Abstract
Knowing how hosts respond to parasite infection is paramount in understanding the effects of parasites on host populations and hence host–parasite co-evolution. Modification of life-history traits in response to parasitism has received less attention than other defence strategies. Life-history theory predicts that parasitised hosts will increase reproductive effort and accelerate reproduction. However, empirical analyses of these predictions are few and mostly limited to animal-parasite systems. We have analysed life-history trait responses in 18 accessions of Arabidopsis thaliana infected at two different developmental stages with three strains of Cucumber mosaic virus (CMV). Accessions were divided into two groups according to allometric relationships; these groups differed also in their tolerance to CMV infection. Life-history trait modification upon virus infection depended on the host genotype and the stage at infection. While all accessions delayed flowering, only the more tolerant allometric group modified resource allocation to increase the production of reproductive structures and progeny, and reduced the length of reproductive period. Our results are in agreement with modifications of life-history traits reported for parasitised animals and with predictions from life-history theory. Thus, we provide empirical support for the general validity of theoretical predictions. In addition, this experimental approach allowed us to quantitatively estimate the genetic determinism of life-history trait plasticity and to evaluate the role of life-history trait modification in defence against parasites, two largely unexplored issues.
Author Summary
Hosts have developed a variety of mechanisms to compensate for the negative impact of parasite infection. Modification of life-history traits in response to parasitism has received less attention than other defence strategies. Life-history theory assumes trade-offs between resource allocation to different fitness components, and predicts that hosts under parasitism will allocate more resources to reproduction, subtracting them from those dedicated to growth and survival. Empirical support for predictions is not abundant, and derives mostly from the analysis of animal-parasite systems. We have analysed the modification of various life-history traits in the plant Arabidopsis thaliana infected by Cucumber mosaic virus. Life-history trait modification upon virus infection depended on the host genotype and on the developmental stage at infection. All plant genotypes delayed flowering, but only the more tolerant ones allocated more resources to reproduction, and reduced the length of reproductive period. These results agree with reports from parasitised animals and with predictions from life-history theory, providing empirical support for the general validity of theoretical predictions. In addition, results allow for the more precise evaluation of the role of life-history trait modification in defence against parasites by taking into account plant–virus interactions where life-history traits were differentially modified.
Introduction
Parasites affect the welfare of humans and of domestic animals and plants, with a high socioeconomic impact. In addition, an increasing number of reports provide evidence of the important role of parasites in ecosystem composition and dynamics [1]. Parasite infection has a negative impact on host fitness, which has been defined as virulence [2]. Consequently, parasites may modulate the dynamics and genetic structure of populations of their hosts, as well as of non-host species by altering inter-specific competition [3],[4]. It has been proposed that, through these effects, parasites may drive biodiversity [5]. Knowing how hosts respond to parasite infection is capital for understanding the role of parasites in shaping host populations and ecosystems. Hosts have developed a variety of mechanisms to compensate for the cost of parasite infection, which may be grouped into four strategies [6]: hosts can modify their behaviour to avoid contact with parasites; hosts may have mechanisms that prevent the establishment of infection and trigger defence responses; hosts may develop immune systems, which in addition to act as barriers to infection may also clear the infection if parasites overcome host defences; and a fourth mechanism to reduce the harm of parasite infection is tolerance, which may involve the alteration of host life-history traits. While literature on the first three strategies is extensive, particularly regarding defence responses and immune systems, tolerance and, particularly, host life-history trait modification, has received comparatively less attention.
Various host life-history traits have been reported to respond to pathogen infection, including pre-reproductive life span [7],[8], reproductive effort [9],[10], and body size [11],[12]. These observations have prompted theoretical analyses aimed to predict optimal host life-history trait responses to parasitism. Life-history theory makes predictions for the evolution of resource investment by organisms, based on the concept that trade-offs exist between resources allocated to different fitness components: reproduction, growth and survival [13]. The optimal pattern of resource allocation may differ depending on environmental conditions, which include parasitism [14]. Thus, parasite infection may modify optimal resource distribution. Inspired by this concept, models for evolution of resource allocation predict that parasitised hosts will allocate more resources to reproduction, subtracting them from those dedicated to growth and survival [15]–[18]. Life-history theory also states that environmental conditions affecting mortality rates modify temporal life-history schedules in order to maximize fitness [19]. Accordingly, models predict that highly virulent parasites will induce shorter host pre-reproductive periods in order to produce progeny before resource depletion, castration or death. In contrast, low virulence will result in a delay in host reproduction, which allows for compensation of parasite damage [20],[21].
If theoretical efforts at understanding the evolution of life-history traits under parasite infection are not abundant, experimental analyses are scarcer and have been mostly limited to animal hosts and highly virulent parasites causing mainly host death or castration. Most experimental results support predictions for the effects of parasitism on age at maturity [7],[22] or on reproductive effort [23],[24]. However, there are also examples that do not fit theoretical predictions and that have been explained as consequence of particular host genetic features [8],[25], environmental conditions [26] or parasite manipulation of the trade-off between growth and fecundity (e.g. gigantism) [11]. Experimental analyses are usually focused on a single host genotype infected by one or various pathogen genotypes, but the role of genotype×genotype interactions, which may affect the outcome of host-parasite interactions [27],[28], has been mostly overlooked. Experimental analyses of the evolution of plant life-history traits under parasitism are rather limited, with the notable exception of analyses of the effects of infection by the fungus Microbotryum violaceum on the perennial plant Silene latifolia [29],[30]. However, studies of plant host-parasite systems are relevant to test the general validity of theoretical predictions, since plants and animals differ widely in organisation, and plant parasites mostly affect growth and reproduction of their host without causing immediate host death.
To analyse the effects of parasitism on plant life-history traits we have chosen the plant-virus system Arabidopsis thaliana L. Heynh. (Brassicaceae)-Cucumber mosaic virus (CMV, Bromoviridae). In the last twenty years, Arabidopsis thaliana (from here on, referred to as Arabidopsis) has arisen as the model organism for the molecular and genetic study of a wide range of plant traits, including resistance patterns against parasite infection [31],[32]. Recently it has been also used in analyses of host-parasite co-evolution [33],[34] and of life-history traits responses to abiotic stress associated with changes in light, nutrients or water availability [35]–[37]. The annual plant Arabidopsis is a typical semelparous species, with two clearly differentiated developmental phases or periods in its post-embryonic life cycle. During the vegetative growth period, a rosette of leaves is produced. Vegetative growth ceases when the vegetative meristem becomes an inflorescence meristem [38]. This is the start of the reproduction period, when the reproductive structure (inflorescence) grows, new flowers are produced continuously and older flowers develop into fruits (siliques). Flower production almost ceases after ripening of the first silique, and most flowers produced later on fail to set fruits and seeds [39],[40]. Plant senescence and death end the reproduction period. Hence, vegetative growth effort, total reproductive effort and progeny production are easily differentiated in Arabidopsis.
