Abstract
In mammals, somatic growth is rapid in early postnatal life but decelerates with age and eventually halts, thus determining the adult body size of the species. This growth deceleration, which reflects declining proliferation, occurs simultaneously in multiple organs yet appears not to be coordinated by a systemic mechanism. We, therefore, hypothesized that growth deceleration results from a growth-limiting genetic program that is common to multiple tissues. Here, we identified a set of 11 imprinted genes that show down-regulation of mRNA expression with age in multiple organs. For these genes, Igf2, H19, Plagl1, Mest, Peg3, Dlk1, Gtl2, Grb10, Ndn, Cdkn1c, and SLC38a4, the declines show a temporal pattern similar to the decline in growth rate. All 11 genes have been implicated in the control of cell proliferation or somatic growth. Thus, our findings suggest that the declining expression of these genes contributes to coordinate growth deceleration in multiple tissues. We next hypothesized that the coordinate decline in expression of these imprinted genes is caused by altered methylation and consequent silencing of the expressed allele. Contrary to this hypothesis, the methylation status of the promoter regions of Mest, Peg3, and Plagl1 did not change with age. Our findings suggest that a set of growth-regulating imprinted genes is expressed at high levels in multiple tissues in early postnatal life, contributing to rapid somatic growth, but that these genes are subsequently downregulated in multiple tissues simultaneously, contributing to coordinate growth deceleration and cessation, thus imposing a fundamental limit on adult body size.
Keywords: imprinting, growth regulation, methylation, Zac1
in mammals, somatic growth is rapid in embryonic and early postnatal life but decelerates dramatically with age, declining toward zero as the organism approaches its adult body size. This growth deceleration appears to occur in all mammalian species, but at differing rates. In some species, such as mice and rats, the deceleration occurs over weeks, resulting in a smaller body size, whereas in other species, such as cows and humans, the deceleration occurs over years, resulting in a larger body size. Thus, the rate of deceleration appears to determine the adult body size of a species.
Somatic growth results from both cell proliferation (hyperplasia) and cell enlargement (hypertrophy). The decline in the somatic growth rate with age is due, in large part, to a decrease in the rate of proliferation (5, 28, 39). In fact, a constant rate of cell proliferation would lead to an exponential increase in weight, and thus, even a linear increase in weight suggests a declining rate of proliferation.
The biological mechanism responsible for this decline in proliferation and consequent growth deceleration is not known. One possible clue regarding the mechanism is that this phenomenon occurs in all major organs, such as kidney, liver, lungs, heart, skeletal muscle, and bone. However, this coordinate growth deceleration does not appear to be caused by a hormonal mechanism. For example, in the late adolescent human, as the somatic growth rate approaches zero, growth hormone levels and circulating IGF-I levels are higher than in the infant when growth is very rapid (27, 41). Furthermore, transplantation experiments suggest that the growth rate of transplanted structures generally depends on the age of the donor animal, rather than the age of the recipient, again arguing against a hormonal or other systemic mechanism (9, 33).
We, therefore, hypothesized that the coordinate growth deceleration in multiple organs results from a local mechanism that is common to multiple tissue types. On the basis of this hypothesis, we sought a gene expression program that occurs as somatic growth decelerates and that is common to multiple organs. Toward this goal, we previously performed microarray analysis to identify changes in gene expression in mice during early postnatal life. We focused our attention on genes that were upregulated or downregulated in multiple organs and thus are more likely to contribute to the putative common program of growth deceleration.
We noticed that some of the genes that showed the greatest changes in expression with age are imprinted, that is, genes that show differential expression from the maternal and paternal alleles. It has previously been observed that some imprinted genes positively regulate fetal growth (11, 12). Our microarray observations raised the possibility that expression of these genes persists into early postnatal life and that their subsequent downregulation is responsible for the dramatic decline in proliferation and somatic growth that determines adult body size.
In the current study, we sought to explore the temporal regulation of imprinted genes during postnatal life and the relationship between the temporal changes and somatic growth deceleration. First, we systematically assessed the age-related changes in gene expression of all known imprinted genes using microarray analysis for kidney, lung, and heart to identify imprinted genes that showed highly consistent changes with age. Second, for the identified genes, we measured the temporal changes in gene expression from the late embryonic period through the postnatal growth period to determine whether their expression declines in a temporal pattern that could account for the declines in growth rate. Third, we searched the previous literature to determine whether targeted ablation of these genes affects somatic growth in mice. Fourth, because allele-specific expression of imprinted genes often involves DNA methylation, we determined whether temporal changes in DNA methylation levels of the differentially methylated regions of these genes might explain the temporal changes in expression levels.
MATERIALS AND METHODS
Animal procedures and tissue processing.
