Abstract
This work represents a maiden effort to systematically screen the transcriptome of the healing wound-edge tissue temporally using high-density GeneChips. Changes during the acute inflammatory phase of murine excisional wounds were characterized histologically. Sets of genes that significantly changed in expression during healing could be segregated into the following five sets: up-early (6–24 h; cytokine-cytokine receptor interaction pathway), up-intermediary (12–96 h; leukocyte-endothelial interaction pathway), up-late (48–96 h; cell-cycle pathway), down-early (6–12 h; purine metabolism) and down-intermediary (12–96 h; oxidative phosphorylation pathway). Results from microarray and real-time PCR analyses were consistent. Results listing all genes that were significantly changed at any specific time point were further mined for cell-type (neutrophils, macrophages, endothelial, fibroblasts, and pluripotent stem cells) specificity. Candidate genes were also clustered on the basis of their functional annotation, linking them to inflammation, angiogenesis, reactive oxygen species (ROS), or extracellular matrix (ECM) categories. Rapid induction of genes encoding NADPH oxidase subunits and downregulation of catalase in response to wounding is consistent with the fact that low levels of endogenous H2O2 is required for wound healing. Angiogenic genes, previously not connected to cutaneous wound healing, that were induced in the healing wound-edge included adiponectin, epiregulin, angiomotin, Nogo, and VEGF-B. This study provides a digested database that may serve as a valuable reference tool to develop novel hypotheses aiming to elucidate the biology of cutaneous wound healing comprehensively.
Keywords: redox, tissue repair
impaired wound healing states in the elderly lead to substantial morbidity and mortality and a cost to the health services of over 9 billion dollars per annum. Recently we have developed a novel approach to specifically laser capture blood vessels from standard 3 mm human wound biopsies such that the captured blood vessel tissue element would lend itself to genomic screening as well as to verification of candidate genes using quantitative PCR (65). In humans, we have also identified stress-sensitive transcripts in wound site neutrophils (63). While results from clinical material are of extraordinary interest, they often pose limitations in the design of studies aimed at gaining mechanistic insight.
Our current understanding of the mechanisms underlying cutaneous wound healing is primarily derived from the study of experimental animal models, which lend themselves to more detailed scrutiny. Murine models are commonly studied because of the availability of a large number of genetically modified mice that help address pointed hypotheses (13). Wound healing represents the outcome of a large number of interrelated biological events that are orchestrated over a temporal sequence in response to injury and its microenvironment. Previous reports have characterized global patterns of gene expression in the burn wound (19). This work represents a maiden effort to screen the transcriptome of the healing wound-edge tissue on a temporal basis using high-density GeneChips. Focus of the study was directed toward examining acute changes in the wound-edge tissue during the inflammatory phase, which is viewed as setting the stage for the consequent phases.
MATERIALS AND METHODS
Secondary-intention excisional dermal wound model.
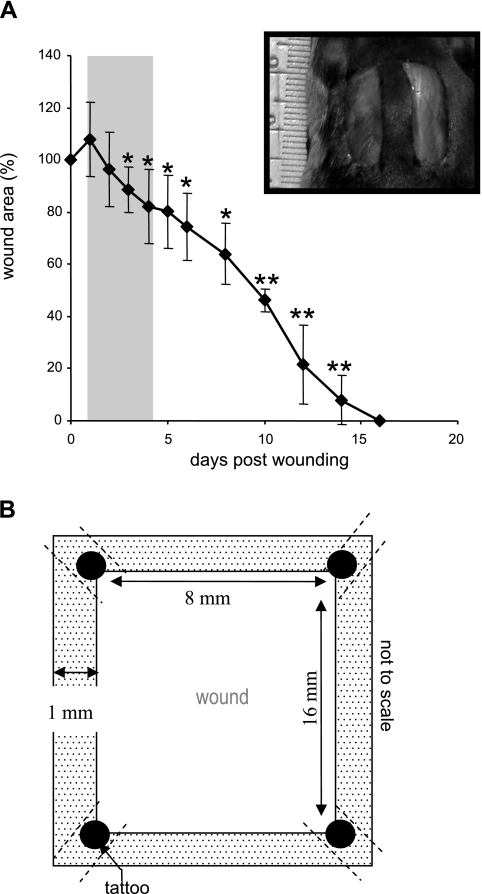
Young (8-wk old) male C57BL/6 mice were used for this study. Two 8 × 16 mm full-thickness excisional wounds (71) were placed on the dorsal skin, equidistant from the midline and adjacent to the four limbs (Fig. 1A, inset). The animals were killed at the indicated time points after wounding and wound-edge (1–2 mm, Fig. 1B) tissues were harvested. All animal studies were performed in accordance with protocols approved by the Laboratory Animal Care and Use Committee of the Ohio State University. During the wounding procedure, mice were anesthetized by low-dose isoflurane inhalation for 5–10 min per standard recommendation (41). While there are no direct data on how 5–10 min isoflurane exposure may influence cutaneous injury following acute excision, it is generally known that isoflurane exposure for 30 min or more may influence hemodynamics and leukocyte rolling velocities in the mesenteric microcirculation during lipopolysaccharide-induced inflammation (30).
Fig. 1.
Secondary-intention excisional dermal wound closure in mice. A: two 16 × 8 mm full-thickness rectangular (inset) excisional wounds were placed on the dorsal skin, equidistant from the midline and adjacent to the 4 limbs. These wounds were left to heal by secondary intention. Wound closure is shown as percentage of area of initial wound determined on the indicated day after wounding. The shaded area indicates time period selected for the study of gene expression profile in wound edge tissue postinjury. Data are means ± SD, n = 4. *P < 0.05; **P < 0.001 compared with 0 h time point postwound. B: definition of wound edge tissue used for microarray analysis. The dotted area represents the wound edge tissue. Tattooing was performed on 4 corners (filled circles) of the wound site to keep track of the edges. To minimize the possible interfering effects of tattoo-related trauma, the procedure was performed 1 wk ahead of the wounding. The tissue containing tattoo dots was excluded from analyses.
Determination of wound area.
Imaging of wounds was performed with a digital camera (Canon PowerShot G6). The wound area was determined using WoundMatrix software as described previously (71).
Histology.
Formalin-fixed paraffin-embedded or OCT-embedded frozen wound-edge specimens were sectioned. The paraffin sections (4 μm) were deparaffinized and stained with hematoxylin & eosin by standard procedures. Immunohistochemical staining of paraffin or frozen sections was performed as described earlier (60) using the following primary antibodies: anti-neutrophil (1:200; AbD Serotec, Raleigh, NC), F4/80 (1:100, AbD Serotec) or CD31 (1:200; BD Pharmingen, San Diego, CA). Secondary antibody detection and counterstaining were performed as described previously (60).
GeneChip probe array analyses.
To identify sets of gene differentially expressed during the temporal course of healing, we utilized the GeneChip approach (59, 61–63, 65). Total RNA was extracted (TRIzol, GIBCO BRL) from wound-edge tissue 0, 6, 12, 24, 48, and 96 h after wounding. Further clean-up of RNA was performed using the RNeasy kit (Qiagen). The quality of RNA thus obtained was examined using the Agilent 2100 Bioanalyzer. Targets were prepared for microarray hybridization according to previously described protocols (59, 61, 62, 64). To assess the quality of hybridization, samples were hybridized for 16 h at 45°C to GeneChip test arrays. Satisfactory samples were hybridized with the Murine Genome Array U74Av2 for the screening of >12,000 probe sets. The arrays were washed, stained with streptavidin-phycoerythrin, and were then scanned with the GeneArray scanner (Affymetrix) in our own facilities as described earlier (59, 61, 65). Data were collected from four mice (n = 4) in each group representing a time point. Tissue from both wounds of each mice were pooled to represent one sample. Each such sample was analyzed using a separate GeneChip resulting in four separate data sets from each group.
Data analyses.
Data acquisition and image processing were performed using GCOS (Gene Chip Operating Software, Affymetrix). Raw data were collected and analyzed using Stratagene ArrayAssist Expression Software v. 5.1 (Stratagene). Additional processing of data was performed using dChip software (v. 1.3, Harvard University) (59). Data normalization and background corrections were performed using GC-RMA. Differentially expressed genes were identified using a two-class t-test where significance level was set at P < 0.05 with Benjamin-Hochberg false discovery rate correction (59, 86). Genes that were >2.0 fold up- or downregulated compared with 0 h intact skin samples were selected. A detailed analysis scheme has been illustrated in Fig. 3. All genes that were significantly changed at any single time point compared with 0 h samples were subjected to hierarchical clustering. Major clusters of genes that changed during the course of healing were identified. The genes from the cluster were subjected to further functional analysis using DAVID (Database for Annotation, Visualization and Integrated Discovery; NIAID, NIH). Major known pathways [Kyoto Encyclopedia of Genes and Genomes (KEGG)] identified in each of the cluster have been shown as supplemental figures.1 In addition, all genes that were significantly changed at any single time point were subjected to specific filters for wound cell-type or processes (see Figs. 7 and 8). Select differentially expressed candidate genes were verified with quantitative real-time PCR.
Reverse transcription and quantitative real-time PCR.
Tissue mRNA was quantified by real-time PCR assay using double-stranded DNA binding dye SYBR Green-I as described previously (62, 65). The primer set used for the individual genes are listed in Supplemental Table S1. GAPDH was used as a reference housekeeping gene.
Statistics.
Results are presented as means ± SD. In these cases, difference between means was tested using Students t-test. Microarray data processing is described above under GeneChip probe array analyses.
RESULTS
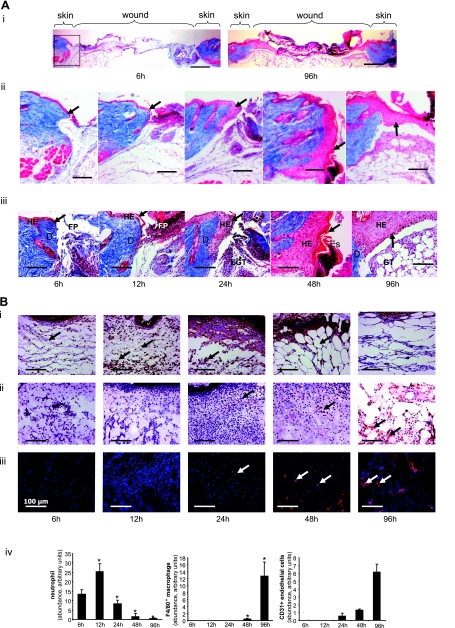
To avoid dilution of the effects at the wound-edge, only 1 mm of the margin was collected as wound-edge tissue (Fig. 1). To maximize the yield of wound-edge tissue we studied a relatively large excisional wound (Fig. 1) as described previously (60, 70, 71). In this model, complete closure is achieved in 2 wk (Fig. 1). In the interest of tight focus, the emphasis of this study was directed at the first 96 h of the repair process primarily covering the acute inflammatory phase (Fig. 1B shaded area, and Fig. 2). During this 96 h period, a modest but statistically significant decrease of wound size was observed. Before profiling the transcriptome of the wound-edge tissue at specific time points after wounding, we sought to histologically characterize the wound-edge tissue at those time points. The Mason trichrome stain (Fig. 2A) provided a reasonable appreciation of cellularity and advancement of the epidermal edge. At 6 h after wounding, the wound-edge tissue was characterized by the presence of abruptly discontinuous epidermal edge at the perimeter of the defect caused by wounding (Fig. 2A, 6 h). At this time point, infiltration of a few neutrophils and macrophages to the wound site was noted (Fig. 2B, 6 h). Overall cellularity in the granulation tissue was low, and CD31+ endothelial cells were not detected (Fig. 2B, 6 h). An additional 6 h later, i.e., 12 h after wounding, the epidermal cells did not advance substantially but were noted to orient themselves for downward advancement along the perimeters of the injured skin (Fig. 2A). Compared with the 6 h wound edge, a larger number of neutrophils were noted at the wound site while the count of macrophages remained unchanged (Fig. 2B, 12 h). Consistent with the sharply increased abundance of neutrophils at the wound site, the cellularity of the 12 h granulation tissue was higher but devoid of endothelial cells (Fig. 2B, 12 h). An additional 12 h later, i.e., 24 h after wounding, cellular proliferation as well as downward advancement of the epidermal cells was noted (Fig. 2A, 24 h). While the abundance of neutrophils subsided, there was a clear increase in the presence of macrophages (Fig. 2B, 24 h). For the first time, a few isolated endothelial cells were noted in the granulation tissue (Fig. 2B, 24 h). One day later, i.e., 48 h after wounding, changes in the epidermal layer of tissue were prominent. In addition to the downward migration and proliferation noted on the previous day, the epidermal layer of cells migrated laterally across the wound to cover the defect caused by wounding (Fig. 2A, 48 h). Remaining neutrophils started getting pushed upward toward the eschar tissue, while the number of macrophages that were noted in day 1 after wounding continued to be present (Fig. 2B, 48 h). On this day 2 after wounding, the presence of both isolated as well as clusters of endothelial cells in the granulation tissue were evident (Fig. 2B, 48 h). On day 4 after wounding, the hyperproliferative epithelium was clearly evident as arching over the defect (Fig. 2A, 96 h). The layer of tissue advanced from both ends of the defect as nascent tissue that is several layers of cell in thickness (not shown). Very few neutrophils were noted, while a significant number of macrophages continued to exist in the granulation tissue (Fig. 2B, 96 h). At this time point, the abundance of endothelial cells in the granulation tissue sharply increased, but very few of such cells seem to encircle a lumen and be a part of functional blood vessel.
Fig. 2.
Histological characterization of healing wound tissue used for gene expression profiling studies. Wound (Fig. 1) tissues from excisional wounds were collected at indicated time postinjury. Formalin-fixed paraffin sections or OCT-embedded frozen sections were stained using Masson Trichrome procedure (A). This procedure results in blue-black nuclei, blue collagen and cytoplasm. Epidermal cells are stained in red. i: Low magnification (×1.25) images showing cross-section of entire wounds at 6 and 96 h postwounding. Boxed area shows the wound edge tissue shown in ii and iii. Wound-edge tissue imaged with ×5 (ii) or ×20 (iii) objectives. Scale bar = 200 μm (for ii) and 50 μm (for iii). D, dermis; EGT, early granulation tissue; Es, Eschar tissue; FP, fibrin plug; HE, hyperproliferative epithelium. The newly forming epithelial tip is shown with an arrow. Alternatively, sections were immunostained as shown in B. B, i: Antineutrophil antibody (brown, shown with black arrow) that recognizes a polymorphic 40 kDa antigen expressed by polymorphonuclear cells. Counterstaining was performed using hematoxylin (blue); ii: F4/80 antibody (brown, shown with black arrow) that recognizes the murine F4/80 antigen, a 160 kDa cell surface glycoprotein expressed on a wide range of mature tissue macrophages. Counterstaining was performed using hematoxylin (blue); iii: Cd31 antibody (red fluorescence, shown with white arrow) that recognizes the mouse CD31, a 140 kDa cell surface glycoprotein that is expressed at high levels on endothelial cells. Counterstaining was performed using DAPI (blue, fluorescence). Scale bar, 50 μm. iv: Relative quantification (arbitrary units) of neutrophils, macrophages, and CD31+ endothelial cells from tissue sections obtained 6–96 h postwound was performed using a image processing tool kit. Data are means ± SD (n = 3). *P < 0.05 compared with 6 h time point postwounding.