CMV is a generalist virus that infects about 1,200 plant species in more than 100 mono- and dicotyledonous families, including natural populations of Arabidopsis (our unpublished observations). CMV is horizontally transmitted by more than 70 species of aphids, and vertically through seeds with rates that vary depending on the genotypes of CMV and host-plant species. CMV has a messenger-sense, single-stranded, three-segmented RNA genome encapsidated in three isometric particles. The structure of the CMV genome and the roles of the five encoded proteins have been extensively characterized. The genetic variability of CMV has also been much analysed and CMV isolates have been classified into two subgroups, subgroup I and subgroup II, based on the nucleotide sequence similarity of their genomes (reviewed in [41],[42]).
In this work, we have tested predictions of life-history evolution theory by analysing the effect of CMV infection on Arabidopsis growth and reproductive effort and on age at maturity and reproductive period span. To test the contribution of genotype×genotype interactions on life-history traits response to virus infection, we challenged 18 wild genotypes (accessions) of Arabidopsis with three CMV strains. A general reduction of growth and reproductive effort was observed after infection as well as a tendency to increase the age at maturity. However, some accessions previously shown to manifest tolerance to CMV infection [34] presented a relative increase of the reproductive effort upon viral infection together with a reduction of the reproductive period. Overall, these life-history trait modifications can be interpreted as host reactions that reduce the impact of infection on plant fitness.
Results
Resource allocation to growth and reproduction in Arabidopsis accessions
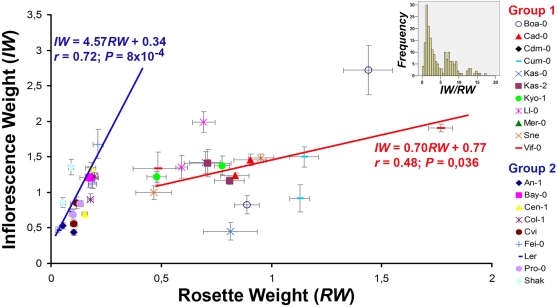
Plant architecture and, consequently, resource allocation to growth and reproductive effort, differ among Arabidopsis accessions and condition responses to viral infection [34]. To properly evaluate the effect of virus infection on different fitness components of the host, we first analysed the relationship between rosette weight (RW), as a measure of growth effort; inflorescence weight (IW) as measure of total reproductive effort; seed weight (SW), as a measure of progeny production [43], and total above-ground biomass (BM) in mock-inoculated plants of eighteen Arabidopsis accessions (see Materials and Methods and Table S1). SW was taken as measure of progeny production since it was previously shown that in these accessions CMV infection did not affect seed size or viability [34]. All traits differed significantly among accessions (P<1×10−5). Rosette weight was positively correlated with inflorescence weight (r = 0.61, P = 1×10−4) and negatively with seed weight (r = −0.36, P = 0.04), which indicates a general positive correlation between growth and reproductive efforts. No significant correlation was found between inflorescence weight and seed weight (r = 0.22, P = 0.21).
The balance between growth and reproductive effort estimated as IW/RW, showed a bimodal distribution across accessions (Figure 1). Thus, two allometric groups of accessions differing significantly in IW/RW (P<1×10−5) were defined: group 1 with IW/RW <5.0 (mean value of 1.75±0.17) including accessions Boa-0, Cad-0, Cum-0, Kas-0, Kas-2, Kyo-1, Ll-0, Sne and Vif-0, and group 2 with IW/RW >5.0 (mean value 6.99±0.88) containing An-1, Bay-0, Cen-1, Col-1, Cvi, Fei-0, Ler, Pro-0 and Shak. A differential linear relationship between RW and IW was found for each group (Figure 1). These two accession groups are the same as those defined in Pagán et al. [34] based on the SW/BM relationship.
Figure 1. Relationship between growth (RW) and reproductive effort (IW) in Arabidopsis accessions.
Correlation between IW and RW for allometry group 1 (red) and allometry group 2 (blue) using mean accession values of mock-inoculated plants. Data are mean±standard error of RW and IW in g. The upper-right panel shows the frequency distribution of the IW/RW relationship of the 18 accessions, based in individual plant values.
For each allometric group, BM was positively correlated with RW, IW and SW (r>0.53, P<0.03); RW was positively correlated with IW (r>0.47, P<0.04) but did not correlate with SW, and IW was positively correlated with SW (r>0.62, P<0.01). Thus, in both allometric groups there is a positive correlation between growth and reproductive efforts and between reproductive effort and progeny production.
Effects of CMV infection on resource allocation to growth and reproduction
Eighteen Arabidopsis were inoculated with three CMV isolates early in the vegetative period (see Materials and Methods). The effect of CMV infection on Arabidopsis growth and reproductive efforts was quantified as the ratios of rosette and inflorescence weights, respectively, between infected and mock-inoculated plants (RWi/RWm and IWi/IWm, i and m denoting infected and mock-inoculated plants respectively) (see Table S2 for statistical parameters of the variables). A general reduction of RW and IW was observed in infected plants, but the effect of CMV infection on both traits depended on the accession, isolate, and the interaction between the two genotypes (P<1×10−5) (see Table S3 for ANOVA parameter values). On average, the effect of infection by Fny-CMV on both RW and IW was about 16% stronger than the effect of infection by LS-CMV and about 38% stronger than the effect of De72-CMV (Figure S1, and Table S2). Broad-sense heritabilities of RWi/RWm ranged from low to moderate (h2b = 0.11–0.56) depending on isolate, while IWi/IWm showed a narrower variation (h2b = 0.39–0.52) (Table S2). Isolates and accessions accounted for a higher fraction of variance of RWi/RWm than the interaction (VC = 16.92, VC = 17.95, VC = 6.15, respectively) but the three components explained similar levels of IWi/IWm variance (VC = 22.22, VC = 19.14, VC = 16.67 for isolate, accession and interaction, respectively) (Table S3). Thus, responses of Arabidopsis on growth and reproductive efforts to CMV infection depend on the host-genotype×parasite-genotype combination.