C57BL/6 male mice (Charles River Laboratory, Wilmington, MA) were maintained and used in accordance with the Guide for the Care and Use of Laboratory Animals (26a). Animals were weighed, and tail lengths were measured before death (15). Mice were killed by carbon dioxide inhalation at embryonic day 19 (E19), and postnatally at 1, 4, and 8 wk of age. Heart, liver, lung, and kidney were excised and weighed. Tibias were excised, separated from adjacent muscle, and their lengths were measured using a digital vernier caliper. For E19 pups, gender was determined by the presence or absence of Sry, assessed by PCR of liver genomic DNA. For consistency, only male pups were used.
Microarray analysis.
Microarray analysis had been performed previously in heart, lung, and kidney at 1, 4, and 8 wk of age (G. P. Finkielstain, P. Forcinito, J. C. Liu, K. M. Barnes, R. Marino, unpublished data). For the current analysis, a heat map was constructed using JMP 7 software (SAS Institute, Cary, NC) to visualize change in expression of all known imprinted genes. Hierarchal clustering of these genes using JMP 7 was performed to group genes with similar expression patterns.
Quantitative real-time RT-PCR.
Total RNA was extracted from heart, lung, and kidney at 1, 4, and 8 wk of age using TRIzol reagent (Invitrogen, Carlsbad, CA), further purified with RNeasy kit (Qiagen, Valencia, CA), and then reverse transcribed using Superscript III Reverse Transcriptase (Invitrogen), according to the manufacturer's instruction. Resulting cDNA was diluted and stored at −20°C. Quantitative real-time RT-PCR was performed using the following assays containing primers and specific intron-spanning FAM-labeled TaqMan probes (Applied Biosystems, Foster City, CA): Cdkn1c, Mm00438170_m1; Dlk1, Mn00494477_m1; Grb10, Mm01180444_m1, Gtl2, Mm00522599_m1; H19, Mm01156721_g1; Igf2, Mm00439564_m1; Mest, Mm00484993_m1; Ndn, Mm02524479_s1; Peg3, Mm01337379_m1; Plagl1, Mm00494250_m1; Slc38a4, Mm00459056_m1; and Sry, Mm00441712_s1. VIC/TAMRA-labeled eukaryotic 18S rRNA endogenous control TaqMan assay (Applied Biosystems) was used for normalization. Reactions were performed in triplicate using cDNA, TaqMan Universal PCR Master Mix (Applied Biosystems), and the ABI Prism 7300 Sequence Detection System (Applied Biosystems), according to manufacturer's instructions with the following thermal cycling conditions: 1 cycle at 50°C for 2 min and 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. The quantity of each mRNA was calculated relative to the amount of starting cDNA using the formula: relative expressioni = (2)CTr/(2)CTi, where r represents 18S rRNA (for internal normalization), i represents the gene of interest, and CT represents the threshold cycle. For convenience, relative expression values were multiplied by 106. Serial 10-fold dilutions of embryonic cDNA were used to confirm near-theoretical efficiencies of amplification.
Determination of methylation by pyrosequencing.
Genomic DNA was isolated from liver at 1, 4, and 8 wk of age using DNeasy tissue kit (Qiagen) and then treated with bisulfite to convert unmethylated cytosine into uracil using the EZ DNA Methylation-Gold Kit (Zymo Research, Orange, CA), according to manufacturer's instructions. Briefly, 300 ng of genomic DNA was incubated with bisulfite reagent in the dark at 98°C for 10 min followed by 64°C for 2.5 h. DNA was then desulfonated, purified, and recovered with a spin column. Differentially methylated regions in Mest, Peg3, and Plagl1 at different ages were then PCR amplified and subjected to pyrosequencing (EpigenDx). Forty-five cycles of PCR were performed using Platinum Taq DNA polymerase (Invitrogen) with denaturation at 95°C, annealing at 50°C (for Peg3 and Plagl1) or 55°C (for Mest), and elongation at 72°C, with MgCl2 concentration of 1.5 mM (for Peg3 and Plagl1) or 3 mM (for Mest). Primers for PCR and pyrosequencing were as follows: Mest, forward PCR primer: 5′ GGT TGG GTT TGG ATA TTG TAA A 3′, reverse PCR primer: 5′ CCC TTA AAA ATC ATC TTT CAC ACC 3′, sequencing primer I: 5′ TTA AAG TTG TAG TAA ATT A 3′, sequencing primer II: 5′ GTG TTT AGG TTG TTA GAA T 3′; Peg3, forward PCR primer: 5′ AGA GAT GTT TAT TTT GGG TTG GTG 3′, reverse PCR primer: 5′ CAC CCC AAA CAC CAT CTA AAC T 3′, sequencing primer: 5′ ATT TTG GCT TGG TGG 3′; and Plagl1, forward PCR primer: 5′ GGT TAG GGT AGG TAA GTA GTG AT 3′, reverse PCR primer: 5′ CCA AAT TCA AAA TTT ATC ACC 3′, sequencing primer: 5′ GGT AGG TAA GTA GTG ATA AT 3′.