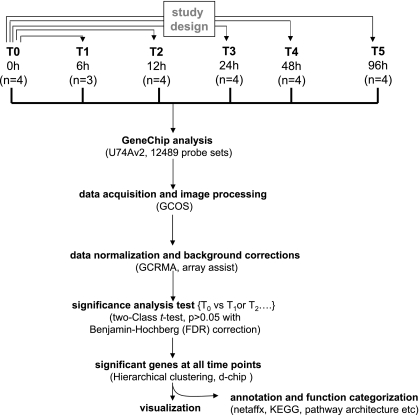
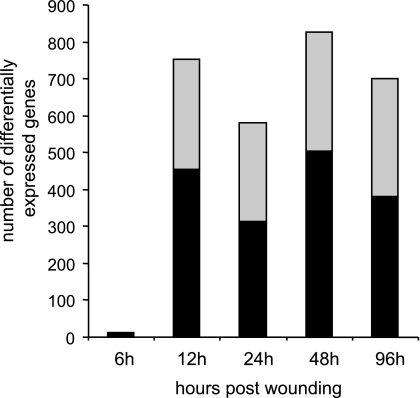
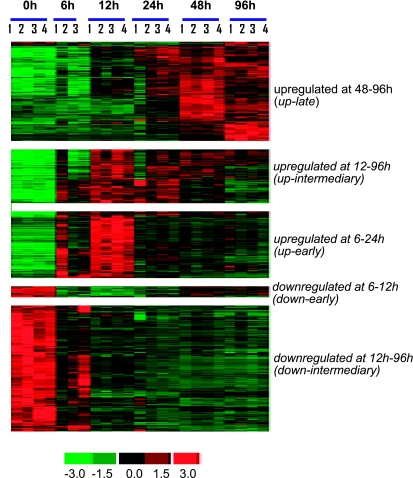
The microarray study and data analysis design is illustrated in Fig. 3 and was modeled on the basis of our previously published studies (57, 59, 65). A total of 12,489 probe sets were screened. Six hours after wounding, the expression of very few genes in the wound-edge tissue was significantly changed. Beyond that time point, a comparable number of genes were significantly changed in response to wounding. Taken together, the upregulated (filled) and downregulated (open) genes accounted for 4–5% of the total number of probe sets screened (Fig. 4). When analyzed temporally, the sets of genes that significantly changed in expression during the healing process could be segregated into the following five sets (Fig. 5): up-early (6–24h), up-intermediary (12–96 h), up-late (48–96 h), down-early (6–12 h), and down-intermediary (12–96 h). Functional categories of each of these sets of genes are itemized in Table 1. The specific genes and their signaling pathways are illustrated in Supplemental Figs S1–S5. Up-early: The genes upregulated in early phase as shown in Fig. 5 were subjected to functional analysis using DAVID (NIAID, NIH). KEGG-based analysis identified the cytokine-cytokine receptor interaction pathway as being primarily affected (Supplemental Fig. S1). Up-intermediary: Using the approach outlined above, we noted the leukocyte-endothelial interaction pathway to be primarily affected at this temporal phase (Supplemental Fig. S2). Up-late: The cell-cycle pathway related genes were most affected at this phase (Supplemental Fig. S3). Down-early: Genes related to purine metabolism were rapidly and transiently downregulated following wounding (Supplemental Fig. S4). Down-intermediary: The expression of genes functionally associated with oxidative phosphorylation was downregulated in the inflammatory phase of healing (Supplemental Fig. S5).
Fig. 3.
GeneChip data analysis scheme used to identify kinetics of differentially expressed genes in dermal wounds. Image acquisition and processing were performed using GCOS (GeneChip operating software, Affymetrix). GC-RMA was applied for data normalization and background correction. ArrayAssist v5.1 software was used to identify significant (P < 0.05; false discovery rate corrected) differentially expressed genes in wound tissue compared with skin (0 h) from the site where wound was created. Details of software and other resources for data analysis have been provided in methods.
Fig. 4.
Number of differentially expressed genes in course of healing. Total number of genes differentially expressed (significant, P < 0.05) at a specific time postwounding. Gray shaded area in each bar shows number of downregulated vs. the solid area that shows number of upregulated genes.
Fig. 5.
Cluster of genes showing specific pattern of expression in wound tissue during healing. All genes that were significantly changed at any single time point were subjected to hierarchical clustering. Five major clusters of genes that change during the course of healing were identified. Major functional categories in each of these clusters were identified and are presented in Table 1.
Table 1.
Functional cluster analysis of data presented in Figure 5. Major clusters at indicated time points are shown
| upregulated at 48–96 h (up-late, 299 probe sets) |
| • Keratinization |
| • Cell cycle* |
| • Histone core |
| • Cytoskeleton |
| • DNA replication |
| • ATP binding |
| • Proteolysis |
| • Hydrolase |
| • Ion transport |
| upregulated at 12–96 h (up-intermediary, 219 probe sets) |
| • Apoptosis |
| • Proteolysis |
| • Cytoskeleton |
| • Immune response* |
| • Kinase |
| • Rac GTPase |
| • Glucose metabolism |
| • ATPase activity |
| • Signal transduction/NADPH oxidase |
| • Transcription factor |
| upregulated at 6–24 h (up-early, 208 probe sets) |
| • Chemotaxis* |
| • Defense response* |
| • Cytokine-receptor interaction* |
| • Transcription factor |
| • Kinase |
| downregulated at 6–12 h (down-early, 32 probe sets) |
| • Cellular metabolism |
| downregulated at 12 h–96 h (down-intermediary, 416 probe sets) |
| • Oxidoreductase* |
| • Muscle protein |
| • Mitochondrion |
| • Calcium ion binding |
| • Mitochondrial membrane |
| • Glucose metabolism |
| • cytoskeleton |
| • Lim domain |
| • Ion transport |
| • Glycoprotein |
| • Ubiquitin cycle |
| • Protein modification |
| • Protein kinase |
See Supplemental Figures S1–S4 for pathways and individual genes involved.
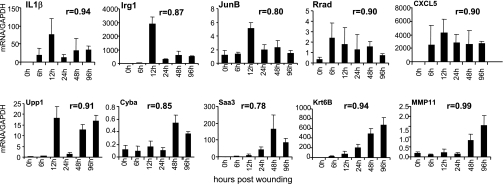
Arranged in the order of fold-change, the top 50 genes that were statistically significantly upregulated or downregulated in the wound-edge tissue during the five time points (6, 12, 24, 48, and 96 h) studied are listed in Supplemental Tables S2–S6. Select genes from these lists were picked for verification of results using quantitative real-time PCR (Fig. 6). Results of all time points obtained from microarray and real-time PCR approaches tightly correlated (r values indicated in each panel of Fig. 6), validating the data mining approach adopted in the current study. Findings related to commonly studied genes are consistent with the literature. The proinflammatory cytokine interleukin-1 beta (IL-1β) was acutely induced (Fig. 6) as previously reported (5). Immune response gene-1 (Irg1) is found in LPS-induced macrophages (3). Acute and transient induction of Irg1 was noted in response to open wound that lends itself to bacterial colonization. JunB is a component of the AP1 transcription factors that regulates epidermal regeneration, wound inflammation, as well as contraction (21, 81). Rapid and transient induction of JunB was observed in the wound-edge tissue. Ras-related associated with diabetes or Rrad is a prototypical member of a family of novel Ras-related GTPases. After muscle injury, Rrad is expressed within the myogenic progenitor cell population during the early phases of regeneration (29). Our observation represents first evidence demonstrating that high levels of the Rrad gene are found in the wound-edge tissue as early as 6 h after wounding. High levels of Rrad mRNA were noted during the entire inflammatory phase (Fig. 6). Similar temporal response was also noted for CXCL5. ELR(+) CXC chemokines such as CXCL5 attract and activate neutrophils, amplify the inflammatory cascade, and stimulate local production of cytokines that have negative inotropic and proapoptotic effects (9). Uridine phosphorylase (Upp) is a cytokine-inducible enzyme that converts the pyrimidine nucleoside uridine into uracil. Upon availability of ribose-1-phosphate, Upp can also catalyze the formation of nucleosides from uracil as well as from 5-fluorouracil, therefore involved in fluoropyrimidine metabolism (6). We observed sustained induction of Upp1 in the wound-edge tissue (Fig. 6). The functional significance of this response remains unclear at this time. Chronic granulomatous disease, a condition associated with impaired wound healing, results from defects in the phagocyte NADPH oxidase, central to which is the membrane-bound cytochrome b-245. The alpha polypeptide of cytochrome b-245 (Cyba) encodes the NADPH oxidase-light chain p22-phox (8), a NADPH oxidase component with significant vascular functions (27). Adipocytes represent a significant source of serum amyloid A3 (Saa3), which is now known to play a role in monocyte recruitment and inflammation (26). Saa3 is known to be an inducible acute-phase protein in the skin (80). We noted early induction of Saa3 in the wound-edge tissue. The induction intensified with time. Keratins are intermediate filament-forming proteins that provide mechanical support and fulfill a variety of additional functions in epithelial cells during wound repair (90). The type II keratin 6 is known to be expressed in the wound edge (43). We observed progressive induction of keratin 6B (Krt6B) in the wound-edge tissue as early as in the inflammatory phase (Fig. 6). Stromelysin-3 (ST3) is a member of the matrix metalloproteinase (MMP) family, MMP-11, which is expressed in the skin during wound healing (89). Typically associated with cutaneous scar formation, we observed that MMP11 was induced in the wound-edge tissue in the latter half of the inflammatory phase (Fig. 6).
Fig. 6.
Real-time PCR validation of GeneChip microarray expression analysis. Expression levels of selected genes identified (Tables S2–S6) using GeneChip analysis were independently determined using real-time PCR. The correlation coefficient (r) of data obtained from microarray analysis vs. real-time PCR is shown for each of the gene. The regression is derived from 4 pairs of data at each time point for a total number of 24. PCR data represent means ± SD, n = 5. The animals used for real-time PCR were a set that was independent of the animals used for microarray analysis. Irg-1, immunoresponsive gene 1; IL-1β, interleukin 1 beta; JunB, Jun-B oncogene; Rrad, Ras-related associated with diabetes; CXCL5, chemokine (C-X-C motif) ligand 5; Upp1, uridine phosphorylase 1; Cyba, cytochrome b-245, alpha polypeptide (p22 phox); Saa3, serum amyloid A 3; Krtb6, keratin 6B; MMP11, matrix metallopeptidase 11.
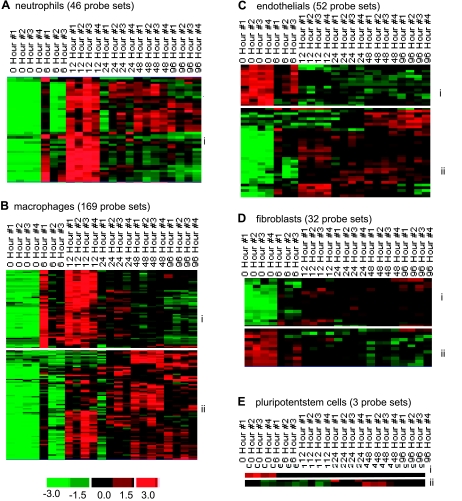
The design of the current study was aimed at examining cell-specific gene expression in the wound-edge tissue as a function of time. All genes that were significantly changed at any specific time point (as shown in Fig. 5) were subjected to further mining for cell-type specific genes. This mining was based on annotation of the genes in the Affymetrix database. The following five major wound-associated types of cells were targeted: neutrophils, macrophages, endothelial, fibroblasts, and pluripotent stem cells. Based on the temporal nature of the expression pattern in response to wounding, subsets of genes were identified within the cell-specific clusters (Fig. 7). The annotations for each cell-specific cluster are listed in Tables 2, A–E. The abundance of neutrophil-specific genes was low in the intact skin. Wounding increased the presence of such genes. The response was clearly noted at 6 h after wounding and peaked at 12 h followed by gradual lowering of the abundance of neutrophil-specific genes at the wound-edge (Fig. 7Ai). These results are consistent with the histological evidence demonstrating a comparable temporal response for neutrophil recruitment into the wound-edge tissue (Fig. 2B). The kinetics of enrichment of macrophage specific genes in the wound-edge tissue was somewhat delayed compared with the pattern for neutrophil-specific genes (Fig. 7, Ai vs. Bii). This result is consistent with the histological observations presented in Fig. 2.
Fig. 7.
Visualization of the expression pattern of candidate genes representing wound-specific cell types in a healing wound. All genes that were significantly changed at any single time point were subjected to specific filtration for wound cell type specific genes. The following 5 major wound cell types were targeted: neutrophils (A), macrophages (B), endothelial cells (C), fibroblasts (D), and pluripotent stem cells (E). Further subsets based on patterns of kinetics within each of the cell type were identified and indicated on margins of each cluster. Annotations of each cell type-specific subclusters (e.g., i and ii) are presented in Tables 2, A–E.
Table 2.