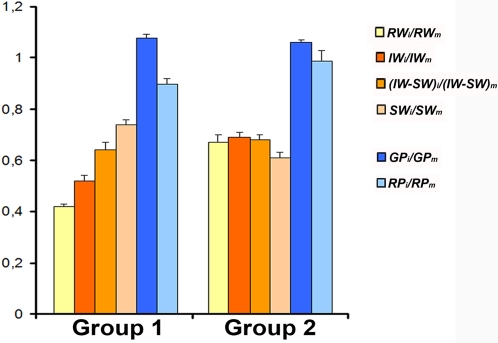
When the two allometric groups of accessions were compared, they differed significantly for RWi/RWm and IWi/IWm (P<0.009) indicating that the effect of virus infection depends on the allometric relationships (Table S4). Isolate and group explained a similar and higher level of RWi/RWm and IWi/IWm variation than the interaction accession×isolate (e.g. VC = 16.92, VC = 11.11, VC = 0.97, respectively, for RWi/RWm). Therefore, both allometric groups were analysed separately. As shown in Figure 2, the effect of infection was much larger for accessions of group 1 (0.42±0.01 and 0.52±0.02 for RW and IW, respectively) than for group 2 (0.67±0.03 and 0.69±0.02 for RW and IW, respectively). For accessions of group 1 the effect of infection on RW was 19% larger than on IW (P<2×10−5), but the effects were similar for accessions of group 2 (P≥0.61) (Figure 2). Thus, the effect of virus infection on growth and reproductive efforts depends on the allometric relationship of the accessions.
Figure 2. Effect of CMV infection on life-history traits for the two allometric groups of accessions.
Effect of viral infection was estimated as the ratio between infected (i) and mock-inoculated (m) plants. Data are mean±standard errors of accession means.
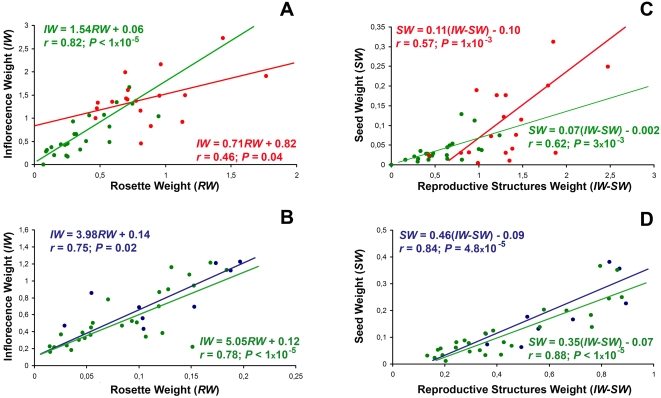
The effect of infection on the relationship between growth and reproductive efforts was further analysed using the ratio (IW/RW)i/(IW/IRW)m. Significant differences were found among allometric groups, isolates and due to the interaction between both factors (P<0.007) (Table S4). Therefore, the effect of CMV infection on IW/RW was analysed for each accession group separately. For allometric group 1, linear regressions of IW on RW significantly differed in slope and intercept between infected and mock-inoculated plants (P<0.01) and the average value of IW/RW was higher for infected than for mock-inoculated plants (2.75±0.02 vs. 2.11±0.16) (Figure 3A). For group 2 of accessions, the regression lines of IW on RW did not differ significantly between mock-inoculated and infected plants (P≥0.37), for which average values of IW/RW were 8.03±0.69 and 7.52±0.47, respectively (Figure 3B). Therefore, infected plants of allometry group 1, but not of group 2, allocated a higher fraction of resources to reproduction than mock-inoculated plants.
Figure 3. Effects of CMV infection on growth/reproduction resource allocation of Arabidopsis accessions.
(A) Effect of infection on IW/RW relationship for allometry group 1. (B) Effect of infection on IW/RW relationship for allometry group 2. (C) Effect of infection on SW/(IW-SW) relationship for allometry group 1. (D) Effect of infection in SW/(IW-SW) relationship for allometry group 2. Relationship in infected plants (green) is compared with that of mock-inoculated plants of allometry group 1 (red) and 2 (blue). Data are mean values of each accession. RW, IW and (IW-SW) units are g.
Effects of CMV infection on resource allocation to reproductive structures and progeny production
CMV effects on the weights of seeds and reproductive structures were quantified as the ratios of infected vs. mock-inoculated plants SWi/SWm and (IW-SW)i/(IW-SW)m, respectively (see Table S2 for statistical parameters). Viral effects on SW and IW-SW differed significantly between isolates and accessions and for the isolate×accession interaction (P<1×10−5; Table S3). Broad-sense heritabilities of SWi/SWm were lower (h2b = 0.19–0.31) than those of (IW-SW)i/(IW-SW)m (h2b = 0.34–0.44) (Table S2). Accession factor explained a higher fraction of the variation of SWi/SWm and (IW-SW)i/(IW-SW)m than isolate or the interaction accession×isolate (e.g. VC = 2.28, VC = 28.92, VC = 3.01, for isolate, accession and interaction for SWi/SWm) (Table S3). In addition, virus infection had the same effect on SW and IW-SW (P = 0.52), average values of SWi/SWm and (IW-SW)i/(IW-SW)m being 0.67±0.01 and 0.66±0.02. Thus, CMV effect on progeny production and on reproductive structures depended on host-genotype×parasite-genotype interaction.
The effect of CMV on SW, but not on (IW-SW), differed significantly between the two allometric groups (P<3×10−4 and P>0.23, respectively). Isolate, group and the interaction accounted for a similar fraction of SWi/SWm variation (e.g. CV = 2.28; CV = 5.51; CV = 1.91 for isolate, group and isolate×group interaction, respectively), whereas isolate and the interaction isolate×group accounted for a similar fraction of (IW-SW)i/(IW-SW)m variance (CV = 3.39; CV = 2.12 for isolate and isolate×group interaction, respectively) (Table S4). Therefore, the two allometric groups were analysed separately. As shown in Figure 2, the effect of virus infection on SW was smaller for group 1 (0.74±0.02) than for group 2 (0.61±0.02), but no significant difference was found for (IW-SW)i/(IW-SW)m (0.68±0.02 and 0.64±0.03, for group 1 and 2, respectively). In addition, for accessions of group 1, SW was significantly less affected by CMV infection than IW-SW (8%, P<3.7×10−2). The opposite was observed for group 2, where viral effects on SW were slightly higher than on IW-SW (3%, P<4.3×10−2). Thus, viral effect on seed and reproductive structures weight also depended on plant architecture.