Statistical analysis.
Data are presented as means ± SE. For real-time PCR data, a one-way ANOVA for the effect of time was performed.
RESULTS AND DISCUSSION
A subset of imprinted genes shows declining expression in postnatal life in multiple organs.
In mammals and other multicellular organisms, exponential growth generally occurs only early in life (14). The growth rate then drops postnatally with age and approaches zero as final body size is attained (22). This decline in growth rate is caused in large part by a decline in the rate of proliferation. Importantly, this growth deceleration occurs coordinately in multiple different organs even though there does not appear to be a hormonal or other systemic mechanism responsible for the coordination. We therefore hypothesized that the coordinate growth deceleration in multiple organs results from a local mechanism that is common to multiple tissue types. On the basis of this hypothesis, we sought a gene expression program that occurs coordinately in multiple organs as somatic growth decelerates. We performed cDNA microarray analysis of kidney, lung, and heart of mice at 1, 4 and 8 wk of age and focused our attention on genes that were coordinately upregulated or downregulated in multiple organs (G. P. Finkielstain, P. Forcinito, J. C. Liu, K. M. Barnes, R. Marino, unpublished data).
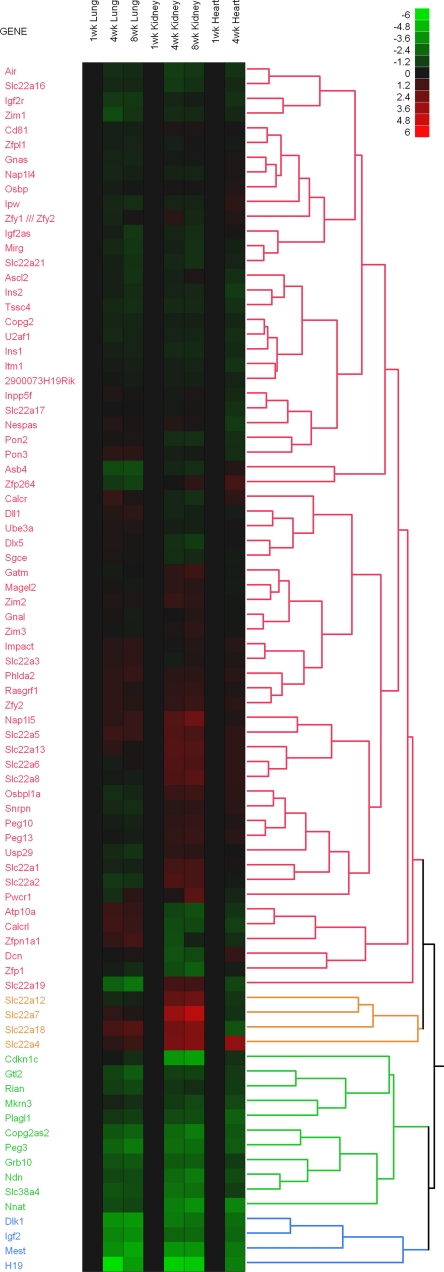
We noted that many of the genes that showed strong changes in expression with age in multiple tissues are imprinted genes. We, therefore, systematically analyzed the expression pattern of all the known imprinted genes in our microarray data set by generating a heat map using hierarchical clustering (Fig. 1). Eighty-two imprinted genes fell into four major clusters, of which two clusters, encompassing 15 genes, showed downregulation with age in all three tissues (Fig. 1, gene designations shown in green and blue). A second, parallel, more stringent analysis was performed (see Supplemental Table 1, which is available in the online version of this article) to identify imprinted genes that met the following three criteria: 1) mRNA expression changed significantly (P < 0.05) in all three organs studied, 2) the direction of the change (increase vs. decrease with age) was consistent in all three organs, and 3) the fold change was large, at least five-fold in one or more organs. Of the 82 imprinted genes, 11 met all three of these criteria: Igf2, H19, Plagl1, Mest, Peg3, Dlk1, Gtl2, Grb10, Ndn, Cdkn1c, and SLC38a4. For all of these genes, expression decreased with age, and they all belonged to the two clusters generated by the hierarchical cluster analysis. There were no imprinted genes that showed increasing expression with age that met these criteria. Thus, the analysis delineated a subset of 11 imprinted genes that were downregulated with age coordinately in multiple organs.
Fig. 1.
Temporal changes in expression of imprinted genes in mice. Changes in gene expression were analyzed by microarray in mouse lung, kidney, and heart from 1 to 8 wk of age. Heat maps were constructed for all known imprinted genes using JMP software ver. 7. Green rectangles represent down-regulated genes, and red rectangles represent up-regulated genes compared with 1-wk-old animals. The color intensity corresponds to the magnitude of the change from baseline (log2 [value at later time point/value at baseline time point]). The dendrogram on the right side of the heat map shows the hierarchal clustering of genes imposed by the software package, which grouped genes with similar expression patterns into 4 major clusters (colored red, green, blue, and yellow).