A. Annotation of neutrophil specific gene cluster shown in Figure 7A
| Probe ID | Gene Description | Symbol |
|---|---|---|
| 102914_s_at | B-cell leukemia/lymphoma 2 related protein A1a | Bcl2a1a |
| 102424_at | chemokine (C-C motif) ligand 3 | Ccl3 |
| 104388_at | chemokine (C-C motif) ligand 9 | Ccl9 |
| 102718_at | chemokine (C-C motif) receptor 5 | Ccr5 |
| 102719_f_at | chemokine (C-C motif) receptor 5 | Ccr5 |
| 161968_f_at | chemokine (C-C motif) receptor 5 | Ccr5 |
| 101160_at | chemokine (C-X-C motif) ligand 2 | Cxcl2 |
| 101436_at | chemokine (C-X-C motif) ligand 9 | Cxcl9 |
| 101728_at | complement component 5a receptor 1 | C5ar1 |
| 162181_f_at | Fc receptor, IgE, high affinity I, gamma polypeptide | Fcer1 g |
| 101793_at | Fc receptor, IgG, high affinity I | Fcgr1 |
| 102879_s_at | Fc receptor, IgG, high affinity I | Fcgr1 |
| 101800_at | formyl peptide receptor, related sequence 2 | Fpr-rs2 |
| 102884_at | inositol polyphosphate-5-phosphatase D | Inpp5d |
| 102353_at | integrin beta 2 | Itgb2 |
| 103486_at | interleukin 1 beta | Il1b |
| 102218_at | interleukin 6 | Il6 |
| 103266_at | leukotriene B4 receptor 1 | Ltb4r1 |
| 160564_at | lipocalin 2 | Lcn2 |
| 102957_at | lymphocyte cytosolic protein 2 | Lcp2 |
| 102430_at | myeloid differentiation primary response gene 88 | Myd88 |
| 102326_at | neutrophil cytosolic factor 2 | Ncf2 |
| 103662_at | neutrophil cytosolic factor 4 | Ncf4 |
| 101554_at | nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha | Nfkbia |
| 104149_at | nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha | Nfkbia |
| 100328_s_at | paired-Ig-like receptor A1 | Lilrb3 |
| 162475_f_at | peptidoglycan recognition protein 1 | Pglyrp1 |
| 102663_at | plasminogen activator, urokinase receptor | Plaur |
| 103579_at | RAS-related C3 botulinum substrate 2 | Rac2 |
| 103448_at | S100 calcium binding protein A8 (calgranulin A) | S100a8 |
| 103887_at | S100 calcium binding protein A9 (calgranulin B) | S100a9 |
| 161650_at | secretory leukocyte peptidase inhibitor | Slpi |
| 104692_at | selectin, platelet | Selp |
| 103488_at | selectin, platelet (p-selectin) ligand | Selplg |
| 102641_at | SFFV proviral integration 1 | Sfpi1 |
| 103839_at | sphingosine kinase 1 | Sphk1 |
| 100425_at | spleen tyrosine kinase | Syk |
| 162206_f_at | suppressor of cytokine signaling 3 | Socs3 |
| 92232_at | suppressor of cytokine signaling 3 | Socs3 |
| 104601_at | thrombomodulin | Thbd |
| 160469_at | thrombospondin 1 /// similar to thrombospondin 1 | Thbs1 |
| 101464_at | tissue inhibitor of metalloproteinase 1 | Timp1 |
| 92369_at | transforming growth factor alpha | Tgfa |
| 100397_at | TYRO protein tyrosine kinase binding protein | Tyrobp |
| B. Annotation of macrophage specific gene cluster shown in Figure 7B | ||
|---|---|---|
| Probe Set ID | Gene Title | Gene Symbol |
| Sub-cluster i (up-regulated, 6–12 h post wounding) | ||
| 101160_at | chemokine (C-X-C motif) ligand 2 | Cxcl2 |
| 101450_at | colony stimulating factor 1 (macrophage) | Csf1 |
| 101554_at | nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha | Nfkbia |
| 101996_at | protein tyrosine phosphatase, nonreceptor type 2 | Ptpn2 |
| 102218_at | interleukin 6 | Il6 |
| 102239_at | B-cell leukemia/lymphoma 3 | BclIII |
| 102362_i_at | Jun-B oncogene | Junb |
| 102363_r_at | Jun-B oncogene | Junb |
| 102424_at | chemokine (C-C motif) ligand 3 | Ccl3 |
| 102430_at | myeloid differentiation primary response gene 88 | Myd88 |
| 102736_at | chemokine (C-C motif) ligand 2 | Ccl2 |
| 103066_at | thymidylate kinase family LPS-inducible member | Tyki |
| 103486_at | interleukin 1 beta | Il1b |
| 103839_at | sphingosine kinase 1 | Sphk1 |
| 104149_at | nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha | Nfkbia |
| 104155_f_at | activating transcription factor 3 | Atf3 |
| 104156_r_at | activating transcription factor 3 | Atf3 |
| 104388_at | chemokine (C-C motif) ligand 9 | Ccl9 |
| 104406_at | prostaglandin E synthase | Ptges |
| 104533_at | proviral integration site 1 | Pim1 |
| 104538_at | prostaglandin I2 (prostacyclin) synthase | Ptgis |
| 104647_at | prostaglandin-endoperoxide synthase 2 | Ptgs2 |
| 104712_at | myelocytomatosis oncogene | Myc |
| 160894_at | CCAAT/enhancer binding protein (C/EBP), delta | Cebpd |
| 160933_at | interferon gamma induced GTPase | Igtp |
| 162206_f_at | suppressor of cytokine signaling 3 | Socs3 |
| 92232_at | suppressor of cytokine signaling 3 | Socs3 |
| 92562_at | nuclear factor, erythroid derived 2, like 2 | Nfe2l2 |
| 92694_at | chitinase 3-like 3 | Chi3l3 |
| 92731_at | pentraxin related gene | Ptx3 |
| 92793_at | tumor necrosis factor receptor superfamily, member 1a | Tnfrsf1a |
| 92830_s_at | zinc finger protein 36 | Zfp36 |
| 92849_at | chemokine (C-C motif) ligand 6 | Ccl6 |
| 92925_at | CCAAT/enhancer binding protein (C/EBP), beta | Cebpb |
| 93328_at | histidine decarboxylase | Hdc |
| 93858_at | chemokine (C-X-C motif) ligand 10 | Cxcl10 |
| 93871_at | interleukin 1 receptor antagonist | Il1rn |
| 93914_at | interleukin 1 receptor, type I | Il1r1 |
| 93956_at | interferon-induced protein with tetratricopeptide repeats 3 | Ifit3 |
| 94140_at | macrophage scavenger receptor 1 | Msr1 |
| 94142_at | colony stimulating factor 3 (granulocyte) | Csf3 |
| 94146_at | chemokine (C-C motif) ligand 4 | Ccl4 |
| 94224_s_at | interferon activated gene 203 | Ifi203 |
| 94246_at | E26 avian leukemia oncogene 2, 3′ domain | Ets2 |
| 94747_at | colony stimulating factor 2 receptor, beta 1, low-affinity (granulocyte-macrophage) | Csf2rb1 |
| 94748_g_at | colony stimulating factor 2 receptor, beta 1, low-affinity (granulocyte-macrophage) | Csf2rb1 |
| 94755_at | interleukin 1 alpha | Il1a |
| 94761_at | chemokine (C-C motif) ligand 7 | Ccl7 |
| 94929_at | protein tyrosine phosphatase, nonreceptor type 1 | Ptpn1 |
| 95348_at | chemokine (C-X-C motif) ligand 1 | Cxcl1 |
| 95349_g_at | chemokine (C-X-C motif) ligand 1 | Cxcl1 |
| 96551_at | C-type lectin domain family 4, member e | Clec4e |
| 96752_at | intercellular adhesion molecule | Icam1 |
| 97106_at | mitogen activated protein kinase kinase kinase 8 | Map3k8 |
| 98088_at | CD14 antigen | Cd14 |
| 98417_at | myxovirus (influenza virus) resistance 1 | Mx1 |
| 98500_at | interleukin 1 receptor-like 1 | Il1rl1 |
| 98501_at | interleukin 1 receptor-like 1 | Il1rl1 |
| 98579_at | early growth response 1 | Egr1 |
| 98773_s_at | immunoresponsive gene 1 | Irg1 |
| 98774_at | immunoresponsive gene 1 | Irg1 |
| 99099_at | signal transducer and activator of transcription 3 | Stat3 |
| 99100_at | signal transducer and activator of transcription 3 | Stat3 |
| 99413_at | chemokine (C-C motif) receptor 1 | Ccr1 |
| Sub-cluster ii (up-regulated, 24–96 h post wounding) | ||
| 102330_at | allograft inflammatory factor 1 | Aif1 |
| 93097_at | arginase 1, liver | Arg1 |
| 101521_at | baculoviral IAP repeat-containing 5 | Birc5 |
| 102914_s_at | B-cell leukemia/lymphoma 2 related protein A1a | Bcl2a1a/ |
| 93869_s_at | B-cell leukemia/lymphoma 2 related protein A1a | Bcl2a1a |
| 100988_at | BCL2-like 11 (apoptosis facilitator) | Bcl2l11 |
| 104023_at | Cd300D antigen | Cd300a |
| 101878_at | CD72 antigen | Cd72 |
| 93454_at | CD93 antigen | Cd93 |
| 160815_at | cDNA sequence BC032204 | BC032204 |
| 98406_at | chemokine (C-C motif) ligand 5 | Ccl5 |
| 93397_at | chemokine (C-C motif) receptor 2 | Ccr2 |
| 102718_at | chemokine (C-C motif) receptor 5 | Ccr5 |
| 102719_f_at | chemokine (C-C motif) receptor 5 | Ccr5 |
| 161968_f_at | chemokine (C-C motif) receptor 5 | Ccr5 |
| 103210_at | colony stimulating factor 2 receptor, beta 2 | Csf2rb2 |
| 101728_at | complement component 5a receptor 1 | C5ar1 |
| 100300_at | cytochrome b-245, beta polypeptide | Cybb |
| 103518_at | cytotoxic T lymphocyte-associated protein 2 beta | Ctla2b |
| 102896_at | docking protein 1 | Dok1 |
| 160901_at | FBJ osteosarcoma oncogene | Fos |
| 162181_f_at | Fc receptor, IgE, high affinity I, gamma polypeptide | Fcer1 g |
| 101793_at | Fc receptor, IgG, high affinity I | Fcgr1 |
| 102879_s_at | Fc receptor, IgG, high affinity I | Fcgr1 |
| 92188_s_at | feline sarcoma oncogene | Fes |
| 94697_at | Gardner-Rasheed feline sarcoma viral (Fgr) oncogene | Fgr |
| 97384_at | glia maturation factor, gamma | Gmfg |
| 99597_at | guanine nucleotide binding protein, alpha inhibiting 2 | Gnai2 |
| 99598_g_at | guanine nucleotide binding protein, alpha inhibiting 2 | Gnai2 |
| 93483_at | hemopoietic cell kinase | Hck |
| 100511_at | histocompatibility (minor) HA-1 | Hmha1 |
| 102884_at | inositol polyphosphate-5-phosphatase D | Inpp5d |
| 98828_at | integrin alpha M | Itgam |
| 104308_at | integrin alpha X | Itgax |
| 102353_at | integrin beta 2 | Itgb2 |
| 104750_at | interferon gamma inducible protein 47 | Ifi47 |
| 93425_at | interferon regulatory factor 5 | Irf5 |
| 104669_at | interferon regulatory factor 7 | Irf7 |
| 98002_at | interferon regulatory factor 8 | Irf8 |
| 103639_at | interferon-induced protein with tetratricopeptide repeats 2 | Ifit2 |
| 100428_at | laminin, gamma 2 | Lamc2 |
| 97507_at | lectin, galactoside-binding, soluble, 3 binding protein | Lgals3bp |
| 98003_at | leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 | Lilrb3 |
| 94278_at | lymphocyte cytosolic protein 1 | Lcp1 |
| 102957_at | lymphocyte cytosolic protein 2 | Lcp2 |
| 99071_at | macrophage expressed gene 1 /// similar to macrophage expressed gene 1 | Mpeg1 |
| 94792_at | macrophage scavenger receptor 1 | Msr1 |
| 97203_at | MARCKS-like 1 | Marcksl1 |
| 99957_at | matrix metallopeptidase 9 | Mmp9 |
| 97763_at | neutrophil cytosolic factor 1 | Ncf1 |
| 103614_at | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2, p49/p100 | Nfkb2 |
| 100328_s_at | paired-Ig-like receptor A1 | Lilrb3 |
| 103866_at | phosphatidylinositol 3-kinase catalytic delta polypeptide | Pik3cd |
| 103299_at | phospholipase D family, member 4 | Pld4 |
| 160801_at | PQ loop repeat containing 1 | Pqlc1 |
| 101881_g_at | procollagen, type XVIII, alpha 1 | Col18a1 |
| 99638_at | procollagen, type XVIII, alpha 1 | Col18a1 |
| 102851_s_at | protein tyrosine phosphatase, nonreceptor type 6 | Ptpn6 |
| 101048_at | protein tyrosine phosphatase, receptor type, C | Ptprc |
| 103451_at | PTK2 protein tyrosine kinase 2 beta | Ptk2b |
| 103579_at | RAS-related C3 botulinum substrate 2 | Rac2 |
| 95612_at | replication factor C (activator 1) 5 | Rfc5 |
| 102649_s_at | retinoic acid early transcript 1, alpha | Raet1a |
| 103448_at | S100 calcium binding protein A8 (calgranulin A) | S100a8 |
| 103887_at | S100 calcium binding protein A9 (calgranulin B) | S100a9 |
| 97519_at | secreted phosphoprotein 1 | Spp1 |
| 103488_at | selectin, platelet (p-selectin) ligand | Selplg |