The relationship between seed weight (SW) and reproductive structure weight (IW-SW) was further analysed using the ratio [SW/(IW-SW)]i/[SW/(IW-SW)]m. This ratio showed strong significant differences among accessions (P<1×10−5; Table S3) and between allometric groups (P = 6×10−4; Table S4) but no or small differences among isolates (Tables S3 and S4). Hence the effect of infection on SW/(IW-SW) was analysed for each accession group separately. For group 1 of accessions, SW to IW-SW regression lines of infected plants (Figure 3C) differed significantly from those of mock-inoculated plants (P = 0.03), the average value of SW/(IW-SW) being 0.07±0.001 for mock-inoculated plants, and 0.09±0.001 for infected plants (Figure 3C). For group 2, regression lines did not differ between infected and mock-inoculated plants (P>0.28), and SW/(IW-SW) showed average values of 0.28±0.02 and 0.25±0.02 for mock-inoculated and infected plants respectively (Figure 3D). Thus, infected plants of allometry group 1, but not of group 2, allocated proportionally more resources to the production of progeny than to reproductive structures than did healthy plants.
Effect of CMV infection on age at maturity and reproductive life span
To analyse if the temporal control of Arabidopsis transition from vegetative growth to reproductive phase may vary in response to CMV infection, we measured the span of growth and reproductive periods (GP and RP, respectively). Both traits differed significantly among accessions in mock-inoculated plants (P<1×10−5). In addition, GP was negatively correlated with RP when all accessions were analysed together and for each allometric group of accessions (r>−0.32, P<0.04). Arabidopsis heritabilities of temporal life-history traits and their CMV responses ranged from low to moderate (h2b = 0.14–0.36) depending on CMV isolate (Table S2).
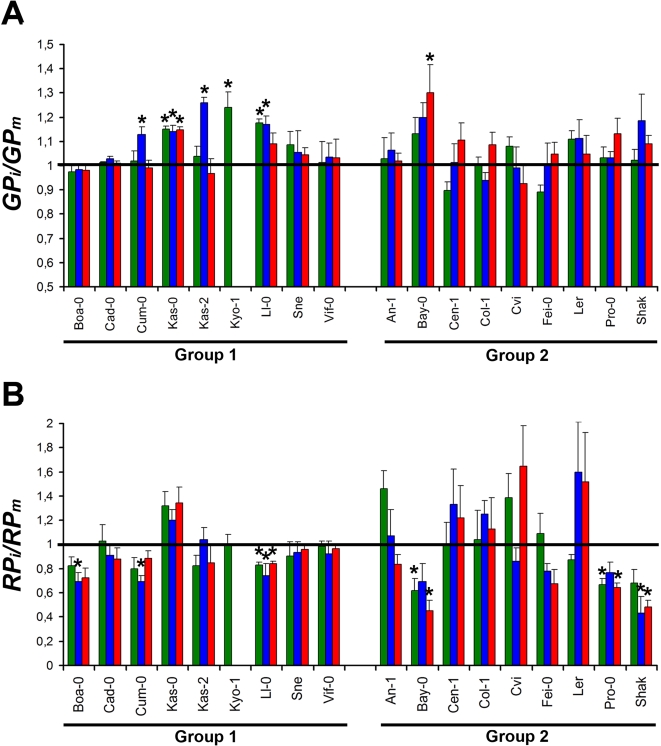
The effect of virus infection on GP and RP was quantified as GPi/GPm and RPi/RPm. Both traits showed significant differences among accessions (P<1×10−5) and due to the interaction accession×isolate (P = 0.01), but not among isolates (P = 0.74). Again, accessions accounted for a higher proportion of the variance than the interaction (e.g. VC = 17.27; VC = 9.39, for accessions and interaction, respectively) (Table S3). However, CMV infection affected differently GP and RP. Infection resulted in an increase of GP in most accessions (16 out of 18), although significant differences were observed in six of them (Cum-0, Kas-0, Kas-2, Kyo-1, Ll-0 and Bay-0) (Figure 4A). In contrast, infection decreased RP in 12 out of 18 accessions, the decrease being significant in six of them (Boa-0, Cum-0, Ll-0, Bay-0, Pro-0 and Shak) (Figure 4B). We further analysed the effect of infection on the time span to seed production (GP+RP), and again significant differences were found among accessions and for the interaction isolate×accession (P<5×10−3; VC = 12.12, VC = 6.06 for accession and interaction, respectively) but not among isolates (P>0.31) (Table S3). Infection resulted in shortening of GP+RP in 11 accessions and elongation in 7 out of 18 accessions, although differences with mock-inoculated controls were significant only for 4 out of 11 (Boa-0, Cum-0, Fei-0 and Shak) and 3 out of 7 accessions (Kas-0, Kyo-1 and Ler) respectively (not shown). All accessions showing reduction of RP also had shorter GP+RP, except Kyo-1.
Figure 4. Effect of viral infection on growth (GP) and reproductive (RP) period span of Arabidopsis accessions.
(A) Effect of CMV infection in GP period span estimated as GPi/GPm, where i and m denote infected and mock-inoculated plants, respectively. (B) Effect of CMV infection in RP period span estimated as described for (A). Data are mean values of accessions infected by each CMV isolates±standard errors. Asterisks indicate significant differences between mock-inoculated and infected plants (P<0.05). The effect of infection is shown for LS-CMV (green), Fny-CMV (blue) and De72-CMV (red). Accessions are divided into allometry groups 1 and 2.
Comparison of CMV effects on temporal life span traits between the two allometric groups of accessions showed significant differences for RPi/RPm (P = 1×10−3). Groups 1 and 2 showed mean values 0.90±0.02 and 0.99±0.04 respectively (Figure 2), allometric group accounting for a 5.1% of RPi/RPm variance (Table S4). No significant difference between groups was found for GPi/GPm (P = 0.29), group 1 and 2 showing mean values of 1.08±0.01 and 1.06±0.01, respectively. On the other hand, (GP+RP)i/(GP+RP)m differed among groups (P = 2×10−3), which showed mean values of 0.98±0.01 and 1.02±0.01 for group 1 and group 2, respectively. Overall, upon CMV infection, most Arabidopsis genotypes tended to increase the age at maturity (growth period). In contrast, the effect on reproductive life span and time to seed production depended on the accession allometric group, both traits being increased in accessions of group 2.
Relationship between the effects of CMV infection on temporal life-history traits and on resource allocation
To explore if the amount of resources allocated to growth and reproduction might condition the span of growth and reproductive periods, we analysed the relationship between both sorts of traits. In mock-inoculated plants, when all accessions were analysed together, duration of growth period was positively correlated with plant biomass and rosette weight (r = 0.37, P = 0.03; r = 0.57, P = 1×10−3); it was negatively correlated with seed weight (r = −0.54, P = 1×10−3); and it did not correlate with inflorescence weight (r = 0.08, P = 0.64). Reproductive period was positively correlated with biomass and inflorescence weight (r = 0.38, P = 0.03; r = 0.43, P = 0.01 respectively), but not with rosette and seed weights (r = 0.27, P = 0.14; r = 0.19, P = 0.28, respectively). In contrast, no significant correlation was found between the effect of CMV infection on the amount of resources allocated to vegetative or reproductive structures (RWi/RWm and IWi/IWm,) and on the time invested in vegetative and reproductive growth (GPi/GPm and RPi/RPm, respectively) when all accessions and isolates were analysed together (r≤−0.13, P≥0.32). Similarly, no significant correlation was found when the various traits were analysed separately for each viral isolate (r≤0.47, P≥0.08).
