significance of a particular gene or gene product is more likely to be in its interaction in a process together with other genes/gene products and with potentially redundant roles for particular genes/proteins, thereby enabling robust function in the system emerging from the network interactions. From the present data set, for example, baroreflexes might be hypothesized to be combinatorially affected by interactions of NK1, 5-HT3, and GABAA receptors as suggested by prior literature (15, 16).
Given the likelihood of redundancy and high degrees of compensatory interaction, reverberations are likewise expected. Our results may reflect this reality, as different genes demonstrate different response dynamics. These results are consistent with previous findings that also showed rapid and different response dynamics among NTS genes responsive to blood pressure (12). It may be useful to explore these complex dynamical relationships in computational models.
Relevance to long-term regulation in hypertension.
Recent reports indicate that the central nervous system plays a significant role in long-term regulation of blood pressure, including the development and maintenance of hypertension, by baroreflex resetting to a higher mean arterial pressure set point (5, 7, 35, 45, 65). The molecular mechanisms underlying these adaptive responses are largely unknown. Our discoveries on receptor and channel differential gene expression may play a significant role in long-term baroreflex resetting by providing the initial phase in a mechanism for neuronal plasticity. This is consistent with the findings of plasticity in the fine structure in the NTS following acute hypertension (9, 14, 23). We also observed that there are parallel changes in the regulation of heart rate and blood pressure, such that it appears that the reflex regulation provides less stable cardiovascular regulation in the same time frame as the observed molecular responses. The present findings of transcriptional regulation may be the initial adaptive process responsive to the loss of homeostatic stability, a kind of homeodynamics.
Our analysis approach identified several receptors as candidates for mediating NTS baroreflex modulation in hypertension, and although many of these are novel, a subset of these is consistent with differential regulation that can be inferred from the previous pharmacological literature. For example, activation of NMDA receptors is known to have bradycardic effects, and hence, one could hypothesize NMDA receptor upregulation when the system is challenged by a hypertensive stimulus. Our results also point to the multivariate complexity of the response by identifying concurrent upregulation of several other receptors that can mediate bradycardic effects (such as the natriuretic peptide and tachykinin receptors in Table 1). These findings relating to NTS adaptive behavior could be important for controlling reflex sensitivity, contributing to control of the operating point and gain of the baroreflexes in health and disease (47). Our physiological transcriptomic data and results provide a framework for further investigation into this central resetting of baroreflex set point in hypertension. Previous studies implicate glutamate (AMPA and NMDA) and GABAA receptors. AMPA receptors are recruited to the dendritic spines during long-term potentiation (LTP) and endocytosed during long-term depression via PKA phosphorylation and calcineurin-protein phosphatase 2B dephosphorylation mediated by NMDA receptor activation and deactivation (20, 57). Additionally, AMPA receptor trafficking is mediated by PI3K pathways activated by calcium influx from NMDA (37) or from l-type calcium channels (6). Finally, AMPA receptor subtypes GluR1 and GluR2L may be differentially trafficked by MAPK and PI3K pathways, respectively (52). It has been suggested that this LTP through AMPA receptor trafficking may be part of a mechanism of neuronal plasticity that may subserve long-term baroreflex resetting (64, 73).
Validation and prediction testing.
While microarrays are inherently noisy, our use of replicates and extensive QC, as described in the methods, allowed us to achieve the desired FDR and statistical significance. We validated these results for 44 neuronal function specific receptor and channel protein genes using qPCR, and this is further proof that we have indeed achieved our desired low FDR. It is also important that our analysis of the coexpression of genes to predict gene network and the specific binding of TFs led to specific predictions. To test these, we have developed an improved method based on carrier chromatin immunoprecipitation (CChIP) that is suitable for analysis of small tissue samples such as the cardiovascular NTS (24). The positive result for CREB and AP-1 binding using our new Fast CChIP approach is indicative that the present observed differential mRNA changes reflect active transcriptional regulatory gene expression changes leading to long-term functional effects (24).