| 160836_at | (semaphorin) 4D | Sema4d |
| 102712_at | serum amyloid A 3 | Saa3 |
| 102641_at | SFFV proviral integration 1 | Sfpi1 |
| 92975_at | SH3-domain binding protein 2 | Sh3bp2 |
| 96562_at | solute carrier family 11 | Slc11a1 |
| 103065_at | solute carrier family 20, member 1 | Slc20a1 |
| 100425_at | spleen tyrosine kinase | Syk |
| 102012_at | src family associated phosphoprotein 2 | Skap2 |
| 103205_at | T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 protein A3 | Tcirg1 |
| 92807_at | thioredoxin 1 | Txn1 |
| 98304_at | toll-like receptor 6 | Tlr6 |
| 101918_at | transforming growth factor, beta 1 | Tgfb1 |
| 94928_at | tumor necrosis factor receptor superfamily, member 1b | Tnfrsf1b |
| 100397_at | TYRO protein tyrosine kinase binding protein | Tyrobp |
| 99564_at | ubiquitin-like, containing PHD and RING finger domains, 1 | Uhrf1 |
| 99799_at | vav 1 oncogene | Vav1 |
| 103349_at | Yamaguchi sarcoma viral (v-yes-1) oncogene homolog | Lyn |
| C. Annotation of endothelial specific gene cluster shown in Figure 7C | ||
|---|---|---|
| Probe Set ID | Gene Description | Symbol |
| Sub-cluster i (down-regulated, 12–96 h post wounding) | ||
| 102703_s_at | aquaporin 4 | Aqp4 |
| 102704_at | aquaporin 4 | Aqp4 |
| 104590_at | myocyte enhancer factor 2C | Mef2c |
| 104591_g_at | myocyte enhancer factor 2C | Mef2c |
| 104592_i_at | myocyte enhancer factor 2C | Mef2c |
| 102720_at | endothelial-specific receptor tyrosine kinase | Tek |
| 102327_at | amine oxidase, copper containing 3 | Aoc3 |
| 100614_at | myoglobin | Mb |
| 100494_at | fibroblast growth factor 1 | Fgf1 |
| 103214_at | heat shock protein 2 | Hspb2 |
| 101178_at | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C | Sema3c |
| 103001_at | vascular endothelial growth factor B | Vegfb |
| 101990_at | lactate dehydrogenase B | Ldhb |
| 103215_g_at | heat shock protein 2 | Hspb2 |
| 100629_at | glutathione S-transferase, mu 5 | Gstm5 |
| 100605_at | tropomyosin 2, beta | Tpm2 |
| 100997_at | desmin | Des |
| 101521_at | baculoviral IAP repeat-containing 5 | Birc5 |
| 100153_at | neural cell adhesion molecule 1 | Ncam1 |
| 100928_at | fibulin 2 | Fbln2 |
| Sub-cluster ii (up-regulated, 12–96 h post wounding) | ||
| 101160_at | chemokine (C-X-C motif) ligand 2 | Cxcl2 |
| 100484_at | matrix metallopeptidase 13 | Mmp13 |
| 102663_at | plasminogen activator, urokinase receptor | Plaur |
| 101972_at | napsin A aspartic peptidase | Napsa |
| 103024_at | a disintegrin and metallopeptidase domain 8 | Adam8 |
| 101464_at | tissue inhibitor of metalloproteinase 1 | Timp1 |
| 102798_at | adrenomedullin | Adm |
| 102025_at | chemokine (C-X-C motif) ligand 13 | Cxcl13 |
| 102957_at | lymphocyte cytosolic protein 2 | Lcp2 |
| 102884_at | inositol polyphosphate-5-phosphatase D | Inpp5d |
| 102914_s_at | B-cell leukemia/lymphoma 2 related protein A1a | Bcl2a1a |
| 101728_at | complement component 5a receptor 1 | C5ar1 |
| 104469_at | podoplanin | Pdpn |
| 104601_at | thrombomodulin | Thbd |
| 103210_at | colony stimulating factor 2 receptor, beta 2, low-affinity (granulocyte-macrophage) | Csf2rb2 |
| 100425_at | spleen tyrosine kinase | Syk |
| 103707_at | complement component 3a receptor 1 | C3ar1 |
| 160101_at | heme oxygenase (decycling) 1 | Hmox1 |
| 104701_at | basic helix-loop-helix domain containing, class B2 | Bhlhb2 |
| 103818_at | solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 | Slc7a7 |
| 100428_at | laminin, gamma 2 | Lamc2 |
| 103839_at | sphingosine kinase 1 | Sphk1 |
| 103451_at | PTK2 protein tyrosine kinase 2 beta | Ptk2b |
| 100308_at | procollagen, type VIII, alpha 1 | Col8a1 |
| 103866_at | phosphatidylinositol 3-kinase catalytic delta polypeptide | Pik3cd |
| 100134_at | endoglin | Eng |
| 100970_at | thymoma viral proto-oncogene 1 | Akt1 |
| 100016_at | matrix metallopeptidase 11 | Mmp11 |
| 100915_at | myosin, heavy polypeptide 9, nonmuscle | Myh9 |
| 103462_at | dedicator of cyto-kinesis 2 | Dock2 |
| 101881_g_at | procollagen, type XVIII, alpha 1 | Col18a1 |
| D. Annotation of fibroblast specific gene cluster shown in Figure 7D | ||
|---|---|---|
| Probe Set ID | Gene Description | Symbol |
| Sub-cluster i (up-regulated, 12–96 h post wounding) | ||
| 100484_at | matrix metallopeptidase 13 | Mmp13 |
| 102712_at | serum amyloid A 3 | Saa3 |
| 101631_at | SRY-box containing gene 11 | Sox11 |
| 101993_at | tenascin C | Tnc |
| 101464_at | tissue inhibitor of metalloproteinase 1 | Timp1 |
| 102218_at | interleukin 6 | Il6 |
| 100016_at | matrix metallopeptidase 11 | Mmp11 |
| 101918_at | transforming growth factor, beta 1 | Tgfb1 |
| 102021_at | interleukin 4 receptor, alpha | Il4ra |
| 100884_at | aldo-keto reductase family 1, member B8 | Akr1b8 |
| 100009_r_at | SRY-box containing gene 2 | Sox2 |
| 100277_at | inhibin beta-A | Inhba |
| 102204_at | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein B (avian) | Mafb |
| 102195_at | mitogen-activated protein kinase kinase kinase kinase 4 | Map4k4 |
| 101881_g_at | procollagen, type XVIII, alpha 1 | Col18a1 |
| 100581_at | cystatin B | Cstb |
| 100507_at | nephroblastoma overexpressed gene | Nov |
| 100019_at | versican | Vcan |
| 100420_at | filaggrin | Flg |
| Sub-cluster ii (down-regulated, 12–96 h post wounding) | ||
| 102774_at | epidermal growth factor | Egf |
| 101028_i_at | actin, alpha, cardiac | Actc1 |
| 100003_at | ryanodine receptor 1, skeletal muscle | Ryr1 |
| 100605_at | tropomyosin 2, beta | Tpm2 |
| 102256_at | T-box 15 | Tbx15 |
| 100494_at | fibroblast growth factor 1 | Fgf1 |
| 100879_at | actinin alpha 3 | Actn3 |
| 100068_at | aldehyde dehydrogenase family 1, subfamily A1 | Aldh1a1 |
| 100997_at | desmin | Des |
| 101892_f_at | thymoma viral proto-oncogene 1 interacting protein | Aktip |
| 101029_f_at | actin, alpha, cardiac | Actc1 |
| 100566_at | insulin-like growth factor binding protein 5 | Igfbp5 |
| E. Annotation of pluripotent stem cell specific gene cluster shown in Figure 7E | ||
|---|---|---|
| Probe Set ID | Gene Description | Symbol |
| Sub-cluster i (down-regulated, 6–96 h post wounding) | ||
| 92722_f_at | sine oculis-related homeobox 1 homolog (Drosophila) | Six1 |
| Sub-cluster ii (up-regulated, 6–96 h post wounding) | ||
| 160069_at | geminin | Gmnn |
| 100009_r_at | SRY-box containing gene 2 | Sox2 |
Footnote to Tables 2A–E: Probe ID, Affymetrix probe identifications.
Wounding is associated with the disruption of vasculature (22). Consistently, we have observed that endothelial cell specific genes which are abundant in the intact skin, sharply disappear in response to wounding (Fig. 7Ci). Although histological visualization does not support the reappearance of endothelial cells in the early phase (6–12 h) after wounding, the more sensitive microarray approach to detect endothelial cell-specific genes demonstrates the appearance of few endothelial cell related genes in the wound-edge tissue during such early phase. Consistent with the histological observations presented in Fig. 2D, the abundance of endothelial cell-specific genes in the wound-edge tissue sharply increased from 24–96h after wounding (Fig. 7Cii).
Dermal fibroblasts represent an integral component of the skin tissue. The abundance of fibroblast specific genes in the intact skin tissue (Fig. 7Dii) was therefore expected. In response to wounding, the abundance of this set of genes in the wound edge tissue was sharply lowered and stayed that way over the 96 h study (Table 2D). Wounding, however, increased the abundance of a distinct set of fibroblast-related genes whose basal abundance in the intact skin tissue is low (Fig. 7Di). The abundance of such genes in the wound-edge tissue increased progressively over time. It remains unknown to what extent induction in the resident cells and infiltration of blood-borne cells following wounding contributes to such increased abundance of this subset of genes (Table 2D).
Based on annotations in the Affymetrix database and on our data mining approach, only three pluripotent stem cell specific genes were noted to change. Six1, implicated in maintenance of the differentiated state of tissues, was noted to be relatively abundant in the intact skin. The abundance of this gene in the wound-edge tissue decreased in response to wounding (Fig. 7Ei, Table 2E). Two other genes in this category, geminin and Sox2, were noted to be present in low abundance in the intact skin but the abundance in the wound-edge tissue increased 48h (up-late, as defined in Fig. 5) after wounding.
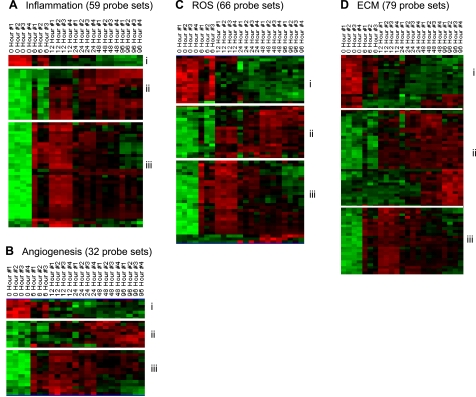
To enable appreciation of our results from the temporal analysis of transcriptome profiling in the context of functional processes that are implicated in wound repair, candidate genes identified were clustered on the basis of their annotation linking them to inflammation, angiogenesis, reactive oxygen species (ROS), and extracellular matrix (ECM) as depicted in Fig. 8. In the category of inflammation (Fig. 8A), two subsets of upregulated genes were identified. A larger subset of genes that was up-early in response to wounding (Fig. 8Aii, Table 3A). The second smaller subset of genes was up-late (Fig. 8Ai, Table 3A). The up-early subset of genes included neutrophil-related genes that are consistent with the infiltration of neutrophils to the wound-edge tissue as depicted in Fig. 2. The up-late subset of genes included macrophage-related genes, and the changes may be explained by the temporal kinetics of macrophage recruitment to the wound-edge tissue as noted in Fig. 2. In the functional category of angiogenesis, three subsets of genes were identified that responded to wounding (Fig. 8B). The smallest subcluster of genes in this category represented those that were found at an elevated level in the intact skin but were rapidly downregulated in response to wounding (Fig. 8Bi, Table 3B). The other two subcluster of genes in this functional category represented genes that were up-early (Fig. 8Biii) and up-late (Fig. 8Bii) in response to wounding (Table 3B).
Fig. 8.
Visualization of the expression pattern of candidate genes representing repair-specific functional categories in a healing wound. All genes that were significantly changed at any single time point were subjected to specific filtration for process/factors-specific genes. The following processes/factors that play a major role in wound healing were targeted: inflammation (A), angiogenesis (B), reactive oxygen species (ROS, C), and extracellular matrix (ECM, D). Further subsets based on patterns of kinetics within each of the processes were identified and indicated on margins of each cluster. Annotations of each process-specific subclusters are presented in Tables 3, A–D.
Table 3.