When these relationships were analysed for each accession separately, four of them (Cad-0, Cum-0 from group 1, and Bay-0 and Shak, from group 2) showed significant negative correlation between RPi/RPm and IWi/IWm (r≥−0.43; P≤0.03); five accessions (An-1, Col-1, Cvi, Fei-0 and Shak all from group 2) presented a significant positive correlation between GPi/GPm and RWi/RWm (r≥0.67; P≤0.01); and in three accessions (Cum-0, Kas-2 and Ll-0 from group 1) GPi/GPm was negatively correlated with IWi/IWm (r≥−0.45; P≤0.02).
The relationships between GPi/GPm or RPi/RPm and SWi/SWm or (IW-SW)i/(IW-SW)m, were also analysed. When all accessions were considered together, viral effects in RP and SW were marginally correlated (r = −0.24; P = 0.07). No other significant correlation was found when considering all accessions (r≤−0.11; P>0.39). When each allometric group of accessions was analysed separately, a marginal negative correlation was found between RPi/RPm and SWi/SWm for group 1 (r = −0.33; P = 0.06) but not for group 2 (r = −0.21; P = 0.37). CMV effect on GP was positively correlated with viral effects on SW and IW-SW in group 2 (r = 0.35; P = 0.04), but not in group 1 (r = 0.15; P = 0.53). RPi/RPm and (IW-SW)i/(IW-SW)m were not correlated for any allometric group (r≤0.18; P≥0.16).
Thus, virus infection disrupted the relationships between resource allocation and temporal life-history traits that occurred in mock-inoculated plants.
Discussion
Plastic modification of life-history traits in response to environmental conditions may be an adaptive mechanism to selection pressures such as abiotic stress, intra- or inter-specific competition or parasitism/disease [15],[44],[45]. Parasites are important ecological agents that can mediate changes in host life-history traits by two sorts of mechanisms. On one hand, parasitic use of host resources can lead to modifications of host resource allocation and developmental time schedules as pathogenic effects of parasitism. Alternatively, life-history modifications may be host responses to compensate for the negative effects of parasitism [17],[18],[20],[21]. The latter are then considered part of tolerance mechanisms, since tolerance is defined as the host ability to reduce the effect of infection on its fitness [46].
Previously, we have reported that within host multiplication of CMV in Arabidopsis does not correlate with virulence due to accession-specific tolerance mechanisms associated with differences in resource allocation patterns [34]. These results prompted to study if Arabidopsis shows plastic responses of life-history traits to CMV parasitism, as a CMV tolerance mechanism. In this work, we have analysed the effects of CMV parasitism on several Arabidopsis life-history traits related with resource allocation and life cycle schedule and found significant plastic modifications of most of them. The relationships among life-history traits has been widely analysed in plants, and correlations between age at maturity and growth effort, and between reproductive period span and reproductive efforts are well documented [47]–[49]. These significant correlations also occurred in our mock-inoculated plants, but not in the infected ones, which indicates that CMV infection not only modifies life-history traits but also alter the relationships between them.
Virus infection had a major effect on resource allocation to growth and reproduction, infection resulting in a general reduction of resources allocated to both traits. However, allocation of resources upon infection was different depending on the allometric features of Arabidopsis genotypes. In accessions of group 1, with a low ratio inflorescence weight (IW) to rosette weight (RW), infection at an early vegetative stage modified the pattern of resource allocation at two levels. First, vegetative growth of infected plants was severely reduced, but a larger fraction of resources was allocated to reproduction than to growth when compared with mock-inoculated plants (Figure 2). Second, infected plants allocated a higher fraction of resources than mock-inoculated ones to progeny production than to production of reproductive structures (Figure 2). In a second experiment, in which Arabidopsis accessions were inoculated at the beginning of the reproductive stage (see Methods), similar results were obtained, although the effect of infection on growth and reproductive efforts was less severe and the IW/RW relationship was not significantly altered. However, the fraction of resources allocated to progeny production was also increased relative to that allocated to reproductive structures (data not shown). In contrast, the effect of infection on accessions of allometric group 2, with a high ratio of inflorescence weight to rosette weight, did not result in significant modifications of resource allocation neither when plants were infected at vegetative stage, nor at the beginning of the reproductive stage (not shown). The shorter life cycles and the higher fraction of reproductive vs. total biomass characteristic of accessions of allometric group 2 [34] could reduce their ability to modify resource allocation upon infection.
Temporal life cycle parameters of Arabidopsis also responded to CMV infection but effects were much smaller than those observed for resource allocation. Vegetative and reproductive span traits behaved differently. Plants infected at vegetative stage tended to increase growth period span (GP) by delaying flowering time, independently of allometric group (Figure 2). On the other hand, changes in reproductive span (RP) differed significantly between the two allometric groups. Early CMV infection of accessions of group 1 resulted in a reduction of RP and total time to seed production (GP+RP) indicating faster reproduction of infected plants. These effects were not observed in plants of accession group 2, which were less tolerant to CMV infection [34].
Modification of life-history traits in parasitised hosts can be part of a host defence response, or may be due to the pathogenic effects of parasitism, either as a manipulation of the host by the parasite, which derives some advantage from it, or a by product of infection [7],[16],[20]. However, causal distinction of life-history modifications is not straightforward. Elongation of GP and/or inability for reproduction has been interpreted in parasitised insects and molluscs as due to parasite manipulation [50]–[52], as a retard/arrest in development would favour parasite transmission (but see also [53]). However, it seems unlikely that the observed increase of GP will be the result of a CMV modification of Arabidopsis life cycle favouring its transmission because aphids that transmit CMV [42] can acquire the virus from any green organ, and the total (GP+RP) was often shortened by infection. It has been shown that Arabidopsis can modulate rosette growth in response to resource availability to maximize reproduction later in development [37]. Since CMV infection results in diminished growth, it can be speculated that plants will delay flowering until a minimum rosette size is attained. Hence, the increase of GP might be interpreted as a by-product of parasitism, although it cannot be discarded that it is part of a general tolerance defence reaction.