The previous literature shows that induced expression of immediate early genes peaks 40 min after the onset of the hypertensive stimulus (13, 34) and thus with our time series from 60 to 120 min, we believe that we have captured the “first wave” of transcriptomic changes to the pulse hypertensive stimulus. In the present study, we consistently observed a widespread and coordinated molecular response, which we hypothesized to be the initial adaptive response to hypertension. The relation of this early response to long-term adaptive states will require further study. However, the present data certainly encourage such study, indicating that there is an extensive molecular remodeling of the NTS during hypertension and that this may contribute to altered baroreflex function in hypertension.
Systematic analysis of the findings in the context of the prior literature.
As seen above, some of the receptor types differentially regulated in the present study have been previously associated with NTS baroreceptor reflex modulation by the available pharmacological and anatomical literature. To place our findings in the context of this literature, we first made a list of receptors, both differentially regulated in the present study and also associated by the previous literature in baroreflex function in the NTS (Table 4, columns 1 and 2). We annotated this list based on the effect on the baroreflex, either bradycardic or tachycardic. We then searched the literature for links between the baroreflex-implicated receptor types and possible modulation of major components of downstream signaling pathways (Table 4, columns 3–6). Finally, we built a graphical representation that matched the intersection of the baroreflex-implicated receptor-associated pathways and our gene expression data.
Development of receptor-centric graphical representations.
The results were collated into two types of graphical representations to promote understanding of the processes and interrelationships in the regulated genes. To develop a graphical representation of each receptor and its downstream elements, we analyzed the literature and created a model pathway structure for each receptor in Table 4, illustrating their downstream intracellular proteins. We began from the literature that describes the pharmacological effects of various receptor agonists and antagonists that linked these receptors to various Na+, K+, and Ca2+ channel types. We also searched the literature that used protein activity blockers to link signaling pathways to receptor activity. Conceptually, our approach is similar to the Reactome (http://www.reactome.org) and Biocyc (http://www.biocyc.org) approaches that largely focus on metabolic pathways and basal transcription, except that our focus is on signaling. Our objective was aided by the availability of many of the relevant signaling pathways as a public resource in the Panther Pathways Database (40), http://www.pantherdb.org. This resource uses the open source software CellDesigner (32) to represent single receptor pathways in the SBML format (http://www.sbml.org).
We then further used CellDesigner to edit and consolidate the pathways into a single graphical representation. Since some receptor-associated pathways were not available in Panther, we also used CellDesigner to curate additional relevant NTS function-related receptor pathways not available in Panther, such as ANG II receptor type 1 (AT1R) and GABA receptor pathways (42, 66).
The results of this effort at the 60-min time point are shown in Fig. 2, containing signaling pathways with known downstream pathway components and connections to selected receptors with differential response to the hypertensive stimulus (enlarged versions and other time points available in the supplement). Notably, the receptors and associated signaling pathways for ionotropic and metabolomic glutamate, serotonin, dopamine, angiotensin, GABA, insulin, endothelin, α-adrenergic, muscarinic and nicotinic ACh, corticotropin, enkephalin, and general GPCR-mediated pathways are represented in the current version of the model. Similar graphical models for these receptors at the later four time points are available in the supplement. These results, together with further studies of these time points, while maintaining elevated blood pressure, can be used to suggest molecular network mechanisms employed by the NTS in the physiological function of blood pressure regulation for further experimental tests.
The CellDesigner-based model shown in Fig. 2 provides a mechanistic structure to enable such computational analyses. For example, we expect that modeling the role of the ANG II receptor AT1 in the observed response to be highly informative. Classically, it has been useful to study signaling pathway behavior starting from receptors, and the AT1R has been shown to mediate its effects through PKC, RAS, and PI3K pathways, all of which we have identified as relevant to the hypertensive response. Further, the AT1 receptor has been proposed to have a push-pull function in set point regulation for blood pressure in the RVLM (20), and the present evidence supports a similar mechanism in NTS. The model predictions of the effects of AT1R activation could be coupled with specific RNA and/or protein inhibitors and measures of physiological responses, as well as gene expression responses.
Pathway enrichment.
