Abstract
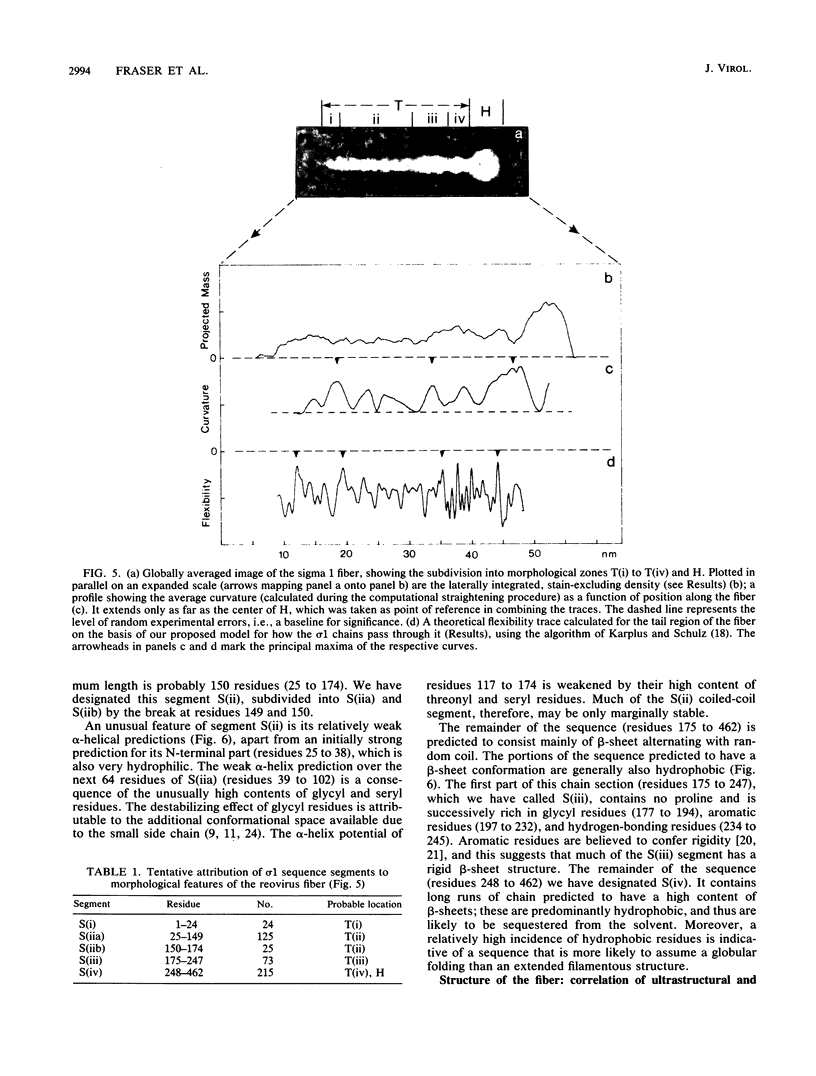
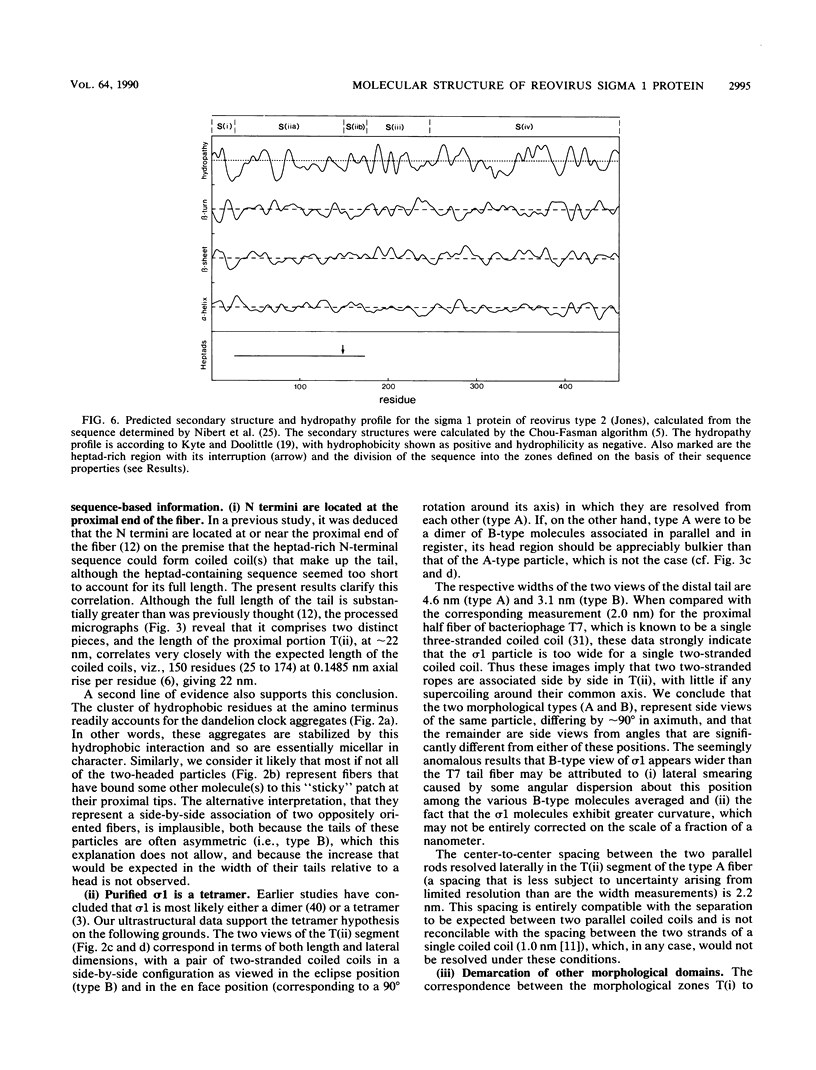
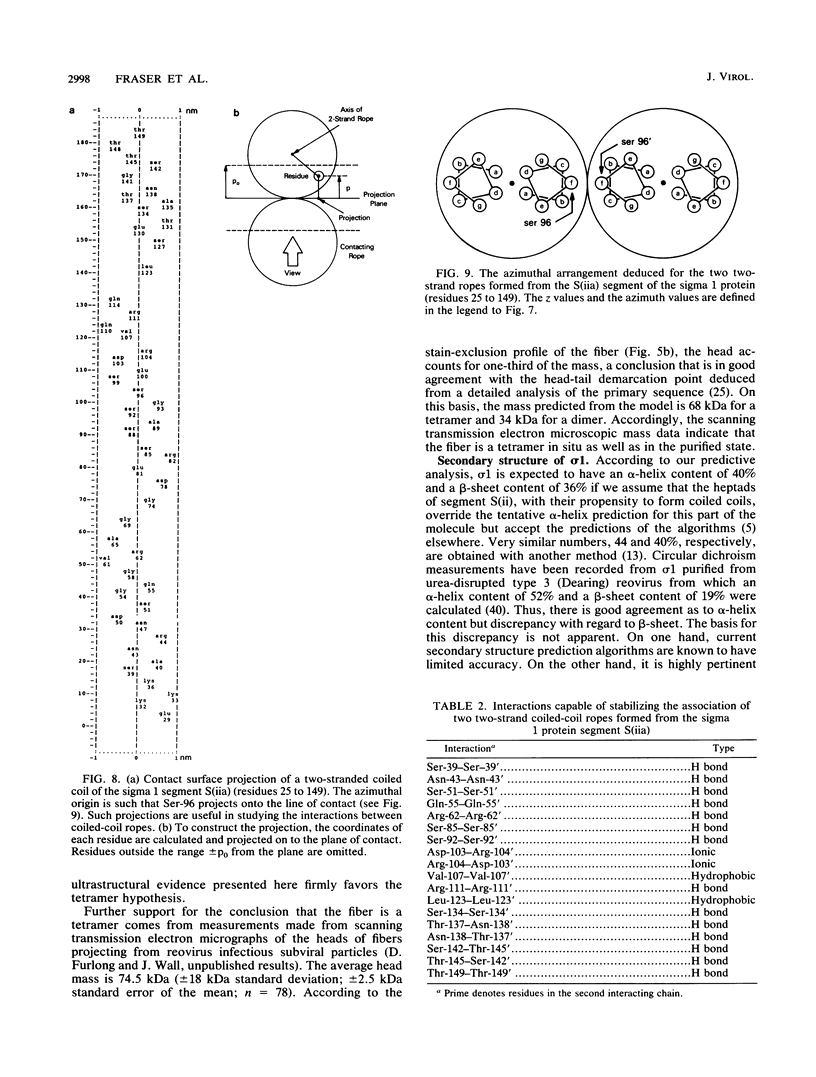
The receptor-recognition interaction that initiates reovirus infection is mediated by the sigma 1 protein, located at the vertices of the icosahedral virion. We have applied computer-based image-averaging techniques to electron micrographs of negatively stained preparations of sigma 1 purified from virions (serotype 2 Jones). Combining these results with inferences based on the amino acid sequence has led to a molecular model in which the overall folding of the chains is described; its conformation embodies motifs, coiled-coil alpha-helices and nodular multichain elements rich in beta-sheets, previously detected in the corresponding proteins of other viruses, but with some novel variations. Sigma 1 is a filamentous lollipop-shaped molecule with an overall length of approximately 48 nm; it has a flexible "tail," approximately 40 nm long by 4 to 6 nm wide, terminating at its distal end in a globular "head," approximately 9.5 nm in diameter. The purified protein is a tetramer (4 by 50 kilodaltons) consisting of two similarly oriented dimers bonded side by side and in register. For each chain, a cluster of hydrophobic residues at its amino terminus resides at the proximal end of the tail; next, an alpha-helical domain (residues 25 to 172) participates in a two-chained coiled coil, 22 nm long, with two such coiled coils pairing laterally to form the proximal half of the tail. The remainder of the tail (residues 173 to approximately 316) is less uniform in width and is expected to be rich in beta-sheet; the interdimer bonding is evidently sustained through this portion of the molecule. Finally, the globular head consists of the carboxy-terminal domains (which contain the receptor-binding sites) folded into compact globular conformations; in appropriate side views, the head is resolved into two subunits, presumably contributed by the respective dimers. This model for how the four sigma 1 polypeptide chains are threaded in parallel through the fiber is supported by the observed match between an empirical curvature profile, which identifies the locations of relatively flexible sites along the tail, and the flexibility profile predicted on the basis of the model. Appraisal of the interactions that stabilize the coiled coils suggests that (i) the alpha-helices are individually only marginally stable, a property that may be of significance with regard to the retracted conformation in which sigma 1 is accommodated in the intact virion, and (ii) the predominant interactions between the two coiled coils are likely to involve hydrogen bonding between patches of uncharged residues.