Our experimental approach do not allow to determine if resource allocation responses of Arabidopsis are a defensive mechanism triggered by the host in order to reduce the impact of CMV infection in its fitness, or an unavoidable consequence of the virus pathogenic effects. These two possibilities could be analysed by mimicking viral infection but avoiding parasite multiplication. It has been reported for several plant species, including Arabidopsis, that expression of different virulence factors in transgenic plants induces viral-like symptoms in the absence of infection (e.g., [54]–[58]). However, life-history trait modifications were not analysed in these transgenic plants, which would determine if the host plant activates compensatory mechanisms in response to virus damage or if resource allocation modifications are due to the viral multiplication. Despite this uncertainty, our results support the hypothesis that life-history trait modifications are a defence mechanism in response to CMV infection. The modification of resource allocation in accession group 1 but not in accession group 2 correlates with the lower virulence of CMV on accession group 1 [34] and might partly explain the tolerance to CMV infection observed in this group of accessions as compared with those of group 2. Thus, when infected late in the life cycle, plants of group 1 suffer less from infection than plants from group 2 (data not shown), but when infected early during vegetative development, the growth of plants from group 1 is more severely reduced than the growth of plants from group 2, although tolerance results in a less severe effect of infection on progeny production. The accession group explained ∼5%–10% of the variance of the effect of infection on RW, IW, RW/IW ratio and SW, and a similar fraction of the variance of virulence (effect of infection on progeny production). These results strongly suggest that the differential CMV tolerance observed between both Arabidopsis allometric groups is due to the distinct plastic responses of resource allocation traits. Faster reproduction of infected plants of group 1, but not of group 2, appears also associated with tolerance to CMV. Ultimately, identification and characterization of the molecular mechanisms involved in the quantitative life-history trait modifications of Arabidopsis triggered by CMV (currently underway in our laboratory) will further shed light on the role of these responses in tolerance.
The increased reproduction investment in infected individuals of accession group 1 conforms to predictions for highly virulent parasites [16]–[18]. For various animal-parasite systems increase in reproductive effort has been reported, estimated as parental care [59],[60], mating effort [24] or progeny production [23],[61]. Increased reproductive effort has also been reported in animals under strong predation pressure [62]–[64]. The few published plant reports also suggest that an increase in reproduction effort is a general response of plants to environmental stress. For instance, Silene latifolia plants infected by the castrating anther fungus Microbotryum violaceum developed a higher number of flowers than healthy ones. Nevertheless, this was interpreted as a response induced by the fungus in order to increase its transmission success rather than as a host tolerance mechanism [29],[30]. Higher resource allocation to reproduction also has been reported in plants under herbivory or abiotic stress [65]–[68]. Accelerated reproduction is also in agreement with theoretical predictions of host responses to minimize fitness losses caused by highly virulent parasites [20],[21]. The reduction of RP in infected plants of accession group 1 is in concordance with experimental evidence from animal host-parasite systems on faster reproduction when parasitised [23],[24]. Faster reproduction reduces exposure time to parasite infection and optimises host fitness, since reproduction success will be higher when the time of exposure to the parasite is shorter [16],[17]. In agreement with this hypothesis, earlier reproduction has been reported for predator-prey systems [62],[69],[70] or in plants under abiotic stress [37],[45],[65],[67]. Nevertheless, the small reduction of RP in infected plants of group 1 suggests that it will play a minor role in their tolerance to CMV infection as compared to modifications of resource allocation.
A major question in the analysis of life-history trait evolution is whether observed plastic responses are genetically determined [7]. Most experimental reports are inconclusive because do not include different host genotypes [6],[7]. We found genotype-specific life-history trait responses to CMV infection with significant genetic variation (heritability), and plant genotypes explained the largest fraction of the observed life-history traits variance. A genetic control of life-history responses has been reported also in plants under abiotic stress [36]. In addition, we observed significant differences in life-history traits between two experiments, where plants were inoculated at different developmental stages. These experiments did not differ for growth and reproduction efforts and life-history schedules in mock-inoculated plants (unpublished data). Therefore, the variation observed between experiments in CMV infected plants further indicates an important effect of the developmental stage at infection on the life-history responses to CMV infection.
In conclusion, our results are in agreement with modifications of life-history traits reported for parasitised animals, and with predictions from life-history theory. Thus, we provide empirical support for the general validity of theoretical predictions. This experimental approach shows that the capacity to modify life histories depends on the host genotype, and allows estimating quantitatively the genetic determinism of life-history trait plasticity. In addition, we were able to evaluate more precisely the role of life-history trait modification in defence against parasites by taking into account plant/virus genotype combinations where life-history traits were differentially modified.
Materials and Methods
Viral isolates, Arabidopsis accessions, and inoculations
CMV isolates Fny-CMV, belonging to subgroup I of CMV strains, and LS-CMV, belonging to subgroup II have been described and were derived from biologically active cDNA clones [71],[72]. De72-CMV, belonging to subgroup I, was initially derived from a field-infected plant of Diplotaxis erucoides (Brassicaceae) [73]. Isolates were multiplied in tobacco plants, virions from tobacco leaves were purified as described in Lot et al. [74] and viral RNA was extracted by virion disruption with phenol and sodium dodecyl sulphate.
Eighteen wild genotypes (accessions) of Arabidopsis, described in Pagan et al. [34] (see Table S1), were selected to include a broad amount of natural genetic variation of the species in Eurasia and in the Iberian Peninsula, which has been suggested as a Pleistocene glacial refuge for Arabidopsis [75]. Accessions were kindly obtained from Maarten Koornneef (Max Planck Institute for Plant breeding, Cologne, Germany) or were kept in the laboratory of Carlos Alonso-Blanco (CNB-CSIC, Madrid, Spain). The 18 accessions were initially multiplied simultaneously under the same greenhouse conditions to minimise maternal effects. For experiments, seeds were sown on filter paper soaked with water in plastic Petri dishes, and stratified in darkness at 4°C for 3 days before transferring for germination to a growth chamber (22°C, 14 h light and 70% relative humidity). Five day-old seedlings were planted in soil containing pots 10.5 cm of diameter, 0.43 l volume and grown in a greenhouse (25/20°C day/night, 16 h light).
The experimental design is described in detail in Pagán et al. [34]. Briefly, each accession was inoculated with the three CMV isolates. Ten individual plants per treatment, including mock-inoculated controls, were grown in a greenhouse in a completely randomised design. Three rosette leaves per plant were mechanically inoculated with 5 µl of a 100 µg/ml suspension of purified CMV RNA when rosettes presented 4–5 leaves (stages 1.04–1.05 in Boyes et al. [40]). In a second experiment, plants were inoculated when the inflorescence started bolting (first flower bud visible, growth stage 5.0/5.1 as in Boyes et al. [40]). Overall results were similar in both experiments and therefore, only the results of the first one are shown.