Enrichment analysis using Fisher's exact test was performed on each pathway at each time point, and statistically significant results are outlined in Table 5. Enrichment of upregulated genes was found in metabotropic and ionotropic glutamate pathways immediately at the 60-min time point, while enrichment was found in angiotensin and insulin pathways at the 90-min time point. In this representation, metabotropic and ionotropic glutamate pathways share the same ionotropic glutamate receptors (namely, NMDA 1, 2A-D, 3A, AMPA 1-4, and kainate 1-5). No metabotropic receptors were upregulated immediately after the 60-min time point, suggesting that the metabotropic enrichment is an artifact of the correlation between the pathways. However, ionotropic glutamate receptor regulation was a prominent response.
Table 5.
Enrichment analysis using Fisher's exact test was performed on each pathway at each time point
| Pathway | Time | Up/Down | P Value |
|---|---|---|---|
| Ionotropic glutamate receptor pathway | 60 | up | 0.004 |
| Metabotropic glutamate receptor group III pathway | 60 | up | 0.006 |
| 5HT2 type receptor-mediated signaling pathway | 60 | down | 0.020 |
| Angiotensin pathway | 90 | up | 0.024 |
| Insulin IGF pathway-protein kinase B signaling cascade | 90 | up | 0.033 |
Pathways from in silico model significantly enriched for up- or down-regulated genes at all time points. P value indicates statistically significant results (P < 0.05). Time after beginning of hypertensive stimulus is given in column 2, and direction of gene expression regulation is given in column 3. A complete list of P values is provided in Supplemental Tables S3–S5.
Contraction of receptor-centric representation into a pathway-centric representation.
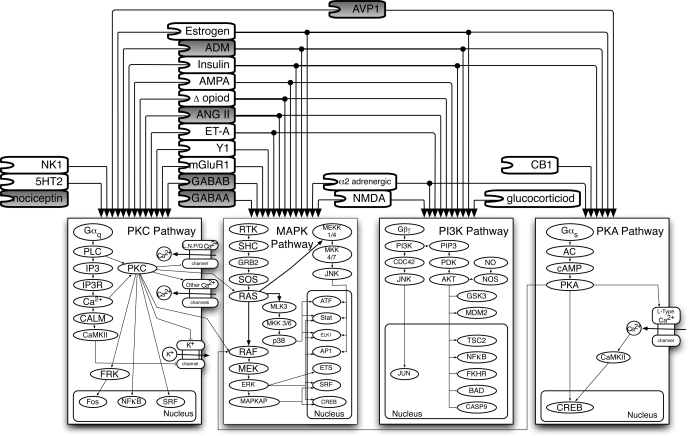
While the above approach is informative by focusing on molecular level details of the individual pathways, its complexity is not suitable for analyzing combinatorial effects on multiple receptors and pathways. Therefore, we simplified the data representation model into a consolidated map amenable to analysis and visualization of combinatorial regulation. Although the graphical model in Fig. 2 is receptor-centric, we collapsed common pathways together to create a pathway-centric view of the data (Fig. 3). Our graphical approach is borrowed from those used in silicon chip circuit design, representing the flow of information in the pathways as circuit designers treat the flow of electrons in wires. Analysis of the curated pathway maps indicates that most of the receptors in the model function through one or more of the following five pathways: PKC, cAMP/PKA, PI3K, MAPK, and soluble N-enthylmalemide sensitive fusion protein attachment receptor. An extensive review of the functional literature with respect to the present pathway results, for each of these five pathways, can be found in the supplemental material in the detailed pathway section of the discussion.
Fig. 3.
Graphical representation of connections between receptors and pathways implicated in NTS baroreflex modulation by pharmacological studies. These pathways are canonical and provide an abstract representation of events downstream from receptors. Although the general structure of each pathway is common to all receptors activating a given canonical pathway, the individual participants and isoforms may vary. Gray-shaded receptors are tachycardic and white receptors are bradycardic. See Table 4, columns 3–6 for references.