A. Inflammation specific gene cluster shown in Figure 8A
| Probe Set ID | Gene Description | Symbol |
|---|---|---|
| Sub-cluster I (down-regulated, 12–96 h post wounding) | ||
| 160519_at | tissue inhibitor of metalloproteinase 3 | Timp3 |
| 93451_at | LIM domain only 7 | Lmo7 |
| 102774_at | epidermal growth factor | Egf |
| 92323_at | mitogen-activated protein kinase 12 | Mapk12 |
| Sub-cluster ii (up-regulated, 12–96 h post wounding) | ||
| 101024_i_at | small proline-rich protein 2A | Sprr2a |
| 101025_f_at | small proline-rich protein 2A | Sprr2a |
| 102914_s_at | B-cell leukemia/lymphoma 2 related protein A1a | Bcl2a1a |
| 103070_at | signal-regulatory protein alpha | Sirpa |
| 93397_at | chemokine (C-C motif) receptor 2 | Ccr2 |
| 93869_s_at | B-cell leukemia/lymphoma 2 related protein A1a | Bcl2a1a |
| 98002_at | interferon regulatory factor 8 | Irf8 |
| 99071_at | macrophage expressed gene 1 | Mpeg1 |
| 99701_f_at | small proline-rich protein 2B | Sprr2b |
| 99957_at | matrix metallopeptidase 9 | Mmp9 |
| Sub-cluster iii (up-regulated, 6–12 h post wounding) | ||
| 99979_at | cytochrome P450, family 1b1 | Cyp1b1 |
| 99413_at | chemokine (C-C motif) receptor 1 | Ccr1 |
| 99387_at | formyl peptide receptor 1 | Fpr1 |
| 98988_at | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta | Nfkbiz |
| 98501_at | interleukin 1 receptor-like 1 | Il1rl1 |
| 98304_at | toll-like receptor 6 | Tlr6 |
| 97519_at | secreted phosphoprotein 1 | Spp1 |
| 96596_at | N-myc downstream regulated gene 1 | Ndrg1 |
| 96562_at | solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 | Slc11a1 |
| 95349_g_at | chemokine (C-X-C motif) ligand 1 | Cxcl1 |
| 95348_at | chemokine (C-X-C motif) ligand 1 | Cxcl1 |
| 94769_at | matrix metallopeptidase 8 | Mmp8 |
| 94761_at | chemokine (C-C motif) ligand 7 | Ccl7 |
| 94146_at | chemokine (C-C motif) ligand 4 | Ccl4 |
| 94140_at | macrophage scavenger receptor 1 | Msr1 |
| 93871_at | interleukin 1 receptor antagonist | Il1rn |
| 93573_at | metallothionein 1 | Mt1 |
| 93425_at | interferon regulatory factor 5 | Irf5 |
| 93328_at | histidine decarboxylase | Hdc |
| 92731_at | pentraxin related gene | Ptx3 |
| 92694_at | chitinase 3-like 3 | Chi3l3 |
| 92232_at | suppressor of cytokine signaling 3 | Socs3 |
| 92217_s_at | glycoprotein 49 A | Gp49a |
| 161968_f_at | chemokine (C-C motif) receptor 5 | Ccr5 |
| 161650_at | secretory leukocyte peptidase inhibitor | Slpi |
| 160469_at | thrombospondin 1 | Thbs1 |
| 104647_at | prostaglandin-endoperoxide synthase 2 | Ptgs2 |
| 104601_at | thrombomodulin | Thbd |
| 103887_at | S100 calcium binding protein A9 (calgranulin B) | S100a9 |
| 103486_at | interleukin 1 beta | Il1b |
| 103448_at | S100 calcium binding protein A8 (calgranulin A) | S100a8 |
| 102957_at | lymphocyte cytosolic protein 2 | Lcp2 |
| 102779_at | growth arrest and DNA-damage-inducible 45 beta | Gadd45b |
| 102736_at | chemokine (C-C motif) ligand 2 | Ccl2 |
| 102719_f_at | chemokine (C-C motif) receptor 5 | Ccr5 |
| 102718_at | chemokine (C-C motif) receptor 5 | Ccr5 |
| 102712_at | serum amyloid A 3 | Saa3 |
| 102663_at | plasminogen activator, urokinase receptor | Plaur |
| 102424_at | chemokine (C-C motif) ligand 3 | Ccl3 |
| 102218_at | interleukin 6 | Il6 |
| 102021_at | interleukin 4 receptor, alpha | Il4ra |
| 101561_at | metallothionein 2 | Mt2 |
| 101554_at | nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha | Nfkbia |
| 101160_at | chemokine (C-X-C motif) ligand 2 | Cxcl2 |
| B. Angiogenesis specific gene cluster shown in Figure 8B | ||
|---|---|---|
| Probe Set ID | Gene Description | Symbol |
| Sub-cluster I (down-regulated, 6–96 h post wounding) | ||
| 99104_at | adiponectin | Adipoq |
| 98802_at | epiregulin | Ereg |
| 95531_at | angiomotin | Amot |
| 160484_at | reticulon 4 | Rtn4 |
| 103001_at | vascular endothelial growth factor B | Vegfb |
| 102720_at | endothelial-specific receptor tyrosine kinase | Tek |
| 100494_at | fibroblast growth factor 1 | fgf1 |
| Sub-cluster ii (up-regulated, 48–96 h post wounding) | ||
| 99957_at | matrix metallopeptidase 9 | Mmp9 |
| 99638_at | procollagen, type XVIII, alpha 1 | Col18a1 |
| 92676_at | runt related transcription factor 2 | Runx2 |
| 92558_at | vascular cell adhesion molecule 1 | Vcam1 |
| 92365_at | c-fos induced growth factor | Figf |
| 92210_at | angiopoietin 2 | Angpt2 |
| 104154_at | transformation related protein 53 | Trp53 |
| 101881_g_at | procollagen, type XVIII, alpha 1 | Col18a1 |
| Sub-cluster iii (up-regulated, 6–24 h post wounding) | ||
| 99100_at | signal transducer and activator of transcription 3 | Stat3 |
| 99099_at | signal transducer and activator of transcription 3 | Stat3 |
| 98629_f_at | hypoxia inducible factor 1, alpha subunit | Hif1a |
| 98628_f_at | hypoxia inducible factor 1, alpha subunit | Hif1a |
| 97519_at | secreted phosphoprotein 1 | Spp1 |
| 96119_s_at | angiopoietin-like 4 | Angptl4 |
| 94147_at | serine (or cysteine) peptidase inhibitor, clade E, member 1 | Serpine1 |
| 92777_at | cysteine rich protein 61 | Cyr61 |
| 92369_at | transforming growth factor alpha | Tgfa |
| 160469_at | thrombospondin 1 /// similar to thrombospondin 1 | Thbs1 |
| 160101_at | heme oxygenase (decycling) 1 | Hmox1 |
| 104712_at | myelocytomatosis oncogene | Myc |
| 104647_at | prostaglandin-endoperoxide synthase 2 | Ptgs2 |
| 104469_at | podoplanin | Pdpn |
| 101464_at | tissue inhibitor of metalloproteinase 1 | Timp1 |
| 100621_at | ribonuclease/angiogenin inhibitor 1 | Rnh1 |
| 100134_at | endoglin | Eng |
| C. ROS-specific gene cluster shown in Figure 8C | ||
|---|---|---|
| Probe Set ID | Gene Description | Symbol |
| Sub-cluster I (down-regulated, 12–96 h post wounding) | ||
| 100614_at | myoglobin | Mb |
| 102382_at | aryl hydrocarbon receptor nuclear translocator-like | Arntl |
| 102720_at | endothelial-specific receptor tyrosine kinase | Tek |
| 160090_f_at | aldolase 1, A isoform | Aldoa |
| 160479_at | catalase | Cat |
| 160481_at | phosphoenolpyruvate carboxykinase 1, cytosolic | Pck1 |
| 160547_s_at | thioredoxin interacting protein | Txnip |
| 160913_at | nebulin-related anchoring protein | Nrap |
| 161889_f_at | Aldolase 1, A isoform | Aldoa |
| 93084_at | solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 | Slc25a4 |
| 93332_at | CD36 antigen | Cd36 |
| 93543_f_at | glutathione S-transferase, mu 1 | Gstm1 |
| 93836_at | BCL2/adenovirus E1B interacting protein 1, NIP3 | Bnip3 |
| 93996_at | cytochrome P450, family 2, subfamily e, polypeptide 1 | Cyp2e1 |
| 95053_s_at | succinate dehydrogenase complex, subunit B, iron sulfur (Ip) | Sdhb |
| 96058_s_at | aldehyde dehydrogenase 2, mitochondrial | Aldh2 |
| 98007_at | ribosomal protein S6 kinase, polypeptide 2 | Rps6ka2 |
| 99104_at | adiponectin, C1Q and collagen domain containing | Adipoq |
| Sub-cluster ii (up-regulated, 48–96 h post wounding) | ||
| 100988_at | BCL2-like 11 (apoptosis facilitator) | Bcl2l11 |
| 101160_at | chemokine (C-X-C motif) ligand 2 | Cxcl2 |
| 101554_at | nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha | Nfkbia |
| 101561_at | metallothionein 2 | Mt2 |
| 102292_at | growth arrest and DNA-damage-inducible 45 alpha | Gadd45a |
| 102362_i_at | Jun-B oncogene | Junb |
| 102363_r_at | Jun-B oncogene | Junb |
| 102430_at | myeloid differentiation primary response gene 88 | Myd88 |
| 102798_at | adrenomedullin | Adm |
| 103887_at | S100 calcium binding protein A9 (calgranulin B) | S100a9 |
| 104149_at | nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha | Nfkbia |
| 104692_at | selectin, platelet | Selp |
| 104701_at | basic helix-loop-helix domain containing, class B2 | Bhlhb2 |
| 104712_at | myelocytomatosis oncogene | Myc |
| 160101_at | heme oxygenase (decycling) 1 | Hmox1 |
| 92562_at | nuclear factor, erythroid derived 2, like 2 | Nfe2l2 |
| 92694_at | chitinase 3-like 3 | Chi3l3 |
| 93573_at | metallothionein 1 | Mt1 |
| 94146_at | chemokine (C-C motif) ligand 4 | Ccl4 |
| 94769_at | matrix metallopeptidase 8 | Mmp8 |
| 94939_at | CD53 antigen | Cd53 |
| 95348_at | chemokine (C-X-C motif) ligand 1 | Cxcl1 |
| 95349_g_at | chemokine (C-X-C motif) ligand 1 | Cxcl1 |
| 96042_at | superoxide dismutase 2, mitochondrial | Sod2 |
| 98010_at | nuclear factor, erythroid derived 2 | Nfe2 |
| 98088_at | CD14 antigen | Cd14 |
| 99985_at | thioredoxin reductase 1 | Txnrd1 |
| Sub-cluster iii (up-regulated, 6–12 h post wounding) | ||
| 100059_at | cytochrome b-245, alpha polypeptide | Cyba (p22phox) |
| 100300_at | cytochrome b-245, beta polypeptide | Cybb (gp91phox) |
| 102353_at | integrin beta 2 | Itgb2 |
| 102389_s_at | growth associated protein 43 | Gap43 |
| 102641_at | SFFV proviral integration 1 | Sfpi1 |
| 103662_at | neutrophil cytosolic factor 4 | Ncf4 (p40 phox) |
| 103707_at | complement component 3a receptor 1 | C3ar1 |
| 92807_at | thioredoxin 1 | Txn1 |
| 93483_at | hemopoietic cell kinase | Hck |
| 93536_at | BclII-associated X protein | Bax |
| 94930_at | thrombospondin 2 | Thbs2 |
| 96085_at | glutathione S-transferase, alpha 4 | Gsta4 |
| 96562_at | solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 | Slc11a1 |
| 97013_f_at | cytochrome b-245, alpha polypeptide | Cyba (p22phox) |
| 97763_at | neutrophil cytosolic factor 1 | Ncf1 (p47phox) |
| 98002_at | interferon regulatory factor 8 | Irf8 |
| 98433_at | BH3 interacting domain death agonist | Bid |
| 98436_s_at | caspase 3 | Casp3 |
| 98828_at | integrin alpha M | Itgam |
| 99810_at | glutathione peroxidase 2 | Gpx2 |
| 99915_at | amphiregulin | Areg |
| D. Extracellular matrix (ECM) specific gene cluster shown in Figure 8D | ||
|---|---|---|
| Probe Set ID | Gene Description | Symbol |
| Sub-cluster i (down-regulated, 12–96 h post wounding) | ||
| 100494_at | fibroblast growth factor 1 | Fgf1 |
| 100597_at | glycogenin | Gyg |
| 100997_at | desmin | Des |
| 101394_at | sarcoglycan, gamma (dystrophin-associated glycoprotein) | Sgcg |
| 102720_at | endothelial-specific receptor tyrosine kinase | Tek |
| 103243_at | epithelial membrane protein 2 | Emp2 |
| 103395_at | sarcoglycan, alpha (dystrophin-associated glycoprotein) | Sgca |
| 103721_at | Nephronectin | Npnt |
| 160319_at | SPARC-like 1 (mast9, hevin) | Sparcl1 |
| 160519_at | tissue inhibitor of metalloproteinase 3 | Timp3 |
| 160631_s_at | sarcoglycan, alpha (dystrophin-associated glycoprotein) | Sgca |
| 161907_s_at | tenascin XB | Tnxb |
| 161984_f_at | procollagen, type III, alpha 1 | Col3a1 |
| 162262_f_at | glycogenin | Gyg |
| 92407_at | myomesin 1 | Myom1 |
| 92441_at | fibroblast activation protein | Fap |
| 93300_at | transforming growth factor, beta 2 | Tgfb2 |
| 93563_s_at | nidogen 2 | Nid2 |
| 94122_at | myocilin | Myoc |
| Sub-cluster ii (up-regulated, 48–96 h post wounding) | ||
| 100019_at | versican | Vcan |
| 100428_at | laminin, gamma 2 | Lamc2 |
| 100484_at | matrix metallopeptidase 13 | Mmp13 |
| 101436_at | chemokine (C-X-C motif) ligand 9 | Cxcl9 |
| 101464_at | tissue inhibitor of metalloproteinase 1 | Timp1 |
| 101918_at | transforming growth factor, beta 1 | Tgfb1 |
| 101993_at | tenascin C | Tnc |
| 102362_i_at | Jun-B oncogene | Junb |
| 102363_r_at | Jun-B oncogene | Junb |
| 102641_at | SFFV proviral integration 1 | Sfpi1 |
| 102663_at | plasminogen activator, urokinase receptor | Plaur |
| 102794_at | chemokine (C-X-C motif) receptor 4 | Cxcr4 |
| 103005_s_at | CD44 antigen | Cd44 |
| 103465_f_at | serum amyloid A 2 | Saa2 |
| 104647_at | prostaglandin-endoperoxide synthase 2 | Ptgs2 |
| 104712_at | myelocytomatosis oncogene | Myc |
| 160094_at | actin related protein 2/3 complex, subunit 4 | Arpc4 |
| 160511_at | chemokine (C-X-C motif) ligand 12 | Cxcl12 |
| 160774_at | ectonucleoside triphosphate diphosphohydrolase 1 | Entpd1 |
| 160901_at | FBJ osteosarcoma oncogene | Fos |
| 162362_f_at | tenascin C | Tnc |
| 92365_at | c-fos induced growth factor | Figf |
| 92369_at | transforming growth factor alpha | Tgfa |
| 92562_at | nuclear factor, erythroid derived 2, like 2 | Nfe2l2 |
| 92676_at | runt related transcription factor 2 | Runx2 |
| 92731_at | pentraxin related gene | Ptx3 |
| 92849_at | chemokine (C-C motif) ligand 6 | Ccl6 |
| 93793_at | LIM and SH3 protein 1 | Lasp1 |
| 94085_at | serglycin | Srgn |
| 94140_at | macrophage scavenger receptor 1 | Msr1 |
| 94147_at | serine (or cysteine) peptidase inhibitor, clade E, member 1 | Serpine1 |
| 94769_at | matrix metallopeptidase 8 | Mmp8 |
| 94792_at | macrophage scavenger receptor 1 | Msr1 |
| 95434_at | actin related protein 2/3 complex, subunit 3 | Arpc3 |
| 95803_at | signal-regulatory protein alpha | Sirpa |
| 95804_g_at | signal-regulatory protein alpha | Sirpa |
| 96603_at | quiescin Q6 | Qscn6 |
| 97519_at | secreted phosphoprotein 1 | Spp1 |
| 97790_s_at | laminin, alpha 3 | Lama3 |
| 98067_at | cyclin-dependent kinase inhibitor 1A (P21) | Cdkn1a |
| 98433_at | BH3 interacting domain death agonist | Bid |
| 98579_at | early growth response 1 | Egr1 |
| 99099_at | signal transducer and activator of transcription 3 | Stat3 |
| 99100_at | signal transducer and activator of transcription 3 | Stat3 |
| 99957_at | matrix metallopeptidase 9 | Mmp9 |
| Sub-cluster iii (up-regulated, 6–24 h post wounding) | ||
| 100016_at | matrix metallopeptidase 11 | Mmp11 |
| 100021_at | cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle) | Chrna1 |
| 100308_at | procollagen, type VIII, alpha 1 | Col8a1 |
| 100928_at | fibulin 2 | Fbln2 |
| 100986_at | four and a half LIM domains 2 | Fhl2 |
| 101881_g_at | procollagen, type XVIII, alpha 1 | Col18a1 |
| 103554_at | a disintegrin and metallopeptidase domain 19 (meltrin beta) | Adam19 |
| 161215_at | cholinergic receptor, nicotinic, gamma polypeptide | Chrng |
| 92701_at | bone morphogenetic protein 1 | Bmp1 |
| 94732_at | repetin | Rptn |
| 94930_at | thrombospondin 2 | Thbs2 |
| 98623_g_at | insulin-like growth factor 2 | Igf2 |
| 99327_at | kallikrein related-peptidase 8 | Klk8 |
| 99638_at | procollagen, type XVIII, alpha 1 | Col18a1 |
Footnote to Tables 3A–D: Probe ID, Affymetrix probe identifications.
At the wound site, ROS are generated at sustained low levels by resident nonphagocytic cells. In the inflammatory phase, copious amounts of ROS are generated by phagocytic cells (60, 69). While excessive ROS may complicate healing, endogenously generated ROS are essential to support the healing process (60). According to annotations in the Affymetrix database ROS-generating, ROS-sensitive, as well as ROS-metabolizing genes are collectively clustered as ROS-related transcripts. Three subsets of ROS-related genes changed significantly in response to wounding. The first subset (Fig. 8Ci, Table 3C), genes abundant in the intact skin were rapidly downregulated in response to wounding. The other two subsets included the up-early (Fig. 8Ciii) and up-late (Fig. 8Cii) genes, the abundance of which in the wound-edge tissue increased in response to wounding (Table 3C). In the functional category of ECM, the subsets of genes represented the same pattern as for the ROS-related and angiogenesis categories. The tables present a complete listing of each of the genes in the subsets described above. Functional aspects of select candidate genes in each category and subset are discussed below.