Quantification of Arabidopsis life-history traits
Plants were harvested at complete senescence stage, and dry weight was determined after plants were maintained at 65°C until constant weight. The weights of rosettes (rosette weight, RW), inflorescence structures including seeds (inflorescence weight, IW) and seeds (seed weight, SW) were measured separately, and the above ground biomass (BM) was estimated as RW plus IW. Following Thompson and Stewart [43], rosette weight was used as an estimate of growth effort, inflorescence weight was taken as an estimate of total reproductive effort (reproductive structures plus seed output) and seed weight was used as an estimator of progeny production.
Two temporal parameters of Arabidopsis life cycle were quantified. Growth period span (GP) was measured as the time (days) elapsed between planting of seedlings on soil and opening of the first flower (stage 6.0 of Boyes et al. [40]). Reproductive period span (RP) was measured as the time (days) from the opening of the first flower to shattering of the first silique, which is the period dedicated to flower production (stage 8.0 of Boyes et al. [40]).
To quantify the effect of CMV infection on life-history traits, the value of each infected plant was divided by the mean value of the mock-inoculated plants of the same genotype.
Statistical analyses
RW, IW, SW and BM and their various transformations, were homocedastic and were analysed using ANOVA. Data on GP and RP showed heterogeneity of variances and therefore differences in GP and RP among CMV isolates or Arabidopsis accessions were also tested by Kruskal-Wallis test. Since ANOVA comparisons of these data led to similar results and conclusions than Kruskal-Wallis test, for simplicity, only ANOVA analyses are shown.
All traits were compared among CMV isolates or Arabidopsis accessions within each experiment by two-way ANOVA using accession and isolate as factors in a complete model. To determine if there are differences in the traits among experiments, a complete three-way ANOVA model was used including accession, isolate and experiment as factors. To test if viral infection affected differentially host life-history traits, a complete three-way ANOVA model was used including accession, isolate, and life-history trait as factors. Differences between allometric groups were analysed by two way ANOVA using isolates and groups as factors. Significance of differences among classes within each factor was determined by Least Significant Difference (LSD) analyses. Accession, isolate, experiment, life-history trait and allometric group were considered as random effect factors in all ANOVAs. For each trait, the percentage of total variance explained by each factor was calculated by variance component (VC) analyses in the corresponding models described above. All of these comparisons were done for the raw untransformed data, and for ratios and differences between values of infected and mock-inoculated plants. The three analyses lead to the same conclusions. As allometric relationships are usually expressed as ratios, we present only the results of analyses using this transformation.
Broad-sense heritability (h2b) of the traits was estimated as the percentage of the total variance accounted by genetic (among accession) variance (h2b = σ2 G /σ2 P, where σ2 G is the genetic variance and σ2 P is the total phenotypic variance). On all plant traits, σ2 P and σ2 G were derived as variance components from univariate analyses for each viral isolate.
Correlations between variables were tested using Pearson coefficients. Analysis involving non-parametric variables were also done using Kendall's robust test and Spearman's correlation test, showing similar results. Linear regression equations were compared using ANOVA to test equality of slopes and intercepts. All statistical analyses were done using the statistical software package SPSS 13.0 (SPSS Inc., Chicago, USA).
Supporting Information
Effect of CMV infection on rosette and inflorescence weight of Arabidopsis accessions. (A) Viral effect on rosette weight of plants estimated as RWi/RWm, where i and m denote infected and mock-inoculated plants, respectively. (B) Viral effect on inflorescence weight estimated as described for (A). Data are mean±standard errors of 10 replicates. The effect of infection is shown for LS-CMV (green), Fny-CMV (blue), and De72-CMV (red). Accessions are divided into allometry groups 1 and 2.
(1.82 KB TIF)
Origin of Arabidopsis thaliana accessions analysed in this work.
(35 KB DOC)
Statistical parameters of analysed host life-history traits.
(90 KB DOC)
Two-way ANOVAs of Arabidopsis life-history traits responses to CMV infection, using accession and virus isolate as factors.
(44 KB DOC)
Two-way ANOVAs of Arabidopsis life-history traits responses to CMV infection, using virus isolate and allometry group as factors.
(44 KB DOC)
Acknowledgments
Leticia Martín, Antolín López Quirós, and Begoña Prieto provided excellent technical assistance.
Footnotes
The authors have declared that no competing interests exist.
This work was supported in part by grants AGL2005-01122 from Plan Nacional de I+D and CAM-UPM CG06-UPM/AGR-429 from Universidad Politécnica de Madrid/Comunidad Autónoma de Madrid, Spain, to FGA. IP was in receipt of a FPI fellowship from Ministerio de Educación y Ciencia, Spain.