The temporal pattern of declining gene expression is similar to the temporal pattern of growth deceleration.
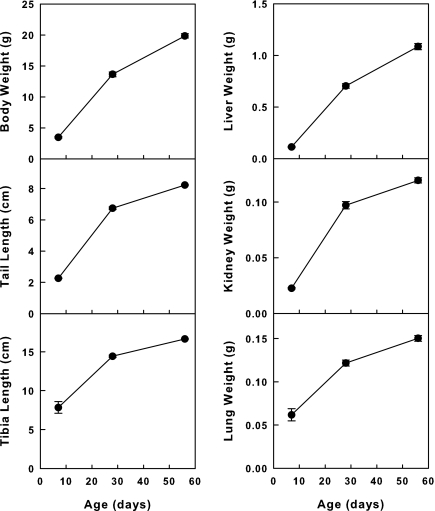
We next sought to define the temporal changes in gene expression more precisely for the 11 genes identified by microarray analysis. We therefore performed quantitative real-time PCR using RNA isolated from kidney, lung, and liver at E19 and 1, 4, and 8 wk of age. We used these three organs because they show similar temporal patterns for the postnatal decline in growth (Fig. 2) Previous studies indicate that this growth deceleration is caused by a marked decline in proliferation by 4 wk of age and more gradual declines thereafter (our unpublished observations). We therefore reasoned that a marked decline in gene expression between 1 and 4 wk of age in these organs would be most consistent with our hypothesis that the declining expression of these genes is responsible for postnatal growth deceleration. If instead, we observed a marked drop in expression between E19 and 1 wk, followed by more gradual subsequent declines, this temporal pattern would suggest that the gene may have a role in earlier embryogenesis and that the postnatal declines merely represent the vestigial remnant expression of a gene that is more important for embryonic development.
Fig. 2.
Temporal changes in organ and whole body size in mice. Weight of liver, kidney, lung, and the whole animal, and length of tail and tibia were measured at 1, 4, and 8 wk of age.
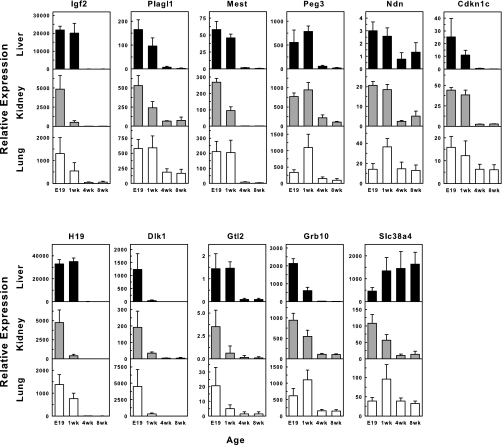
Measurement of mRNA levels by real-time PCR showed that the expression of all 11 genes declined with postnatal age in all organs studied with one exception, Slc38a4 in the liver [which may be due to its critical role in the liver by encoding a transporter of both cationic and neutral amino acids (8, 20)] (Fig. 3). In general, the most consistent and pronounced declines in expression occurred between 1 and 4 wk of age, the time period during which somatic growth rate decreases rapidly. This rapid decline in expression was usually followed by a continued, but less steep, decline from 4 to 8 wks. The changes from E19 to 1 wk were less consistent (upregulation from E19 to 1 wk was observed in some cases). Only for Dlk1 did the greatest declines occur between E19 and 1 wk in all tissues. For many of the genes studied, mRNA expression was still detectable even at 8 wk of age. We speculate that in some cases, residual expression may continue to serve a biological function in adulthood, for example, to maintain a low level of proliferation to compensate for cell death.
Fig. 3.
Concordant temporal changes in a set of imprinted genes in multiple organs. Expression of Plagl1, Mest, Peg3, Dlk1, Gtl2, Igf2, H19, Ndn, Grb10, Cdkn1c, and Slc38a4, were determined by real-time PCR in liver, kidney, and lung at four different time points. Except Slc38a4 in the liver, there is a strikingly consistent decrease in expression from 1 to 4 wk of age in all three organs. E19, embryonic day 19.
We chose to study lung, liver, and kidney as examples of organs in which proliferation slows with age during early postnatal life. Our findings probably would not apply to other organs, such as intestinal epithelium, epidermis, and hematopoietic tissue, in which proliferation continues into adulthood to replace the continual loss of terminally differentiated cells.
The identified subset of imprinted genes affects somatic growth.
It has previously been observed that a number of imprinted genes affect fetal growth (30). To explore the functional relationships between the 11 imprinted genes identified in the current study and somatic growth, we searched electronic databases for reports describing targeted ablation of these genes in mice. Published reports were available for all 11 genes. Interestingly, change in body size/body weight was observed in 10 out of 11 genes (except Ndn), suggesting that these genes play a major role in regulating somatic growth.