For each pathway, we identified the genes corresponding to each pathway component and collated the gene expression data in a tabular form for further analysis (for all components in each pathway, see Supplemental Tables S6–S10, which may be found at http://www.dbi.tju.edu/ajp-hypertension and on the American Journal of Physiology—Regulatory, Comparative and Integrative Physiology Web site). Our analysis combines a population of genes coding for any of the isoforms of a protein or subunits of a protein complex into individual pathway components that are queried for coordinate regulation. Our analysis of these pathways revealed coordinated transcriptional regulation among the upstream components within signaling pathways. Table 2 shows the coordinately regulated upstream components of each pathway. Note that most of the changes are between 20% and 100%. Because our statistical test for differential expression only validates the direction and not the actual amplitude of the change, we analyze the differential expression results as upregulation or downregulation (“direction”) at a given time point. It is not clear whether coordinate regulation further downstream in signaling networks will be observed when we extend the study to longer time points and longer hypertensive stimuli.
Perspectives and Significance
The complexity and the time-dependent coordination of the present gene responses are a first evidence for a very extensive molecular adaptive response in the NTS to the physiological disturbance of acute hypertension and associated increase in baroreceptor-afferent drive. Reasonable speculation as to the broad implications of the study suggests that these NTS adaptive responses are the initial aspects of a functional remodeling that will affect baroreflex function. The potential is that this kind of remodeling of function may participate in the recently described role of baroreflexes in long-term regulation of the blood pressure, contributing to some cases of essential hypertension. Future directions of the work will include additional, longer time points of more sustained exposure to hypertension, parallel description of changes in baroreflex function, and association of specific molecular mechanisms that contribute to these changes.
GRANTS
This work was supported by National Institutes of Health/National Heart, Lung, and Blood Institute awards R33HL087361 and R01HL54194 to J. S. Schwaber, National Institute on Alcohol Abuse and Alcoholism (NIAAA) and National Institute of General Medical Sciences awards R01AA13204 and R01GM076495 to J. S. Schwaber, and by NIAAA training grant and Greater Philadelphia Bioinformatics Alliance Fellowship support of R. L. Khan.
Supplementary Material
Acknowledgments
We thank Hester Hui for microarray fabrication and technical support. We thank Greg Gonye, Haiping Hao, and Greg Miller for discussions on the experimental approach and suggestions on the manuscript.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Allen AM, McKinley MJ, Oldfield BJ, Dampney RA, Mendelsohn FA. Angiotensin II receptor binding and the baroreflex pathway. Clin Exp Hypertens A 10 Suppl 1: 63–78, 1988. [DOI] [PubMed] [Google Scholar]
- 2.Augustine GJ, Santamaria F, Tanaka K. Local calcium signaling in neurons. Neuron 40: 331–346, 2003. [DOI] [PubMed] [Google Scholar]
- 3.Bading H, Ginty DD, Greenberg ME. Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science 260: 181–186, 1993. [DOI] [PubMed] [Google Scholar]
- 4.Baroudi G, Qu Y, Ramadan O, Chahine M, Boutjdir M. Protein kinase C activation inhibits Cav1.3 calcium channel at NH2-terminal serine 81 phosphorylation site. Am J Physiol Heart Circ Physiol 291: H1614–H1622, 2006. [DOI] [PubMed] [Google Scholar]
- 5.Barrett CJ, Malpas SC. Problems, possibilities, and pitfalls in studying the arterial baroreflexes' influence over long-term control of blood pressure. Am J Physiol Regul Integr Comp Physiol 288: R837–R845, 2005. [DOI] [PubMed] [Google Scholar]
- 6.Baxter AW, Wyllie DJ. Phosphatidylinositol 3 kinase activation and AMPA receptor subunit trafficking underlie the potentiation of miniature EPSC amplitudes triggered by the activation of l-type calcium channels. J Neurosci 26: 5456–5469, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooks VL, Sved AF. Pressure to change? Re-evaluating the role of baroreceptors in the long-term control of arterial pressure. Am J Physiol Regul Integr Comp Physiol 288: R815–R818, 2005. [DOI] [PubMed] [Google Scholar]