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerjea A. C., Brechling K. A., Ray C. A., Erikson H., Pickup D. J., Joklik W. K. High-level synthesis of biologically active reovirus protein sigma 1 in a mammalian expression vector system. Virology. 1988 Dec;167(2):601–612. [PubMed] [Google Scholar]
- Bassel-Duby R., Jayasuriya A., Chatterjee D., Sonenberg N., Maizel J. V., Jr, Fields B. N. Sequence of reovirus haemagglutinin predicts a coiled-coil structure. 1985 May 30-Jun 5Nature. 315(6018):421–423. doi: 10.1038/315421a0. [DOI] [PubMed] [Google Scholar]
- Bassel-Duby R., Nibert M. L., Homcy C. J., Fields B. N., Sawutz D. G. Evidence that the sigma 1 protein of reovirus serotype 3 is a multimer. J Virol. 1987 Jun;61(6):1834–1841. doi: 10.1128/jvi.61.6.1834-1841.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashdollar L. W., Chmelo R. A., Wiener J. R., Joklik W. K. Sequences of the S1 genes of the three serotypes of reovirus. Proc Natl Acad Sci U S A. 1985 Jan;82(1):24–28. doi: 10.1073/pnas.82.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974 Jan 15;13(2):211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C., Goldberg E. B., Crowther R. A. The distal half of the tail fibre of bacteriophage T4. Rigidly linked domains and cross-beta structure. J Mol Biol. 1979 Jul 25;132(1):101–113. doi: 10.1016/0022-2836(79)90498-4. [DOI] [PubMed] [Google Scholar]
- Fraser R. D., Harrap B. S., Macrae T. P., Stewart F. H., Suzuki E. Effect of glycyl residues on the stability of the alpha-helix. Biopolymers. 1967 Mar;5(3):251–257. doi: 10.1002/bip.1967.360050303. [DOI] [PubMed] [Google Scholar]
- Fraser R. D., MacRae T. P., Suzuki E. Structure of the alpha-keratin microfibril. J Mol Biol. 1976 Dec;108(2):435–452. doi: 10.1016/s0022-2836(76)80129-5. [DOI] [PubMed] [Google Scholar]
- Furlong D. B., Nibert M. L., Fields B. N. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J Virol. 1988 Jan;62(1):246–256. doi: 10.1128/jvi.62.1.246-256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Geisler N., Kaufmann E., Weber K. Antiparallel orientation of the two double-stranded coiled-coils in the tetrameric protofilament unit of intermediate filaments. J Mol Biol. 1985 Mar 5;182(1):173–177. doi: 10.1016/0022-2836(85)90035-x. [DOI] [PubMed] [Google Scholar]