For 7 of the 11 genes of interest, Igf2, Plagl1, Mest, Peg3, Dlk1, Gtl2, and Slc38a4, targeted ablation in mice causes somatic growth inhibition, suggesting that these genes promote growth, and therefore, the subsequent postnatal declines in expression may contribute to growth deceleration. Deficiencies of Igf2, Plagl1, Mest, Peg3, Dlk1, Gtl2, and Slc38a4 all resulted in reduced body size at birth or earlier, implying fetal growth retardation. Postnatal growth retardation was also observed in mice lacking functional Dlk1, Gtl2, Igf2, and Mest. For Plagl1, there is conflicting evidence about its role in growth regulation. Plagl1 was first reported to be a tumor suppressor that induces apoptosis and cell cycle arrest in cancer cell lines (32), suggesting that it suppresses growth. However, knockout models showed decreased fetal body weight by 11% at E16.5 and 23% at birth (38).
Four of the 11 genes on our list, Grb10, H19, Cdkn1c, and Ndn, are apparently growth inhibitory. Targeted ablation of Grb10 causes overgrowth of the embryo and placenta (6, 29). Similarly, germline deletions involving H19 in mice cause overgrowth. However, H19 mRNA is not translated, and the targeted deletions appear to induce this phenotype by causing upregulation of the neighboring Igf2 gene (18). Cdkn1c also appears primarily to inhibit growth. It encodes a cyclin-dependent kinase inhibitor p57KIP2 that can inhibit proliferation (23, 24). In humans, loss-of-function mutations can cause Beckwith-Wiedemann syndrome, which is characterized by somatic overgrowth and an increased risk of embryonal cancers (4, 35). In mice, targeted ablation caused increased body weight at E13.5 (3), although limb size and body length may be decreased (36, 40, 42). Conversely, overexpression in mice causes somatic growth inhibition (3). Ndn acts as a growth suppressor in vitro (37), but ablation of the Ndn has not been reported to affect body size (13, 26). The reason why these growth-inhibiting genes would show decreasing expression as somatic growth slows is not clear. We speculate that this imprinted gene network may contribute to a complex regulatory system that induces rapid but controlled growth in early life. This overall system, which includes both positive and negative regulatory components, may be gradually phased out over time, thus contributing to somatic growth deceleration.
Other phenotypes of these imprinted genes include neonatal and/or postnatal lethality for Cdkn1c, Dlk1, Gtl2, H19, Igf2, Mest, Ndn, and Plagl1, and developmental defects for various organs (bone formation: Cdkn1c, Dlk1, H19, Igf2, and Plagl1; liver: Dlk1 and Grb10; lung: Igf2 and Plagl1), abnormal placental size (increased: Grb10, H19; decreased: Mest), and abnormal nurturing behavior (Mest and Peg3).
The evolutionary reason that these growth-regulatory genes are imprinted might be related to the conflict hypothesis (25). By regulating proliferation, these genes may indirectly affect nutritional requirements of the offspring and therefore resource allocation between the mother and offspring. It has been suggested that genes that affect such resource allocation tend to be imprinted because the paternal evolutionary interest is to increase resource transfer to the pups, and thus maximize their chance of survival, while the maternal interest is to moderate resource transfer and thus not jeopardize her own survival.
It has previously been observed that some imprinted genes regulate fetal growth (11, 12). However, our findings point to a new concept; many imprinted genes that regulate fetal growth (both stimulatory and inhibitory) are subsequently down-regulated in postnatal life in a temporal pattern that matches the decline in the somatic growth rate, and that this down-regulation occurs concurrently in multiple organs. Therefore our findings suggest that the decline in expression of these imprinted genes serve to downregulate growth coordinately in multiple tissues. Although the findings suggest that these imprinted genes contribute to the normal decline in proliferation and thus somatic growth deceleration, there are likely to be other genes, including nonimprinted genes that also contribute to the mechanisms that limit somatic growth.
The identified subset of imprinted genes are all members of the Zac1 gene network.
The similarity in temporal expression profile of these imprinted genes suggests that they are regulated by a common mechanism. All 11 of the imprinted genes that show a coordinated decline in expression with age belong to a recently described group of genes termed the Zac1-regulated imprinted gene network (38). This network was described by Varrault and colleagues based on a meta-analysis to identify genes whose expression shows a strong correlation with Plagl1/Zac1 in available microarray data sets. Their work also suggests that Plagl1 may act as the “master switch” regulating the expression of other genes in the network. The fact that Plagl1 encodes a transcription factor makes it an attractive hypothesis. Consistent with this hypothesis, overexpression of Plagl1 in a cell line caused upregulation of some genes in the network and knockout of Plagl1 in mice caused downregulation of some genes. However, not all genes in the network were affected, and some of the observed effects were modest in magnitude, suggesting that other regulatory factors, apart from Plagl1, such as other transcription factors or epigenetic modifications, could contribute to the coregulation of these genes.