DISCUSSION
For secondary-intention excisional wounds, advancement of wound edge represents a major mechanism by which a wound defect is closed (91). Changes in the wound-edge tissue are therefore of outstanding interest (20, 66). This work represents the maiden effort temporally characterizing the acute inflammatory phase of the murine excisional cutaneous wound using high-density microarray. In the acute inflammatory phase, chemotaxis is the primary mechanism by which cell movements are directed in response to wounding. Chemotaxis involves a complex cascade of events including formation of signaling complexes via receptor-cytokine interactions (31). In this study, chemotaxis and cytokine-receptor interaction pathways represented the early upregulated genes in response to wounding. Interactions of leukocytes with endothelial cells represent early-intermediate events in acute inflammation wound repair (44). Consistently we noted that components of this pathway were upregulated during the intermediary phase (12–96 h) after wounding. Genes upregulated in response to wounding in the late phase (48–96 h) represented the cell cycle pathway, which is known to be implicated in additional cell supply during the process of tissue repair (14, 68). Cutaneous wound repair is associated with changes in purine metabolism (58). We noted that wounding induced early (6–12 h) downregulation of enzymes of the purine metabolism pathway. Specifically, nucleoside diphosphate kinase and ADP-ribose diphosphatase were downregulated in the wound-edge tissue in response to wounding. Nucleoside diphosphate kinase phosphorylates a nucleoside diphosphate to the corresponding triphosphate at the expense of ATP. Downregulation of this enzyme may be aimed at conserving ATP in the wound-edge tissue for fueling tissue repair. ADP-ribose facilitates neutrophil chemotaxis (54). Downregulation of ADP-ribose diphosphatase would attenuate the hydrolysis of ADP-ribose to AMP, making ADP-ribose available to support wound angiogenesis (12). During the interval 12–96 h after wounding, genes encoding proteins of the oxidative phosphorylation pathway were downregulated. Candidate genes in this category included NADH dehydrogenases, component of the succinate dehydrogenase complex, cytochrome c-1, cytochrome c-oxidase, and ATP synthase. This response may explain wound tissue ATP deficiency that is recognized as a common limiting factor in cutaneous wound healing (7, 11).
Changes in the expression of functionally categorized genes on one hand lent support to currently proposed hypotheses while on the other hand generated novel insight into the acute inflammatory phase of tissue repair. Inflammation-related early upregulated genes included known players such as several chemokine receptors and ligands. Recruitment of leukocyte subtypes to the wound site is tightly regulated by chemokines. Moreover, the presence of chemokine receptors on resident cells (e.g., keratinocytes, endothelial cells) indicates that chemokines also contribute to the regulation of epithelialization, tissue remodeling, and angiogenesis. Thus, chemokines are in an exclusive position to integrate inflammatory events and reparative processes and are important modulators of wound healing (24). Interleukin-6 (IL-6) is a pleiotropic cytokine that is produced by normal constituents of the skin, including epidermal cells, fibroblasts, and dermal endothelial cells. The inflammatory response that occurs after cutaneous wounding is a prerequisite for healing, and inflammatory cytokines such as IL-6 are involved in this process (23). Consistently, we noted that IL-6 represented an early upregulated gene in response to wounding. Metallothioneins (Mt) are a class of ubiquitously occurring low-molecular-weight cysteine- and metal-rich proteins containing sulfur-based metal clusters that are inducible by inflammation (16). Expression of the Mt gene is upregulated in the skin in regions of high mitotic activity. This induction of Mt in the wound margin may reflect its role in promoting cell proliferation and re-epithelialization. The action of Mt in these processes may result from the large number of Zinc-dependent and copper-dependent enzymes required for cell proliferation and matrix remodeling (38). We noted that Mt1 and Mt2 are rapidly induced in response to wounding.
Thrombospondin 1 (TSP1) null mice suffer from impaired healing of cutaneous wounds. Immunohistochemical analyses have demonstrated that the granulation tissue of TSP1-null mice is not well vascularized. TSP1-nulls also exhibit impaired macrophage recruitment to the injury site (2). Our observation that TSP1 is rapidly induced in response to wounding is consistent with these findings. More recent studies have demonstrated that platelets, TSP1, and dermal fibroblasts cooperate for stimulation of endothelial cell tubulogenesis through VEGF and PAI-1 regulation (35). TSP-1 also promotes proliferative healing through stabilization of PDGF (37). Thrombomodulin is a cell surface anticoagulant that is expressed by endothelial cells and epidermal keratinocytes. In both human and murine wounds, thrombomodulin is absent in keratinocytes at the leading edge of the neoepidermis but is expressed strongly by stratifying keratinocytes within the neoepidermis (55). We observed that thrombomodulin was rapidly induced in response to cutaneous wounding as demonstrated previously.
The inflammation-related genes that were induced 12–96 h after wounding included matrix included MMP9. Previous gelatin zymography studies showed that MMP9 is expressed from day 1 to day 5 after healing (53). Also known as gelatinase B, MMP9 coordinates and effects epithelial regeneration (46). Degradation of the ECM, which is an indispensable step in tissue remodeling processes such as embryonic development and wound healing of the skin, has been attributed to collagenolytic activity of MMPs. Macrophage expressed gene (Mpeg1) represented another gene in this category that has never heretofore been known in the context of wound healing. Mpeg1 expression in mouse cells and cell lines is restricted to mature macrophage and macrophage-like cells, with increased Mpeg1 expression detected as progenitor cells differentiated into macrophages (77).
In the functional category of angiogenesis, the three subsets of genes identified include those that were induced early (6–24 h), induced late (48–96 h), and those that were downregulated in the time frame of 6–96 h. Genes encoding transcriptional regulators such as Stat3 and HIF1α were induced rapidly following wounding. Signal transducer and activator of transcription 3 (Stat3) is one of a family of cytoplasmic proteins that participate in normal cellular responses to cytokines and growth factors as transcription factors. Stat3 modulates various physiological functions including cell survival, cell-cycle regulation, and angiogenesis through regulation of gene expression. Stat3 activation contributes to skin wound healing, keratinocyte migration, and hair follicle growth (67). PDGF induces fibroblast migration under the control of STAT3-SOCS3 (48). Disrupted vasculature at the wound site causes tissue hypoxia and induces the HIF transcriptional machinery (18). The angiogenic factor Cyr61 activates a genetic program for wound healing in human skin fibroblasts (10). Recently we have noted that Cyr61 protein was markedly overexpressed in blood vessels laser captured from the chronic human wound compared with blood vessels obtained from the intact skin (65). Consistent with the observation in patients, Cyr61 was observed to be rapidly induced in the wound-edge tissue following injury. Transforming growth factor-α (TGF-α) plays a significant early role in wound epithelialization in vivo (36). In humans, TGF-α represents a major human serum factor that promotes human keratinocyte migration (40). Our results indicate that TGF-α is rapidly induced in response to cutaneous wounding. The broad role of the TGF-β signaling pathway in vascular development, homeostasis, and repair is well appreciated. Endoglin (CD105) is emerging as a novel modifier of TGF-β receptor signaling at the ligand and receptor activation level. Direct effects of endoglin on cell adhesion and migration are known. The emerging roles for endoglin in the determination of stem cell fate and tissue patterning make it an interesting candidate in the biology of wound healing (4). Although it has been recognized that endoglin plays a crucial role in blood cell-mediated vascular repair (85), its significance in cutaneous wound repair remains to be addressed. This work presents first evidence that cutaneous wounding rapidly induces endoglin in the wound-edge tissue.
Genes related to the angiogenesis functional category that were induced in the late 48–96 h time period primarily represented the extracellular matrix and adhesion molecules. In addition to MMP9, which has been discussed above in the inflammation category, the gene encoding procollagen type XVIII α1 was included in this subset. Loss of this basement membrane proteoglycan enhances angiogenesis and vascular permeability during atherosclerosis by distinct gene dose-dependent mechanisms (47). The significance of this proteoglycan in cutaneous wound healing remains to be understood. The injury-inducible gene angiopoietin-1 (Ang-2) (33) was upregulated in the wound-edge tissue. A ligand for Tie2, Ang-2, is produced by angiogenic vessels and is a chemoattractant for Tie2-expressing monocytes (39). Vascular cell adhesion cell molecule-1 (VCAM1, CD106) was also observed to be induced following 48–96 h of wounding. Injury-activated vascular endothelium expresses VCAM-1, a member of the adhesion molecule superfamily, to which monocytes and lymphocytes can bind. These inflammatory cells can then move through the endothelium by diapedesis and release cytokines and enzymes, important components in the progression of the lesion. The specific significance of VCAM1 in cutaneous wound healing remains to be examined.
Functional angiogenic outcomes do not depend solely on any single gene but on the net balance of angiogenesis-related genes. Thus, an understanding of the genes that are downregulated in response to injury is necessary to appreciate the situation as a whole. This work identified a set of angiogenesis-related genes that were downregulated in response to injury. Although it is known that these gene products are functionally related to angiogenesis, their significance to cutaneous wound healing remains unknown. Adiponectin is an adipocyte-specific adipocytokine with antiatherogenic and antidiabetic properties, and its plasma levels are reduced in association with obesity-linked diseases. Adiponectin stimulates angiogenesis in response to ischemic stress, as known to occur in wounds, by promoting AMP-activated kinase signaling (72). Epiregulin, a member of the epidermal growth factor family, is an autocrine growth factor in normal human keratinocytes (74). Epiregulin plays a critical role in immune/inflammatory-related responses of keratinocytes and macrophages at the barrier from the outside milieu (75). Angiomotin is an angiostatin binding protein that facilitates endothelial cell migration and tube formation. It plays an important role in growth factor-induced migration of endothelial cells (1). Angiomotin is important for endothelial polarization during migration, and it controls Rac1 activity in endothelial and epithelial cells (1). Therapeutic antibodies targeting angiomotin inhibit angiogenesis in vivo (84). As an angiogenic factor, VEGF-A has received most attention, as it is a potent stimulator of vascular growth. Results in clinical trials of VEGF-A as a therapeutic agent have fallen short of high expectations because of serious edematous side effects caused by its activity in promoting vascular permeability. VEGF-B, a related factor, binds some of the VEGF-A receptors but not to VEGF receptor 2, which is implicated in the vascular permeability promoting activity of VEGF-A. Recent data support the use of VEGF-B as a therapeutic agent to promote vascular growth, in part, by potentiating angiogenesis. Furthermore, the lack of vascular permeability activity associated with either transgenic overexpression of the VEGF-B gene in endothelial cells or application of VEGF-B protein to the skin of mice indicates that use of VEGF-B as a therapy should not be associated with edematous side effects (83). Reticulons are a group of integral membrane proteins that have a uniquely conserved COOH-terminal domain named RHD. In mammalian genomes, transcripts are produced from four genes, rtn1 to rtn4, under the regulation of tissue or cell type-specific expression. Although this family exists in almost all eukaryotes, only the rtn4 gene product, Nogo (reticulon 4), has gained relatively more in-depth attention. Despite predominant localization in the endoplasmic reticulum, Nogo on the cell surface appears to play a critical role as an inhibitory molecule for axonal growth and regeneration in humans and rodents. Biological functions of reticulon 4 or Nogo include inhibition of neurite growth from the cell surface via specific receptors, intracellular trafficking, cell division, and apoptosis (87). Significance of the observed downregulation of the reticulon 4 in the wound-edge tissue remains unclear.
The current study provides a comprehensive account of the temporal changes in the expression of genes encoding ECM proteins during the acute inflammatory phase. The three subsets of genes identified include those that were induced early (6–24 h), induced late (48–96 h), and those that were downregulated in the time frame of 12–96 h. While some of the results confirmed previous findings, a majority of the findings represent changes in genes previously never studied in the context of wound healing. The rapidly induced genes are discussed first. MMP11, also known as stromelysin-3, was rapidly induced by injury. MMP11 represents zinc-dependent endopeptidases involved in matrix degradation and tissue remodeling. Despite intense efforts since its first characterization 15 yr ago, its role and target substrates in different diseases remain largely unknown (42). Fibulin-2 is an ECM protein belonging to the five-member fibulin family, of which two members have been shown to play essential roles in elastic fiber formation during development. Fibulin-2 interacts with two major constituents of elastic fibers, tropoelastin and fibrillin-1, in vitro and localizes to elastic fibers in many tissues in vivo. The protein is prominently expressed during morphogenesis of the heart and aortic arch vessels and at early stages of cartilage development (76). The role of fibulin-2 in cutaneous wound healing remains to be examined. Repetin was identified as a novel member of the “fused gene” subgroup within the S100 gene family encoding a murine epidermal differentiation protein. The “fused” gene family comprises profilaggrin, trichohyalin, repetin, hornerin, the profilaggrin-related protein, and a protein encoded by c1orf10. Functionally, these proteins are associated with keratin intermediate filaments and partially crosslinked to the cell envelope. Repetin expression is scattered in the normal epidermis but strong in the acrosyringium and in the inner hair root sheath. Ultrastructurally, repetin is a component of cytoplasmic nonmembrane “keratohyalin” F-granules in the stratum granulosum of normal epidermis, similar to profilaggrin (32).
The second wave (48–96 h) of ECM-related genes induced by injury was more numerous than the first (6–24 h) wave. Candidates in this late-responding group were mostly related to cytokines, chemokines, and specific MMPs. The observed induction of TGF-β1 and TIMP1 in the late inflammatory phase is consistent with prior literature (78, 93). MMPs 8, 9, and 13 were induced in this time frame as well. MMP-8 (collagenase-2) is a neutrophil-derived highly effective type I collagenase, recently indicated to be important for acute wound healing (56).MMP9 (gelatinase B) coordinates and effects epithelial regeneration (46). The observed induction of MMP13 in the wound-edge tissue warrants revisiting the hypothesis that cutaneous wound healing is independent of MMP13 (28). Versican is a major hyaluronan-binding component in the dermis that is induced in response to injury. Interestingly, the hyaluronan receptor CD44 is also concurrently induced. The high-molecular-weight polysaccharide hyaluronan is a major component of the ECM between the vital cells of human skin epidermis. The levels of hyaluronan and those of the hyaluronan receptor CD44 and the hyaluronan binding proteoglycan versican correlate with the aggressiveness of different human carcinomas of epithelial origin (34). However, their significance in wound healing remains unknown.