- Green N. M., Wrigley N. G., Russell W. C., Martin S. R., McLachlan A. D. Evidence for a repeating cross-beta sheet structure in the adenovirus fibre. EMBO J. 1983;2(8):1357–1365. doi: 10.1002/j.1460-2075.1983.tb01592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber R., Bennett W. S., Jr Functional significance of flexibility in proteins. Biopolymers. 1983 Jan;22(1):261–279. doi: 10.1002/bip.360220136. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Manjula B. N., Trus B. L., Fischetti V. A. Presence of two distinct regions in the coiled-coil structure of the streptococcal Pep M5 protein: relationship to mammalian coiled-coil proteins and implications to its biological properties. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1064–1068. doi: 10.1073/pnas.82.4.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan A. D., Karn J. Periodic features in the amino acid sequence of nematode myosin rod. J Mol Biol. 1983 Mar 15;164(4):605–626. doi: 10.1016/0022-2836(83)90053-0. [DOI] [PubMed] [Google Scholar]
- Nagata L., Masri S. A., Mah D. C., Lee P. W. Molecular cloning and sequencing of the reovirus (serotype 3) S1 gene which encodes the viral cell attachment protein sigma 1. Nucleic Acids Res. 1984 Nov 26;12(22):8699–8710. doi: 10.1093/nar/12.22.8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata L., Masri S. A., Pon R. T., Lee P. W. Analysis of functional domains on reovirus cell attachment protein sigma 1 using cloned S1 gene deletion mutants. Virology. 1987 Sep;160(1):162–168. doi: 10.1016/0042-6822(87)90056-0. [DOI] [PubMed] [Google Scholar]
- Nibert M. L., Dermody T. S., Fields B. N. Structure of the reovirus cell-attachment protein: a model for the domain organization of sigma 1. J Virol. 1990 Jun;64(6):2976–2989. doi: 10.1128/jvi.64.6.2976-2989.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry D. A. Letter: Double helix of tropomyosin. Nature. 1975 Jul 24;256(5515):346–347. doi: 10.1038/256346b0. [DOI] [PubMed] [Google Scholar]
- SCHERAGA H. A., NEMETHY G., STEINBERG I. Z. The contribution of hydrophobic bonds to the thermal stability of protein conformations. J Biol Chem. 1962 Aug;237:2506–2508. [PubMed] [Google Scholar]