The declines in gene expression do not appear to be caused by methylation of the paternal allele.
Imprinted genes are differentially expressed from the paternal or the maternal alleles. Differentially methylated regions (DMRs) in the promoter regions of imprinted genes contribute to this differential expression (21). In particular, Mest, Peg3, and Plagl1 all contain DMRs that are methylated and transcriptionally silenced on the maternal alleles. We therefore hypothesized that the decline in gene expression that we had observed with age was caused by increasing methylation and therefore transcriptional silencing of the paternal alleles. This hypothesized mechanism of silencing would be the reverse of the transcriptional activation that can be induced in vitro by demethylation of H19, Cdkn1c, Peg3, and Plagl1 (10). Such transcriptional activation of a silenced allele has also been described in a variety of human cancers (2, 16, 17, 34).
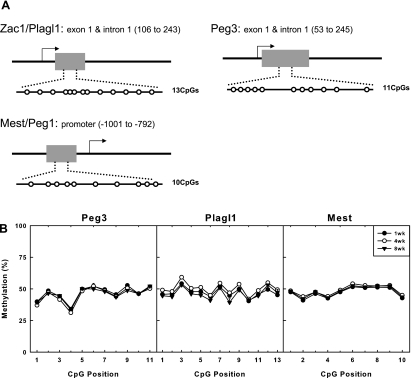
To test the hypothesis that the declining expression with age is due to altered methylation, we determined whether the methylation status of DMRs of Mest, Peg3, and Plagl1 changed with age in the liver. We assessed methylation status by bisulfite treatment followed by pyrosequencing because it provides a reliable and reproducible quantitative method to measure the percentage of methylation at each CpG dinucleotide within a defined region (1, 7). The method was applied to study 10 to 13 CpG dinucleotides within the DMR of each gene (19, 31) (Fig. 4A) in mouse liver at 1, 4, and 8 wk of age (n = 6 mice per time point). For all three genes studied, the DMRs remained ∼50% methylated (Fig. 4B), suggesting that only the maternal allele is methylated. Our findings argue strongly against the hypothesis that increasing methylation of the paternal allele is responsible for the decreasing expression of these three genes. It remains possible that changes in DNA methylation may be occurring in other imprinted genes, but our data indicate that altered imprinting is not a general mechanism responsible for the concurrent declines in expression of this network.
Fig. 4.
DNA methylation status of three different imprinted genes did not change with time. A: schematic diagram depicting the relative position of CpGs studied in the DMRs of Plagl1, Mest, and Peg3. B: percent methylation at CpG dinucleotides within the DMRs of Plagl1, Mest, and Peg3 in liver at different ages determined by bisulfite sequencing. There is ∼50% methylation at each site. Although there is some variability among different positions, values at each position show minimal change with age.
Therefore, the mechanisms responsible for coregulation of this gene network remain unclear. Possibilities include common regulatory elements and transcription factors (including Plagl1) and changes in histone modification with age.
Perspectives and Significance
In multicellular organisms, a constant rate of cell proliferation would lead to exponential growth. Consequently, mechanisms are required to slow proliferation, causing somatic growth to slow with age and, in many organisms, cease. This growth deceleration appears to occur in all mammalian species, but at differing rates, accounting for the enormous variability in adult body size among different species. Furthermore, to maintain body proportions, this decline in proliferation must occur coordinately in multiple organs. The mechanisms responsible for this decline in proliferation and somatic growth have not been determined previously. We have identified a subset of 11 imprinted genes within the Zac1 gene network that are widely expressed in late embryonic and early postnatal life but then are downregulated coordinately in multiple organs. The temporal pattern of this declining gene expression is similar to the temporal pattern of growth deceleration. Furthermore, there is evidence that these genes regulate cell proliferation and/or somatic growth. Taken together, the findings support the hypothesis that, in late embryonic and early postnatal life, this network of imprinted genes is expressed at high levels, which promotes rapid somatic growth, whereas, in later postnatal life, downregulation of the network in multiple tissues causes growth of multiple organs to slow coordinately and eventually halt, thus imposing a fundamental limit on adult body size.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the National Institute of Child Health and Human Development, National Institutes of Health.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Ahmadian A, Ehn M, Hober S. Pyrosequencing: history, biochemistry and future. Clin Chim Acta 363: 83–94, 2006. [DOI] [PubMed] [Google Scholar]
- 2.Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res 58: 5489–5494, 1998. [PubMed] [Google Scholar]