- 8.Chan JY, Chen WC, Lee HY, Chang TJ, Chan SH. Phosphorylation of transcription factor cyclic-AMP response element binding protein mediates c-fos induction elicited by sustained hypertension in rat nucleus tractus solitarii. Neuroscience 88: 1199–1212, 1999. [DOI] [PubMed] [Google Scholar]
- 9.Chan RK, Jarvina EV, Sawchenko PE. Effects of selective sinoaortic denervations on phenylephrine-induced activational responses in the nucleus of the solitary tract. Neuroscience 101: 165–178, 2000. [DOI] [PubMed] [Google Scholar]
- 10.Chan RK, Sawchenko PE. Differential time- and dose-related effects of haemorrhage on tyrosine hydroxylase and neuropeptide Y mRNA expression in medullary catecholamine neurons. Eur J Neurosci 10: 3747–3758, 1998. [DOI] [PubMed] [Google Scholar]
- 11.Chan RK, Sawchenko PE. Hemodynamic regulation of tyrosine hydroxylase messenger RNA in medullary catecholamine neurons: a c-fos-guided hybridization histochemical study. Neuroscience 66: 377–390, 1995. [DOI] [PubMed] [Google Scholar]
- 12.Chan RK, Sawchenko PE. Organization and transmitter specificity of medullary neurons activated by sustained hypertension: implications for understanding baroreceptor reflex circuitry. J Neurosci 18: 371–387, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan RK, Sawchenko PE. Spatially and temporally differentiated patterns of c-fos expression in brainstem catecholaminergic cell groups induced by cardiovascular challenges in the rat. J Comp Neurol 348: 433–460, 1994. [DOI] [PubMed] [Google Scholar]
- 14.Chan SH, Chang KF, Ou CC, Chan JY. Nitric oxide regulates c-fos expression in nucleus tractus solitarii induced by baroreceptor activation via cGMP-dependent protein kinase and cAMP response element-binding protein phosphorylation. Mol Pharmacol 65: 319–325, 2004. [DOI] [PubMed] [Google Scholar]
- 15.Comet MA, Laguzzi R, Hamon M, and Sevoz-Couche C. Functional interaction between nucleus tractus solitarius NK1 and 5-HT3 receptors in the inhibition of baroreflex in rats. Cardiovasc Res 65: 930–939, 2005. [DOI] [PubMed] [Google Scholar]
- 16.Comet MA, Sevoz-Couche C, Hanoun N, Hamon M, Laguzzi R. 5-HT-mediated inhibition of cardiovagal baroreceptor reflex response during defense reaction in the rat. Am J Physiol Heart Circ Physiol 287: H1641–H1649, 2004. [DOI] [PubMed] [Google Scholar]
- 17.Covarrubias MY, Khan RL, Vadigepalli R, Hoek JB, Schwaber JS. Chronic alcohol exposure alters transcription broadly in a key integrative brain nucleus for homeostasis: the nucleus tractus solitarius. Physiol Genomics 24: 45–58, 2005. [DOI] [PubMed] [Google Scholar]
- 18.Dampney RA, Li YW, Hirooka Y, Potts P, Polson JW. Use of c-fos functional mapping to identify the central baroreceptor reflex pathway: advantages and limitations. Clin Exp Hypertens 17: 197–208, 1995. [DOI] [PubMed] [Google Scholar]
- 19.Dampney RA, Polson JW, Potts PD, Hirooka Y, Horiuchi J. Functional organization of brain pathways subserving the baroreceptor reflex: studies in conscious animals using immediate early gene expression. Cell Mol Neurobiol 23: 597–616, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dell'Acqua ML, Smith KE, Gorski JA, Horne EA, Gibson ES, Gomez LL. Regulation of neuronal PKA signaling through AKAP targeting dynamics. Eur J Cell Biol 85: 627–633, 2006. [DOI] [PubMed] [Google Scholar]
- 21.Dinu I, Potter JD, Mueller T, Liu Q, Adewale AJ, Jhangri GS, Einecke G, Famulski KS, Halloran P, Yasui Y. Improving gene set analysis of microarray data by SAM-GS. BMC Bioinformatics 8: 242, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doerner D, Pitler TA, Alger BE. Protein kinase C activators block specific calcium and potassium current components in isolated hippocampal neurons. J Neurosci 8: 4069–4078, 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glass MJ, Chan J, Frys KA, Oselkin M, Tarsitano MJ, Iadecola C, Pickel VM. Changes in the subcellular distribution of NADPH oxidase subunit p47phox in dendrites of rat dorsomedial nucleus tractus solitarius neurons in response to chronic administration of hypertensive agents. Exp Neurol 205: 383–395, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hao H, Liu H, Gonye G, Schwaber J. A fast carrier chromatin immunoprecipitation method applicable to microdissected tissue samples. J Neurosci Methods In Press. [DOI] [PMC free article] [PubMed]
- 25.Hille B Ion Channels of Excitable Membranes. Sunderland, MA: Sinauer Associates, 2001.