- Smith R. E., Zweerink H. J., Joklik W. K. Polypeptide components of virions, top component and cores of reovirus type 3. Virology. 1969 Dec;39(4):791–810. doi: 10.1016/0042-6822(69)90017-8. [DOI] [PubMed] [Google Scholar]
- Steven A. C., Trus B. L., Maizel J. V., Unser M., Parry D. A., Wall J. S., Hainfeld J. F., Studier F. W. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J Mol Biol. 1988 Mar 20;200(2):351–365. doi: 10.1016/0022-2836(88)90246-x. [DOI] [PubMed] [Google Scholar]
- Takahashi S., Ooi T. Three-alpha-helical coiled-coil, as a proposed model for a thin rod segment of bacteriophage T3 tail fibers. Biochem Biophys Res Commun. 1988 Feb 15;150(3):1244–1250. doi: 10.1016/0006-291x(88)90762-0. [DOI] [PubMed] [Google Scholar]
- Unser M., Trus B. L., Steven A. C. Normalization procedures and factorial representations for classification of correlation-aligned images: a comparative study. Ultramicroscopy. 1989 Jul-Aug;30(3):299–310. doi: 10.1016/0304-3991(89)90058-2. [DOI] [PubMed] [Google Scholar]
- Valentine R. C., Green N. M. Electron microscopy of an antibody-hapten complex. J Mol Biol. 1967 Aug 14;27(3):615–617. doi: 10.1016/0022-2836(67)90063-0. [DOI] [PubMed] [Google Scholar]
- Varghese J. N., Laver W. G., Colman P. M. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 A resolution. Nature. 1983 May 5;303(5912):35–40. doi: 10.1038/303035a0. [DOI] [PubMed] [Google Scholar]
- Willey R. L., Smith D. H., Lasky L. A., Theodore T. S., Earl P. L., Moss B., Capon D. J., Martin M. A. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988 Jan;62(1):139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I. A., Skehel J. J., Wiley D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981 Jan 29;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- Yeung M. C., Gill M. J., Alibhai S. S., Shahrabadi M. S., Lee P. W. Purification and characterization of the reovirus cell attachment protein sigma 1. Virology. 1987 Feb;156(2):377–385. doi: 10.1016/0042-6822(87)90417-x. [DOI] [PubMed] [Google Scholar]
- de Groot R. J., Luytjes W., Horzinek M. C., van der Zeijst B. A., Spaan W. J., Lenstra J. A. Evidence for a coiled-coil structure in the spike proteins of coronaviruses. J Mol Biol. 1987 Aug 20;196(4):963–966. doi: 10.1016/0022-2836(87)90422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M., Frank J. Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy. 1981;6(2):187–194. doi: 10.1016/0304-3991(81)90059-0. [DOI] [PubMed] [Google Scholar]