- 3.Andrews SC, Wood MD, Tunster SJ, Barton SC, Surani MA, John RM. Cdkn1c (p57Kip2) is the major regulator of embryonic growth within its imprinted domain on mouse distal chromosome 7. BMC Dev Biol 7: 53, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhuiyan ZA, Yatsuki H, Sasaguri T, Joh K, Soejima H, Zhu X, Hatada I, Morisaki H, Morisaki T, Mukai T. Functional analysis of the p57KIP2 gene mutation in Beckwith-Wiedemann syndrome. Hum Genet 104: 205–210, 1999. [DOI] [PubMed] [Google Scholar]
- 5.Chailler P, Briere N. Integration of proliferation and differentiation phenomena during rodent kidney ontogeny. Growth Dev Aging 55: 11–18, 1991. [PubMed] [Google Scholar]
- 6.Charalambous M, Smith FM, Bennett WR, Crew TE, Mackenzie F, Ward A. Disruption of the imprinted Grb10 gene leads to disproportionate overgrowth by an Igf2-independent mechanism. Proc Natl Acad Sci USA 100: 8292–8297, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal pyrosequencing methylation analysis of CpG sites. Biotechniques 35: 146–150, 2003. [DOI] [PubMed] [Google Scholar]
- 8.Desforges M, Lacey HA, Glazier JD, Greenwood SL, Mynett KJ, Speake PF, Sibley CP. SNAT4 isoform of system A amino acid transporter is expressed in human placenta. Am J Physiol Cell Physiol 290: C305–C312, 2006. [DOI] [PubMed] [Google Scholar]
- 9.Drzewiecki AE, Randolph MA, Hotchkiss RH, Weiland AJ. Vascularized growth-plate transplantation: a comparative study in the rat. J Reconstr Microsurg 8: 93–100, 1992. [DOI] [PubMed] [Google Scholar]
- 10.El Kharroubi A, Piras G, Stewart CL. DNA demethylation reactivates a subset of imprinted genes in uniparental mouse embryonic fibroblasts. J Biol Chem 276: 8674–8680, 2001. [DOI] [PubMed] [Google Scholar]
- 11.Fowden AL, Sibley C, Reik W, Constancia M. Imprinted genes, placental development and fetal growth. Horm Res 65 Suppl 3: 50–58, 2006. [DOI] [PubMed] [Google Scholar]
- 12.Gabory A, Dandolo L. Epigenetics and development: genomic imprinting. Med Sci 21: 390–395, 2005. [DOI] [PubMed] [Google Scholar]
- 13.Gerard M, Hernandez L, Wevrick R, Stewart CL. Disruption of the mouse necdin gene results in early post-natal lethality. Nat Genet 23: 199–202, 1999. [DOI] [PubMed] [Google Scholar]
- 14.Goedbloed JF The embryonic and postnatal growth of rat and mouse. I. The embryonic and early postnatal growth of the whole embryo A model with exponential growth and sudden changes in growth rate. Acta Anat (Basel) 82: 305–306, 1972. [PubMed] [Google Scholar]
- 15.Hughes PC, Tanner JM. A longitudinal study of the growth of the black-hooded rat: methods of measurement and rates of growth for skull, limbs, pelvis, nose-rump and tail lengths. J Anat 106: 349–370, 1970. [PMC free article] [PubMed] [Google Scholar]
- 16.Issa JP, Vertino PM, Boehm CD, Newsham IF, Baylin SB. Switch from monoallelic to biallelic human IGF2 promoter methylation during aging and carcinogenesis. Proc Natl Acad Sci USA 93: 11757–11762, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jelinic P, Shaw P. Loss of imprinting and cancer. J Pathol 211: 261–268, 2007. [DOI] [PubMed] [Google Scholar]
- 18.Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375: 34–39, 1995. [DOI] [PubMed] [Google Scholar]
- 19.Lucifero D, Mertineit C, Clarke HJ, Bestor TH, Trasler JM. Methylation dynamics of imprinted genes in mouse germ cells. Genomics 79: 530–538, 2002. [DOI] [PubMed] [Google Scholar]
- 20.Mackenzie B, Erickson JD. Sodium-coupled neutral amino acid (System N/A) transporters of the SLC38 gene family. Pflügers Arch 447: 784–795, 2004. [DOI] [PubMed] [Google Scholar]
- 21.Mann JR, Szabo PE, Reed MR, Singer-Sam J. Methylated DNA sequences in genomic imprinting. Crit Rev Eukaryot Gene Expr 10: 241–257, 2000. [DOI] [PubMed] [Google Scholar]
- 22.Marino R, Hegde A, Barnes KM, Schrier L, Emons JA, Nilsson O, Baron J. Catch-up growth after hypothyroidism is caused by delayed growth plate senescence. Endocrinology 149: 1820–1828, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuoka S, Edwards MC, Bai C, Parker S, Zhang P, Baldini A, Harper JW, Elledge SJ. p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev 9: 650–662, 1995. [DOI] [PubMed] [Google Scholar]
- 24.Matsuoka S, Thompson JS, Edwards MC, Bartletta JM, Grundy P, Kalikin LM, Harper JW, Elledge SJ, Feinberg AP. Imprinting of the gene encoding a human cyclin-dependent kinase inhibitor, p57KIP2, on chromosome 11p15. Proc Natl Acad Sci USA 93: 3026–3030, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet 7: 45–49, 1991. [DOI] [PubMed] [Google Scholar]
- 26.Muscatelli F, Abrous DN, Massacrier A, Boccaccio I, Le MM, Cau P, Cremer H. Disruption of the mouse Necdin gene results in hypothalamic and behavioral alterations reminiscent of the human Prader-Willi syndrome. Hum Mol Genet 9: 3101–3110, 2000. [DOI] [PubMed] [Google Scholar]
- 26a.Institute of Laboratory Animal Resources, National Research Council. Guide for the Care and Use of Laboratory Animals. Washington D.C.: National Academy Press, 1996.