A small set of genes were observed to be downregulated acutely in response to wounding. The significance of these changes in wound healing remains obscure. Glycogen synthesis in the wound-edge tissue seems to be turned down by lowering of glycogenin expression. Glycogenin initiates glycogen synthesis in an autocatalytic reaction in which individual glucose residues are covalently linked to tyrosine 194 to form a short priming chain of glucose residues that is a substrate for glycogen synthase, which, combined with the branching enzyme, catalyzes the bulk synthesis of glycogen (88). Another gene downregulated in response to injury was nidogen-2. Nidogens represent classical linkers joining laminin and collagen IV networks in basement membranes. Nidogen-2 is equivalent to nidogen-1, and both help develop the basement membrane (50). The significance of lowered nidogen-2 expression in wound healing remains unclear.
ROS, generated at low concentrations during the acute inflammatory phase, are known to be required for wound healing (25, 52, 60, 69, 71). Excess ROS, however, may complicate the healing process. Thus, we sought to functional cluster all ROS-related genes and analyze their changes in response to injury. The three subsets of genes identified include those that were induced early (6–12 h), induced late (48–96 h), and those that were downregulated in the time frame of 12–96 h. These findings are completely novel and provide key insight into the redox regulation of wound healing (73). Of outstanding interest, several components of the NADPH oxidase system such as gp91phox, p22phox, and p47phox were induced rapidly in response to wounding. These components represent critical elements that help drive redox signaling relevant to wound healing (69). Mice deficient in gp91phox and p47phox are known to suffer from impaired healing (60). Defect in p47phox in humans causes chronic granulomatous disease, which is associated with impaired wound healing.
In the 48–96 h period after injury, ROS-sensitive genes were noted to be upregulated. This could be viewed as a consequence of the generation of ROS that is known to occur shortly after wounding (51, 60). ROS-inducible genes in this subset included Mt (49), myc (15), selectin (82), and heme oxygenase (45). Several other products of genes in this list are known to possess antioxidant functions (17, 79, 92). This response may directed toward defending the wound tissue against the threat of excess ROS. The functional significance of the genes downregulated in response to injury remains unclear. Low levels of H2O2 are known to be required for wound healing (51, 60, 69). Injury-induced lowering of catalase gene expression may be viewed as an attempt to slow H2O2 decomposition at a time when H2O2 is required as a second messenger for wound repair.
In summary, the current work presents a detailed analysis of the acute inflammatory phase of murine excisional wound healing from a functional genomics standpoint. While some of the observations confirm previous findings reported in the literature, the vast majority of the results of this study provide new evidence toward novel hypotheses that should help elucidate the biology of cutaneous wound healing comprehensively.
GRANTS
Supported by National Institutes of Health RO1 awards GM-069589, GM-077185, and partly by DK-076566.
Supplementary Material
Address for reprint requests and other correspondence: C. K. Sen, 473 West 12th Ave., 512 DHLRI, The Ohio State Univ. Medical Center, Columbus, OH 43210 (e-mail: Chandan.Sen@osumc.edu).
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Aase K, Ernkvist M, Ebarasi L, Jakobsson L, Majumdar A, Yi C, Birot O, Ming Y, Kvanta A, Edholm D, Aspenstrom P, Kissil J, Claesson-Welsh L, Shimono A, Holmgren L. Angiomotin regulates endothelial cell migration during embryonic angiogenesis. Genes Dev 21: 2055–2068, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agah A, Kyriakides TR, Lawler J, Bornstein P. The lack of thrombospondin-1 (TSP1) dictates the course of wound healing in double-TSP1/TSP2-null mice. Am J Pathol 161: 831–839, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basler T, Jeckstadt S, Valentin-Weigand P, Goethe R. Mycobacterium paratuberculosis, Mycobacterium smegmatis, and lipopolysaccharide induce different transcriptional and post-transcriptional regulation of the IRG1 gene in murine macrophages. J Leukoc Biol 79: 628–638, 2006. [DOI] [PubMed] [Google Scholar]
- 4.Bernabeu C, Conley BA, Vary CP. Novel biochemical pathways of endoglin in vascular cell physiology. J Cell Biochem 102: 1375–1388, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryan DL, Hart P, Forsyth K, Gibson R. Modulation of respiratory syncytial virus-induced prostaglandin E2 production by n-3 long-chain polyunsaturated fatty acids in human respiratory epithelium. Lipids 40: 1007–1011, 2005. [DOI] [PubMed] [Google Scholar]
- 6.Cao D, Pizzorno G. Uridine phosophorylase: an important enzyme in pyrimidine metabolism and fluoropyrimidine activation. Drugs Today (Barc) 40: 431–443, 2004. [DOI] [PubMed] [Google Scholar]
- 7.Carney SA, Hall M, Ricketts CR. The adenosine triphosphate content and lactic acid production of guinea-pig skin after mild heat damage. Br J Dermatol 94: 291–294, 1976. [DOI] [PubMed] [Google Scholar]
- 8.Cepica S, Musilova P, Stratil A, Jurakova M, Rubes J. Assignment of porcine CYBA gene encoding NADPH oxidase-light chain (p22-phox) to chromosome region 6p15. Mamm Genome 8: 299–300, 1997. [DOI] [PubMed] [Google Scholar]
- 9.Chandrasekar B, Melby PC, Sarau HM, Raveendran M, Perla RP, Marelli-Berg FM, Dulin NO, Singh IS. Chemokine-cytokine cross-talk. The ELR+ CXC chemokine LIX (CXCL5) amplifies a proinflammatory cytokine response via a phosphatidylinositol 3-kinase-NF-kappa B pathway. J Biol Chem 278: 4675–4686, 2003. [DOI] [PubMed] [Google Scholar]
- 10.Chen CC, Mo FE, Lau LF. The angiogenic factor Cyr61 activates a genetic program for wound healing in human skin fibroblasts. J Biol Chem 276: 47329–47337, 2001. [DOI] [PubMed] [Google Scholar]
- 11.Chiang B, Essick E, Ehringer W, Murphree S, Hauck MA, Li M, Chien S. Enhancing skin wound healing by direct delivery of intracellular adenosine triphosphate. Am J Surg 193: 213–218, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Constant JS, Feng JJ, Zabel DD, Yuan H, Suh DY, Scheuenstuhl H, Hunt TK, Hussain MZ. Lactate elicits vascular endothelial growth factor from macrophages: a possible alternative to hypoxia. Wound Repair Regen 8: 353–360, 2000. [DOI] [PubMed] [Google Scholar]
- 13.Cooper L, Johnson C, Burslem F, Martin P. Wound healing and inflammation genes revealed by array analysis of ‘macrophageless’ PU.1 null mice. Genome Biol 6: R5, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Souza SJ, Vespa A, Murkherjee S, Maher A, Pajak A, Dagnino L. E2F-1 is essential for normal epidermal wound repair. J Biol Chem 277: 10626–10632, 2002. [DOI] [PubMed] [Google Scholar]
- 15.Dang CV, Li F, Lee LA. Could MYC induction of mitochondrial biogenesis be linked to ROS production and genomic instability? Cell Cycle 4: 1465–1466, 2005. [DOI] [PubMed] [Google Scholar]
- 16.DiSilvestro RA, Carlson GP. Inflammation, an inducer of metallothionein, inhibits carbon-tetrachloride-induced hepatotoxicity in rats. Toxicol Lett 60: 175–181, 1992. [DOI] [PubMed] [Google Scholar]
- 17.Eckenroth B, Harris K, Turanov AA, Gladyshev VN, Raines RT, Hondal RJ. Semisynthesis and characterization of mammalian thioredoxin reductase. Biochemistry 45: 5158–5170, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elson DA, Ryan HE, Snow JW, Johnson R, Arbeit JM. Coordinate up-regulation of hypoxia inducible factor (HIF)-1alpha and HIF-1 target genes during multi-stage epidermal carcinogenesis and wound healing. Cancer Res 60: 6189–6195, 2000. [PubMed] [Google Scholar]
- 19.Feezor RJ, Paddock HN, Baker HV, Varela JC, Barreda J, Moldawer LL, Schultz GS, Mozingo DW. Temporal patterns of gene expression in murine cutaneous burn wound healing. Physiol Genomics 16: 341–348, 2004. [DOI] [PubMed] [Google Scholar]
- 20.Firth JD, Putnins EE. Keratinocyte growth factor 1 inhibits wound edge epithelial cell apoptosis in vitro. J Invest Dermatol 122: 222–231, 2004. [DOI] [PubMed] [Google Scholar]
- 21.Florin L, Knebel J, Zigrino P, Vonderstrass B, Mauch C, Schorpp-Kistner M, Szabowski A, Angel P. Delayed wound healing and epidermal hyperproliferation in mice lacking JunB in the skin. J Invest Dermatol 126: 902–911, 2006. [DOI] [PubMed] [Google Scholar]
- 22.Fries RB, Wallace WA, Roy S, Kuppusamy P, Bergdall V, Gordillo GM, Melvin WS, Sen CK. Dermal excisional wound healing in pigs following treatment with topically applied pure oxygen. Mutat Res 579: 172–181, 2005. [DOI] [PubMed] [Google Scholar]
- 23.Gallucci RM, Sugawara T, Yucesoy B, Berryann K, Simeonova PP, Matheson JM, Luster MI. Interleukin-6 treatment augments cutaneous wound healing in immunosuppressed mice. J Interferon Cytokine Res 21: 603–609, 2001. [DOI] [PubMed] [Google Scholar]
- 24.Gillitzer R, Goebeler M. Chemokines in cutaneous wound healing. J Leukoc Biol 69: 513–521, 2001. [PubMed] [Google Scholar]
- 25.Gordillo GM, Sen CK. Revisiting the essential role of oxygen in wound healing. Am J Surg 186: 259–263, 2003. [DOI] [PubMed] [Google Scholar]
- 26.Han CY, Subramanian S, Chan CK, Omer M, Chiba T, Wight TN, Chait A. Adipocyte-derived serum amyloid A3 and hyaluronan play a role in monocyte recruitment and adhesion. Diabetes 56: 2260–2273, 2007. [DOI] [PubMed] [Google Scholar]
- 27.Hanna IR, Hilenski LL, Dikalova A, Taniyama Y, Dikalov S, Lyle A, Quinn MT, Lassegue B, Griendling KK. Functional association of nox1 with p22phox in vascular smooth muscle cells. Free Radic Biol Med 37: 1542–1549, 2004. [DOI] [PubMed] [Google Scholar]
- 28.Hartenstein B, Dittrich BT, Stickens D, Heyer B, Vu TH, Teurich S, Schorpp-Kistner M, Werb Z, Angel P. Epidermal development and wound healing in matrix metalloproteinase 13-deficient mice. J Invest Dermatol 126: 486–496, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hawke TJ, Kanatous SB, Martin CM, Goetsch SC, Garry DJ. Rad is temporally regulated within myogenic progenitor cells during skeletal muscle regeneration. Am J Physiol Cell Physiol 290: C379–C387, 2006. [DOI] [PubMed] [Google Scholar]
- 30.Hayes JK, Havaleshko DM, Plachinta RV, Rich GF. Isoflurane pretreatment supports hemodynamics and leukocyte rolling velocities in rat mesentery during lipopolysaccharide-induced inflammation. Anesth Analg 98: 999–1006, 2004. [DOI] [PubMed] [Google Scholar]
- 31.Heit B, Kubes P. Measuring chemotaxis and chemokinesis: the under-agarose cell migration assay. Sci STKE 2003: PL5, 2003. [DOI] [PubMed] [Google Scholar]
- 32.Huber M, Siegenthaler G, Mirancea N, Marenholz I, Nizetic D, Breitkreutz D, Mischke D, Hohl D. Isolation and characterization of human repetin, a member of the fused gene family of the epidermal differentiation complex. J Invest Dermatol 124: 998–1007, 2005. [DOI] [PubMed] [Google Scholar]
- 33.Kampfer H, Pfeilschifter J, Frank S. Expressional regulation of angiopoietin-1 and -2 and the tie-1 and -2 receptor tyrosine kinases during cutaneous wound healing: a comparative study of normal and impaired repair. Lab Invest 81: 361–373, 2001. [DOI] [PubMed] [Google Scholar]
- 34.Karvinen S, Kosma VM, Tammi MI, Tammi R. Hyaluronan, CD44 and versican in epidermal keratinocyte tumours. Br J Dermatol 148: 86–94, 2003. [DOI] [PubMed] [Google Scholar]
- 35.Kellouche S, Mourah S, Bonnefoy A, Schoevaert D, Podgorniak MP, Calvo F, Hoylaerts MF, Legrand C, Dosquet C. Platelets, thrombospondin-1 and human dermal fibroblasts cooperate for stimulation of endothelial cell tubulogenesis through VEGF and PAI-1 regulation. Exp Cell Res 313: 486–499, 2007. [DOI] [PubMed] [Google Scholar]
- 36.Kim I, Mogford JE, Chao JD, Mustoe TA. Wound epithelialization deficits in the transforming growth factor-alpha knockout mouse. Wound Repair Regen 9: 386–390, 2001. [DOI] [PubMed] [Google Scholar]
- 37.Krishnaswami S, Ly QP, Rothman VL, Tuszynski GP. Thrombospondin-1 promotes proliferative healing through stabilization of PDGF. J Surg Res 107: 124–130, 2002. [DOI] [PubMed] [Google Scholar]
- 38.Lansdown AB Metallothioneins: potential therapeutic aids for wound healing in the skin. Wound Repair Regen 10: 130–132, 2002. [DOI] [PubMed] [Google Scholar]
- 39.Lewis CE, De Palma M, Naldini L. Tie2-expressing monocytes and tumor angiogenesis: regulation by hypoxia and angiopoietin-2. Cancer Res 67: 8429–8432, 2007. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Fan J, Chen M, Li W, Woodley DT. Transforming growth factor-alpha: a major human serum factor that promotes human keratinocyte migration. J Invest Dermatol 126: 2096–2105, 2006. [DOI] [PubMed] [Google Scholar]
- 41.Ludders JW Advantages and guidelines for using isoflurane. Vet Clin North Am Small Anim Pract 22: 328–331, 1992. [DOI] [PubMed] [Google Scholar]