- 26.Hogenesch JB, Panda S, Kay S, Takahashi JS. Circadian transcriptional output in the SCN and liver of the mouse. Novartis Found Symp 253: 171–180; discussion 152–175, 102–179, 180–173 passim, 2003. [PubMed] [Google Scholar]
- 27.Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, Kidd MJ, King AM, Meyer MR, Slade D, Lum PY, Stepaniants SB, Shoemaker DD, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend SH. Functional discovery via a compendium of expression profiles. Cell 102: 109–126, 2000. [DOI] [PubMed] [Google Scholar]
- 28.Jhamandas JH, Harris KH, Petrov T, Yang HY, Jhamandas KH. Activation of neuropeptide FF neurons in the brainstem nucleus tractus solitarius following cardiovascular challenge and opiate withdrawal. J Comp Neurol 402: 210–221, 1998. [PubMed] [Google Scholar]
- 29.Kamp TJ, Hell JW. Regulation of cardiac L-type calcium channels by protein kinase A and protein kinase C. Circ Res 87: 1095–1102, 2000. [DOI] [PubMed] [Google Scholar]
- 30.Khan RL, Gonye GE, Gao G, Schwaber JS. A universal reference sample derived from clone vector for improved detection of differential gene expression. BMC Genomics 7: 109, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan RL, Vadigepalli R, Gao G, Schwaber JS. A windowed local fdr estimator providing higher resolution androbust thresholds [Online]. QM, arXiv:q-bio/0702044v1, 2007.
- 32.Kitano H, Funahashi A, Matsuoka Y, Oda K. Using process diagrams for the graphical representation of biological networks. Nat Biotechnol 23: 961–966, 2005. [DOI] [PubMed] [Google Scholar]
- 33.Li YW, Dampney RA. Expression of c-fos protein in the medulla oblongata of conscious rabbits in response to baroreceptor activation. Neurosci Lett 144: 70–74, 1992. [DOI] [PubMed] [Google Scholar]
- 34.Li YW, Dampney RA. Expression of Fos-like protein in brain following sustained hypertension and hypotension in conscious rabbits. Neuroscience 61: 613–634, 1994. [DOI] [PubMed] [Google Scholar]
- 35.Lohmeier TE, Hildebrandt DA, Warren S, May PJ, Cunningham JT. Recent insights into the interactions between the baroreflex and the kidneys in hypertension. Am J Physiol Regul Integr Comp Physiol 288: R828–R836, 2005. [DOI] [PubMed] [Google Scholar]
- 36.Maclean RC Pleiotropy and GAL pathway degeneration in yeast. J Evol Biol 20: 1333–1338, 2007. [DOI] [PubMed] [Google Scholar]
- 37.Man HY, Wang Q, Lu WY, Ju W, Ahmadian G, Liu L, D'Souza S, Wong TP, Taghibiglou C, Lu J, Becker LE, Pei L, Liu F, Wymann MP, MacDonald JF, Wang YT. Activation of PI3-kinase is required for AMPA receptor insertion during LTP of mEPSCs in cultured hippocampal neurons. Neuron 38: 611–624, 2003. [DOI] [PubMed] [Google Scholar]
- 38.Matsumoto A, Ukai-Tadenuma M, Yamada RG, Houl J, Uno KD, Kasukawa T, Dauwalder B, Itoh TQ, Takahashi K, Ueda R, Hardin PE, Tanimura T, Ueda HR. A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev 21: 1687–1700, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Munch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res 31: 374–378, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, Kitano H, Thomas PD. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res 33: D284–D288, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miura M, Takayama K, Okada J. Neuronal expression of Fos protein in the rat brain after baroreceptor stimulation. J Auton Nerv Syst 50: 31–43, 1994. [DOI] [PubMed] [Google Scholar]