- 27.Nunez SB, Municchi G, Barnes KM, Rose SR. Insulin-like growth factor I (IGF-I) and IGF-binding protein-3 concentrations compared with stimulated and night growth hormone in the evaluation of short children—a clinical research center study. J Clin Endocrinol Metab 81: 1927–1932, 1996. [DOI] [PubMed] [Google Scholar]
- 28.Post J, Hoffman J. Changes in the replication times and patterns of the liver cell during the life of the rat. Exp Cell Res 36: 111–123, 1964. [DOI] [PubMed] [Google Scholar]
- 29.Riedel H Grb10 exceeding the boundaries of a common signaling adapter. Front Biosci 9: 603–618, 2004. [DOI] [PubMed] [Google Scholar]
- 30.Smith FM, Garfield AS, Ward A. Regulation of growth and metabolism by imprinted genes. Cytogenet Genome Res 113: 279–291, 2006. [DOI] [PubMed] [Google Scholar]
- 31.Smith RJ, Arnaud P, Konfortova G, Dean WL, Beechey CV, Kelsey G. The mouse Zac1 locus: basis for imprinting and comparison with human ZAC. Gene 292: 101–112, 2002. [DOI] [PubMed] [Google Scholar]
- 32.Spengler D, Villalba M, Hoffmann A, Pantaloni C, Houssami S, Bockaert J, Journot L. Regulation of apoptosis and cell cycle arrest by Zac1, a novel zinc finger protein expressed in the pituitary gland and the brain. EMBO J 16: 2814–2825, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stevens DG, Boyer MI, Bowen CV. Transplantation of epiphyseal plate allografts between animals of different ages. J Pediatr Orthop 19: 398–403, 1999. [PubMed] [Google Scholar]
- 34.Sullivan MJ, Taniguchi T, Jhee A, Kerr N, Reeve AE. Relaxation of IGF2 imprinting in Wilms tumours associated with specific changes in IGF2 methylation. Oncogene 18: 7527–7534, 1999. [DOI] [PubMed] [Google Scholar]
- 35.Swanger WJ, Roberts JM. p57KIP2 targeted disruption and Beckwith-Wiedemann syndrome: is the inhibitor just a contributor? Bioessays 19: 839–842, 1997. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi K, Nakayama K, Nakayama K. Mice lacking a CDK inhibitor, p57Kip2, exhibit skeletal abnormalities and growth retardation. J Biochem (Tokyo) 127: 73–83, 2000. [DOI] [PubMed] [Google Scholar]
- 37.Taniura H, Taniguchi N, Hara M, Yoshikawa K. Necdin, a postmitotic neuron-specific growth suppressor, interacts with viral transforming proteins and cellular transcription factor E2F1. J Biol Chem 273: 720–728, 1998. [DOI] [PubMed] [Google Scholar]
- 38.Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, Severac D, Chotard L, Kahli M, Le DA, Pavlidis P, Journot L. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell 11: 711–722, 2006. [DOI] [PubMed] [Google Scholar]
- 39.Winick M, Noble A. Quantitative changes in DNA, RNA, and protein during prenatal and postnatal growth in the rat. Dev Biol 12: 451–466, 1965. [DOI] [PubMed] [Google Scholar]
- 40.Yan Y, Frisen J, Lee MH, Massague J, Barbacid M. Ablation of the CDK inhibitor p57Kip2 results in increased apoptosis and delayed differentiation during mouse development. Genes Dev 11: 973–983, 1997. [DOI] [PubMed] [Google Scholar]
- 41.Zapf J, Walter H, Froesch ER. Radioimmunological determination of insulinlike growth factors I and II in normal subjects and in patients with growth disorders and extrapancreatic tumor hypoglycemia. J Clin Invest 68: 1321–1330, 1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang P, Liegeois NJ, Wong C, Finegold M, Hou H, Thompson JC, Silverman A, Harper JW, DePinho RA, Elledge SJ. Altered cell differentiation and proliferation in mice lacking p57KIP2 indicates a role in Beckwith-Wiedemann syndrome. Nature 387: 151–158, 1997. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.