- 42.Matziari M, Dive V, Yiotakis A. Matrix metalloproteinase 11 (MMP-11; stromelysin-3) and synthetic inhibitors. Med Res Rev 27: 528–552, 2007. [DOI] [PubMed] [Google Scholar]
- 43.Mazzalupo S, Wong P, Martin P, Coulombe PA. Role for keratins 6 and 17 during wound closure in embryonic mouse skin. Dev Dyn 226: 356–365, 2003. [DOI] [PubMed] [Google Scholar]
- 44.McIntyre TM, Prescott SM, Weyrich AS, Zimmerman GA. Cell-cell interactions: leukocyte-endothelial interactions. Curr Opin Hematol 10: 150–158, 2003. [DOI] [PubMed] [Google Scholar]
- 45.McNally SJ, Harrison EM, Ross JA, Garden OJ, Wigmore SJ. Curcumin induces heme oxygenase 1 through generation of reactive oxygen species, p38 activation and phosphatase inhibition. Int J Mol Med 19: 165–172, 2007. [PubMed] [Google Scholar]
- 46.Mohan R, Chintala SK, Jung JC, Villar WV, McCabe F, Russo LA, Lee Y, McCarthy BE, Wollenberg KR, Jester JV, Wang M, Welgus HG, Shipley JM, Senior RM, Fini ME. Matrix metalloproteinase gelatinase B (MMP-9) coordinates and effects epithelial regeneration. J Biol Chem 277: 2065–2072, 2002. [DOI] [PubMed] [Google Scholar]
- 47.Moulton KS, Olsen BR, Sonn S, Fukai N, Zurakowski D, Zeng X. Loss of collagen XVIII enhances neovascularization and vascular permeability in atherosclerosis. Circulation 110: 1330–1336, 2004. [DOI] [PubMed] [Google Scholar]
- 48.Nagai H, Tokumaru S, Sayama K, Shirakata Y, Hanakawa Y, Hirakawa S, Dai X, Tohyama M, Yang L, Hashimoto K. Suppressor of cytokine signaling 3 negative regulation of signal transducer and activator of transcription 3 in platelet-derived growth factor-induced fibroblast migration. J Dermatol 34: 523–530, 2007. [DOI] [PubMed] [Google Scholar]
- 49.Nakano H, Ikenaga S, Aizu T, Kaneko T, Matsuzaki Y, Tsuchida S, Hanada K, Arima Y. Human metallothionein gene expression is upregulated by beta-thujaplicin: possible involvement of protein kinase C and reactive oxygen species. Biol Pharm Bull 29: 55–59, 2006. [DOI] [PubMed] [Google Scholar]
- 50.Nischt R, Schmidt C, Mirancea N, Baranowsky A, Mokkapati S, Smyth N, Woenne EC, Stark HJ, Boukamp P, Breitkreutz D. Lack of nidogen-1 and -2 prevents basement membrane assembly in skin-organotypic coculture. J Invest Dermatol 127: 545–554, 2007. [DOI] [PubMed] [Google Scholar]
- 51.Ojha N, Roy S, He G, Biswas S, Velayutham M, Khanna S, Kuppusamy P, Zweier JL, Sen CK. Assessment of wound-site redox environment and the significance of Rac2 in cutaneous healing. Free Radic Biol Med, 2007. [DOI] [PMC free article] [PubMed]
- 52.Ojha N, Roy S, He G, Biswas S, Velayutham M, Khanna S, Kuppusamy P, Zweier JL, Sen CK. Assessment of wound-site redox environment and the significance of Rac2 in cutaneous healing. Free Radic Biol Med 44: 682–691, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okuse T, Chiba T, Katsuumi I, Imai K. Differential expression and localization of WNTs in an animal model of skin wound healing. Wound Repair Regen 13: 491–497, 2005. [DOI] [PubMed] [Google Scholar]
- 54.Partida-Sanchez S, Gasser A, Fliegert R, Siebrands CC, Dammermann W, Shi G, Mousseau BJ, Sumoza-Toledo A, Bhagat H, Walseth TF, Guse AH, Lund FE. Chemotaxis of mouse bone marrow neutrophils and dendritic cells is controlled by adp-ribose, the major product generated by the CD38 enzyme reaction. J Immunol 179: 7827–7839, 2007. [DOI] [PubMed] [Google Scholar]
- 55.Peterson JJ, Rayburn HB, Lager DJ, Raife TJ, Kealey GP, Rosenberg RD, Lentz SR. Expression of thrombomodulin and consequences of thrombomodulin deficiency during healing of cutaneous wounds. Am J Pathol 155: 1569–1575, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pirila E, Korpi JT, Korkiamaki T, Jahkola T, Gutierrez-Fernandez A, Lopez-Otin C, Saarialho-Kere U, Salo T, Sorsa T. Collagenase-2 (MMP-8) and matrilysin-2 (MMP-26) expression in human wounds of different etiologies. Wound Repair Regen 15: 47–57, 2007. [DOI] [PubMed] [Google Scholar]
- 57.Rink C, Roy S, Khanna S, Rink T, Bagchi D, Sen CK. Transcriptome of the subcutaneous adipose tissue in response to oral supplementation of type 2 Leprdb obese diabetic mice with niacin-bound chromium. Physiol Genomics 27: 370–379, 2006. [DOI] [PubMed] [Google Scholar]
- 58.Rossomando EF, Bertolami CN. Alterations in purine salvage and hypoxanthine levels in granulation tissue during skin wound repair. J Surg Res 35: 259–263, 1983. [DOI] [PubMed] [Google Scholar]
- 59.Roy S, Khanna S, Kuhn DE, Rink C, Williams WT, Zweier JL, Sen CK. Transcriptome analysis of the ischemia-reperfused remodeling myocardium: temporal changes in inflammation and extracellular matrix. Physiol Genomics 25: 364–374, 2006. [DOI] [PubMed] [Google Scholar]
- 60.Roy S, Khanna S, Nallu K, Hunt TK, Sen CK. Dermal wound healing is subject to redox control. Mol Ther 13: 211–220, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roy S, Khanna S, Shah H, Rink C, Phillips C, Preuss H, Subbaraju GV, Trimurtulu G, Krishnaraju AV, Bagchi M, Bagchi D, Sen CK. Human genome screen to identify the genetic basis of the anti-inflammatory effects of Boswellia in microvascular endothelial cells. DNA Cell Biol 24: 244–255, 2005. [DOI] [PubMed] [Google Scholar]
- 62.Roy S, Khanna S, Wallace WA, Lappalainen J, Rink C, Cardounel AJ, Zweier JL, Sen CK. Characterization of perceived hyperoxia in isolated primary cardiac fibroblasts and in the reoxygenated heart. J Biol Chem 278: 47129–47135, 2003. [DOI] [PubMed] [Google Scholar]
- 63.Roy S, Khanna S, Yeh PE, Rink C, Malarkey WB, Kiecolt-Glaser J, Laskowski B, Glaser R, Sen CK. Wound site neutrophil transcriptome in response to psychological stress in young men. Gene Expression 12: 273–287, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roy S, Lado BH, Khanna S, Sen CK. Vitamin E sensitive genes in the developing rat fetal brain: a high-density oligonucleotide microarray analysis. FEBS Lett 530: 17–23, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roy S, Patel D, Khanna S, Gordillo GM, Biswas S, Friedman A, Sen CK. Transcriptome-wide analysis of blood vessels laser captured from human skin and chronic wound-edge tissue. Proc Natl Acad Sci USA 104: 14472–14477, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salaycik KJ, Fagerstrom CJ, Murthy K, Tulu US, Wadsworth P. Quantification of microtubule nucleation, growth and dynamics in wound-edge cells. J Cell Sci 118: 4113–4122, 2005. [DOI] [PubMed] [Google Scholar]
- 67.Sano S, Chan KS, Digiovanni J. Impact of Stat3 activation upon skin biology: A dichotomy of its role between homeostasis and diseases. J Dermatol Sci 50: 1–14, 2007. [DOI] [PubMed] [Google Scholar]
- 68.Seah CC, Phillips TJ, Howard CE, Panova IP, Hayes CM, Asandra AS, Park HY. Chronic wound fluid suppresses proliferation of dermal fibroblasts through a Ras-mediated signaling pathway. J Invest Dermatol 124: 466–474, 2005. [DOI] [PubMed] [Google Scholar]
- 69.Sen CK The general case for redox control of wound repair. Wound Repair Regen 11: 431–438, 2003. [DOI] [PubMed] [Google Scholar]
- 70.Sen CK, Khanna S, Babior BM, Hunt TK, Ellison EC, Roy S. Oxidant-induced vascular endothelial growth factor expression in human keratinocytes and cutaneous wound healing. J Biol Chem 277: 33284–33290, 2002. [DOI] [PubMed] [Google Scholar]
- 71.Sen CK, Khanna S, Venojarvi M, Trikha P, Ellison EC, Hunt TK, Roy S. Copper-induced vascular endothelial growth factor expression and wound healing. Am J Physiol Heart Circ Physiol 282: H1821–H1827, 2002. [DOI] [PubMed] [Google Scholar]
- 72.Shibata R, Ouchi N, Kihara S, Sato K, Funahashi T, Walsh K. Adiponectin stimulates angiogenesis in response to tissue ischemia through stimulation of amp-activated protein kinase signaling. J Biol Chem 279: 28670–28674, 2004. [DOI] [PubMed] [Google Scholar]
- 73.Shilo S, Roy S, Khanna S, Sen CK. Evidence for the involvement of miRNA in redox regulated angiogenic response of human microvascular endothelial cells. Arterioscler Thromb Vasc Biol 28: 471–477, 2008. [DOI] [PubMed] [Google Scholar]
- 74.Shirakata Y, Komurasaki T, Toyoda H, Hanakawa Y, Yamasaki K, Tokumaru S, Sayama K, Hashimoto K. Epiregulin, a novel member of the epidermal growth factor family, is an autocrine growth factor in normal human keratinocytes. J Biol Chem 275: 5748–5753, 2000. [DOI] [PubMed] [Google Scholar]
- 75.Shirasawa S, Sugiyama S, Baba I, Inokuchi J, Sekine S, Ogino K, Kawamura Y, Dohi T, Fujimoto M, Sasazuki T. Dermatitis due to epiregulin deficiency and a critical role of epiregulin in immune-related responses of keratinocyte and macrophage. Proc Natl Acad Sci USA 101: 13921–13926, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sicot FX, Tsuda T, Markova D, Klement JF, Arita M, Zhang RZ, Pan TC, Mecham RP, Birk DE, Chu ML. Fibulin-2 is dispensable for mouse development and elastic fiber formation. Mol Cell Biol 28: 1061–1067, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Spilsbury K, O'Mara MA, Wu WM, Rowe PB, Symonds G, Takayama Y. Isolation of a novel macrophage-specific gene by differential cDNA analysis. Blood 85: 1620–1629, 1995. [PubMed] [Google Scholar]
- 78.Stevenson TJ, Vinarsky V, Atkinson DL, Keating MT, Odelberg SJ. Tissue inhibitor of metalloproteinase 1 regulates matrix metalloproteinase activity during newt limb regeneration. Dev Dyn 235: 606–616, 2006. [DOI] [PubMed] [Google Scholar]
- 79.Suemori S, Shimazawa M, Kawase K, Satoh M, Nagase H, Yamamoto T, Hara H. Metallothionein, an endogenous antioxidant, protects against retinal neuron damage in mice. Invest Ophthalmol Vis Sci 47: 3975–3982, 2006. [DOI] [PubMed] [Google Scholar]
- 80.Sugerman PB, Faber SB, Willis LM, Petrovic A, Murphy GF, Pappo J, Silberstein D, van den Brink MR. Kinetics of gene expression in murine cutaneous graft-versus-host disease. Am J Pathol 164: 2189–2202, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Szabowski A, Maas-Szabowski N, Andrecht S, Kolbus A, Schorpp-Kistner M, Fusenig NE, Angel P. c-Jun and JunB antagonistically control cytokine-regulated mesenchymal-epidermal interaction in skin. Cell 103: 745–755, 2000. [DOI] [PubMed] [Google Scholar]
- 82.Takano M, Meneshian A, Sheikh E, Yamakawa Y, Wilkins KB, Hopkins EA, Bulkley GB. Rapid upregulation of endothelial P-selectin expression via reactive oxygen species generation. Am J Physiol Heart Circ Physiol 283: H2054–H2061, 2002. [DOI] [PubMed] [Google Scholar]
- 83.Trompezinski S, Berthier-Vergnes O, Denis A, Schmitt D, Viac J. Comparative expression of vascular endothelial growth factor family members, VEGF-B, -C and -D, by normal human keratinocytes and fibroblasts. Exp Dermatol 13: 98–105, 2004. [DOI] [PubMed] [Google Scholar]
- 84.Troyanovsky B, Levchenko T, Mansson G, Matvijenko O, Holmgren L. Angiomotin: an angiostatin binding protein that regulates endothelial cell migration and tube formation. J Cell Biol 152: 1247–1254, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Van Laake LW, van den Driesche S, Post S, Feijen A, Jansen MA, Driessens MH, Mager JJ, Snijder RJ, Westermann CJ, Doevendans PA, van Echteld CJ, ten Dijke P, Arthur HM, Goumans MJ, Lebrin F, Mummery CL. Endoglin has a crucial role in blood cell-mediated vascular repair. Circulation 114: 2288–2297, 2006. [DOI] [PubMed] [Google Scholar]
- 86.Verducci JS, Melfi VF, Lin S, Wang Z, Roy S, Sen CK. Microarray analysis of gene expression: considerations in data mining and statistical treatment. Physiol Genomics 25: 355–363, 2006. [DOI] [PubMed] [Google Scholar]
- 87.Wang M, Han Y, Zhang XP, Lu YP. Nogo, a star protein in reticulon family. Neurosci Bull 22: 183–186, 2006. [PubMed] [Google Scholar]
- 88.Wilson RJ, Gusba JE, Robinson DL, Graham TE. Glycogenin protein and mRNA expression in response to changing glycogen concentration in exercise and recovery. Am J Physiol Endocrinol Metab 292: E1815–E1822, 2007. [DOI] [PubMed] [Google Scholar]
- 89.Wolf C, Chenard MP, Durand de Grossouvre P, Bellocq JP, Chambon P, Basset P. Breast-cancer-associated stromelysin-3 gene is expressed in basal cell carcinoma and during cutaneous wound healing. J Invest Dermatol 99: 870–872, 1992. [DOI] [PubMed] [Google Scholar]
- 90.Wong P, Coulombe PA. Loss of keratin 6 (K6) proteins reveals a function for intermediate filaments during wound repair. J Cell Biol 163: 327–337, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Woo K, Ayello EA, Sibbald RG. The edge effect: current therapeutic options to advance the wound edge. Adv Skin Wound Care 20: 99–117; quiz 118–119, 2007. [DOI] [PubMed] [Google Scholar]
- 92.Yoshimoto T, Fukai N, Sato R, Sugiyama T, Ozawa N, Shichiri M, Hirata Y. Antioxidant effect of adrenomedullin on angiotensin II-induced reactive oxygen species generation in vascular smooth muscle cells. Endocrinology 145: 3331–3337, 2004. [DOI] [PubMed] [Google Scholar]
- 93.Zhu KQ, Engrav LH, Tamura RN, Cole JA, Muangman P, Carrougher GJ, Gibran NS. Further similarities between cutaneous scarring in the female, red Duroc pig and human hypertrophic scarring. Burns 30: 518–530, 2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.