- 42.Moss SJ, Smart TG. Constructing inhibitory synapses. Nat Rev Neurosci 2: 240–250, 2001. [DOI] [PubMed] [Google Scholar]
- 43.Murphy AZ, Ennis M, Shipley MT, Behbehani MM. Directionally specific changes in arterial pressure induce differential patterns of fos expression in discrete areas of the rat brainstem: a double-labeling study for Fos and catecholamines. J Comp Neurol 349: 36–50, 1994. [DOI] [PubMed] [Google Scholar]
- 44.Ohta H, Talman WT. Both NMDA and non-NMDA receptors in the NTS participate in the baroreceptor reflex in rats. Am J Physiol Regul Integr Comp Physiol 267: R1065–R1070, 1994. [DOI] [PubMed] [Google Scholar]
- 45.Osborn JW, Jacob F, Guzman P. A neural set point for the long-term control of arterial pressure: beyond the arterial baroreceptor reflex. Am J Physiol Regul Integr Comp Physiol 288: R846–R855, 2005. [DOI] [PubMed] [Google Scholar]
- 46.Panda S, Hogenesch JB. It's all in the timing: many clocks, many outputs. J Biol Rhythms 19: 374–387, 2004. [DOI] [PubMed] [Google Scholar]
- 47.Paton JF, Li YW, Schwaber JS. Response properties of baroreceptive NTS neurons. Ann NY Acad Sci 940: 157–168, 2001. [DOI] [PubMed] [Google Scholar]
- 48.Paton JF, Rogers WT, Schwaber JS. Tonically rhythmic neurons within a cardiorespiratory region of the nucleus tractus solitarii of the rat. J Neurophysiol 66: 824–838, 1991. [DOI] [PubMed] [Google Scholar]
- 49.Pavlidis P Using ANOVA for gene selection from microarray studies of the nervous system. Methods 31: 282–289, 2003. [DOI] [PubMed] [Google Scholar]
- 50.Polson JW, Dampney RA, Boscan P, Pickering AE, Paton JF. Differential baroreflex control of sympathetic drive by angiotensin II in the nucleus tractus solitarii. Am J Physiol Regul Integr Comp Physiol 293: R1954–R1960, 2007. [DOI] [PubMed] [Google Scholar]
- 51.Potts PD, Polson JW, Hirooka Y, Dampney RA. Effects of sinoaortic denervation on Fos expression in the brain evoked by hypertension and hypotension in conscious rabbits. Neuroscience 77: 503–520, 1997. [DOI] [PubMed] [Google Scholar]
- 52.Qin Y, Zhu Y, Baumgart JP, Stornetta RL, Seidenman K, Mack V, van Aelst L, Zhu JJ. State-dependent Ras signaling and AMPA receptor trafficking. Genes Dev 19: 2000–2015, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reuter H Diversity and function of presynaptic calcium channels in the brain. Curr Opin Neurobiol 6: 331–337, 1996. [DOI] [PubMed] [Google Scholar]
- 54.Rinaman L, Card JP, Schwaber JS, Miselis RR. Ultrastructural demonstration of a gastric monosynaptic vagal circuit in the nucleus of the solitary tract in rat. J Neurosci 9: 1985–1996, 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rogers RF, Paton JF, Schwaber JS. NTS neuronal responses to arterial pressure and pressure changes in the rat. Am J Physiol Regul Integr Comp Physiol 265: R1355–R1368, 1993. [DOI] [PubMed] [Google Scholar]
- 56.Serra R, Villani M, Graudenzi A, Kauffman SA. Why a simple model of genetic regulatory networks describes the distribution of avalanches in gene expression data. J Theor Biol 246: 449–460, 2007. [DOI] [PubMed] [Google Scholar]
- 57.Sheng M, Lee SH. AMPA receptor trafficking and the control of synaptic transmission. Cell 105: 825–828, 2001. [DOI] [PubMed] [Google Scholar]
- 58.Shih CD, Chan SH, Chan JY. Participation of Fos protein at the nucleus tractus solitarius in inhibitory modulation of baroreceptor reflex response in the rat. Brain Res 738: 39–47, 1996. [DOI] [PubMed] [Google Scholar]
- 59.Springell DA, Costin NS, Pilowsky PM, Goodchild AK. Hypotension and short-term anaesthesia induce ERK1/2 phosphorylation in autonomic nuclei of the brainstem. Eur J Neurosci 22: 2257–2270, 2005. [DOI] [PubMed] [Google Scholar]
- 60.Spyer KM Central nervous mechanisms responsible for cardio-respiratory homeostasis. Adv Exp Med Biol 381: 73–79, 1995. [DOI] [PubMed] [Google Scholar]
- 61.Storey JD The positive false discovery rate: A Bayesian interpretation and the q-value. Ann Stat 31: 2013–2035, 2003. [Google Scholar]
- 62.Talman WT Glutamatergic transmission in the nucleus tractus solitarii: from server to peripherals in the cardiovascular information superhighway. Braz J Med Biol Res 30: 1–7, 1997. [DOI] [PubMed] [Google Scholar]
- 63.Tan PS, Potas JR, Killinger S, Horiuchi J, Goodchild AK, Pilowsky PM, Dampney RA. Angiotensin II evokes hypotension and renal sympathoinhibition from a highly restricted region in the nucleus tractus solitarii. Brain Res 1036: 70–76, 2005. [DOI] [PubMed] [Google Scholar]
- 64.Thiagarajan TC, Lindskog M, Malgaroli A, Tsien RW. LTP and adaptation to inactivity: Overlapping mechanisms and implications for metaplasticity. Neuropharmacology 52: 156–175, 2007. [DOI] [PubMed] [Google Scholar]
- 65.Thrasher TN Arterial baroreceptor input contributes to long-term control of blood pressure. Curr Hypertens Rep 8: 249–254, 2006. [DOI] [PubMed] [Google Scholar]
- 66.Veerasingham SJ, Raizada MK. Brain renin-angiotensin system dysfunction in hypertension: recent advances and perspectives. Br J Pharmacol 139: 191–202, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang W, Cherry JM, Nochomovitz Y, Jolly E, Botstein D, Li H. Inference of combinatorial regulation in yeast transcriptional networks: a case study of sporulation. Proc Natl Acad Sci USA 102: 1998–2003, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wei Q, Lu XY, Liu L, Schafer G, Shieh KR, Burke S, Robinson TE, Watson SJ, Seasholtz AF, Akil H. Glucocorticoid receptor overexpression in forebrain: a mouse model of increased emotional lability. Proc Natl Acad Sci USA 101: 11851–11856, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weiner N Norepinephrine, epinephrine and the sympathomimetic amines. In: The Pharmacological Basis of Therapeutics, edited by Goodman AG, Goodman LS, Rall TW, and Murad F. New York: MacMillan, 1985, p. 145–180.
- 70.Weston M, Wang H, Stornetta RL, Sevigny CP, Guyenet PG. Fos expression by glutamatergic neurons of the solitary tract nucleus after phenylephrine-induced hypertension in rats. J Comp Neurol 460: 525–541, 2003. [DOI] [PubMed] [Google Scholar]
- 71.Zhang J, Mifflin SW. Differential roles for NMDA and non-NMDA receptor subtypes in baroreceptor afferent integration in the nucleus of the solitary tract of the rat. J Physiol 511: 733–745, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou Y, Spangler R, Ho A, Kreek MJ. Increased CRH mRNA levels in the rat amygdala during short-term withdrawal from chronic ‘binge’ cocaine. Brain Res Mol Brain Res 114: 73–79, 2003. [DOI] [PubMed] [Google Scholar]
- 73.Zhou Z, Champagnat J, Poon CS. Phasic and long-term depression in brainstem nucleus tractus solitarius neurons: differing roles of AMPA receptor desensitization. J Neurosci 17: 5349–5356, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.