References
- 1.Tompkins DM, Greenman JV, Robertson PA, Hudson PJ. The role of shared parasites in the exclusion of wildlife hosts: Heterakis gallinarum in the ring-necked pheasant and the grey partridge. J Anim Ecol. 2000;69:829–840. doi: 10.1046/j.1365-2656.2000.00439.x. [DOI] [PubMed] [Google Scholar]
- 2.Read AF. The evolution of virulence. Trends Microbiol. 1994;2:73–76. doi: 10.1016/0966-842x(94)90537-1. [DOI] [PubMed] [Google Scholar]
- 3.Dobson AP, Hudson PJ. Parasites, disease and the structure of ecological communities. Trends Ecol Evol. 1986;1:11–15. doi: 10.1016/0169-5347(86)90060-1. [DOI] [PubMed] [Google Scholar]
- 4.Mitchell CE, Power AG. Pathogen spillover in disease epidemics. Am Nat. 2004;164:S79–S89. doi: 10.1086/424610. [DOI] [PubMed] [Google Scholar]
- 5.Hudson PJ, Dobson AP, Lafferty KD. Is a healthy ecosystem one that is rich in parasites? Trends Ecol Evol. 2006;21:381–385. doi: 10.1016/j.tree.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Agnew P, Koella JC, Michalakis Y. Host life-history responses to parasitism. Microbes Infect. 2000;2:891–896. doi: 10.1016/s1286-4579(00)00389-0. [DOI] [PubMed] [Google Scholar]
- 7.Michalakis Y, Hochberg ME. Parasitic effects on host life-history traits: a review of recent studies. Parasite. 1994;1:291–294. doi: 10.1051/parasite/1994014291. [DOI] [PubMed] [Google Scholar]
- 8.Koella JC, Agnew P. A correlated response of a parasite's virulence and life cycle to selection on its host's life-history. J Evol Biol. 1999;12:70–79. [Google Scholar]
- 9.Christie P, Richner H, Oppliger A. Begging, food provisioning, and nestling competition in great tit broods infested with ectoparasites. Behav Ecol. 1996;7:127–131. [Google Scholar]
- 10.Sorci G, Morand S, Hugot J-P. Host-parasite coevolution: comparative evidence for covariation of life history traits in primates and oxyurid parasites. Proc R Soc Lond B. 1997;264:285–289. doi: 10.1098/rspb.1997.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorensen RE, Minchella DJ. Parasite influences on host life-history: Echinostoma revolutum parasitism of Lymnaea elodes snails. Oecologia. 1998;115:188–195. doi: 10.1007/s004420050507. [DOI] [PubMed] [Google Scholar]
- 12.Arnott SA, Barber I, Huntingford FA. Parasite-associated growth enhancement in a fish-cestode system. Proc R Soc Lond B. 2000;267:657–663. doi: 10.1098/rspb.2000.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stearns SC. Life-history tactics: A review of the ideas. Q Rev Biol. 1976;51:3–47. doi: 10.1086/409052. [DOI] [PubMed] [Google Scholar]
- 14.van Noordwijk AJ, de Jong G. Acquisition and allocation of resources: their influence on variation in life-history tactics. Am Nat. 1986;128:137–142. [Google Scholar]
- 15.Williams GC. Princeton: Princeton University Press; 1966. Adaptation and natural selection. p. 320. [Google Scholar]
- 16.Minchella DJ. Host life-history variation in response to parasitism. Parasitology. 1985;90:205–216. [Google Scholar]
- 17.Forbes MRL. Parasitism and host reproductive effort. Oikos. 1993;67:444–450. [Google Scholar]
- 18.Perrin N, Christe P. On host life-history response to parasitism. Oikos. 1996;75:317–320. [Google Scholar]
- 19.Williams GC. Pleiotropy, natural selection, and the evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]
- 20.Hochberg ME, Michalakis Y, de Meeus T. Parasitism as a constraint on the rate of life-history evolution. J Evol Biol. 1992;5:491–504. [Google Scholar]
- 21.Gandon S, Agnew P, Michalakis Y. Coevolution between parasite virulence and host life-history traits. Am Nat. 2002;160:374–388. doi: 10.1086/341525. [DOI] [PubMed] [Google Scholar]
- 22.Fredensborg BL, Poulin R. Parasitism shaping host life-history evolution: adaptive responses in a marine gastropod to infection by trematodes. J Anim Ecol. 2006;75:44–53. doi: 10.1111/j.1365-2656.2005.01021.x. [DOI] [PubMed] [Google Scholar]
- 23.Chadwick W, Little T. A parasite-mediated life-history shift in Daphnia magna. Proc R Soc Lond B. 2005;272:505–509. doi: 10.1098/rspb.2004.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polak M, Starmer WT. Parasite-induced risk of mortality elevates reproductive effort in male Drosophila. Proc R Soc Lond B. 1998;265:2197–2201. doi: 10.1098/rspb.1998.0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agnew P, Bedhomme S, Haussy C, Michalakis Y. Age and size at maturity of the mosquito Culex pipiens infected by the microsporidian parasite Vavraia culicis. Proc R Soc Lond B. 1999;266:947–952. [Google Scholar]
- 26.Thompson SN, Redak RA, Wang L-W. Nutrition interacts with parasitism to influence growth and physiology of the insect Manduca sexta L. J Exp Biol. 2005;208:611–623. doi: 10.1242/jeb.01404. [DOI] [PubMed] [Google Scholar]
- 27.Restif O, Koella JC. Concurrent evolution of resistance and tolerance to pathogens. Am Nat. 2004;164:90–102. doi: 10.1086/423713. [DOI] [PubMed] [Google Scholar]
- 28.Lambrechts L, Fellous S, Koella JC. Coevolutionary interactions between host and parasite genotypes. Trends Parasitol. 2006;22:12–16. doi: 10.1016/j.pt.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 29.Shykoff JA, Kaltz O. Effects of the anther smut fungus Microbotryum violaceum on host life-history patterns in Silene latifolia (Caryophyllaceae). Int J Plant Sci. 1997;158:164–171. [Google Scholar]
- 30.Shykoff JA, Kaltz O. Phenotypic changes in host plants diseased by Microbotryum violaceum: parasite manipulation, side effects, and trade-offs. Int J Plant Sci. 1998;159:236–243. [Google Scholar]
- 31.Somerville C, Koornneef M. A fortunate choice: The history of Arabidopsis as a model plant. Nat Rev Genet. 2002;3:883–889. doi: 10.1038/nrg927. [DOI] [PubMed] [Google Scholar]
- 32.Mysore KS, Ryu C-M. Nonhost resistance: how much do we know? Trends Plant Sci. 2004;9:97–104. doi: 10.1016/j.tplants.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Salvaudon L, Héraudet V, Shykoff J. Parasite-host fitness trade-offs change with parasite identity: genotype-specific interactions in a plant-pathogen system. Evolution. 2005;59:2518–2524. [PubMed] [Google Scholar]
- 34.Pagán I, Alonso-Blanco C, García-Arenal F. The relationship of within-host multiplication and virulence in a plant-virus system. PLoS ONE. 2007;2:e786. doi: 10.1371/journal.pone.0000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonser S, Aarssen LW. Allometry and plasticity of meristem allocation throughout development in Arabidopsis thaliana. J Ecol. 2001;89:72–79. [Google Scholar]
- 36.Pigliucci M, Kolodynska A. Phenotypic plasticity to light intensity in Arabidopsis thaliana: invariance of reaction norms and phenotypic integration. Evol Ecol. 2002;16:27–47. [Google Scholar]
- 37.Pigliucci M, Kolodynska A. Phenotypic integration and response to stress in Arabidopsis thaliana: a path analytical approach. Evol Ecol Res. 2006;8:415–433. [Google Scholar]
- 38.Ausin I, Alonso-Blanco C, Martínez-Zapater JM. Environmental regulation of flowering. Int J Dev Biol. 2005;49:689–705. doi: 10.1387/ijdb.052022ia. [DOI] [PubMed] [Google Scholar]
- 39.Diggle PK. Architectural effects and the interpretation of patterns of fruit and seed development. Annu Rev Ecol Syst. 1995;26:531–552. [Google Scholar]
- 40.Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, et al. Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell. 2001;13:1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palukaitis P, Roossinck MJ, Dietzgen RG, Francki RIB. Cucumber mosaic virus. . Adv Virus Res. 1992;41:281–348. doi: 10.1016/s0065-3527(08)60039-1. [DOI] [PubMed] [Google Scholar]
- 42.Palukaitis P, García-Arenal F. Cucumoviruses. Adv Virus Res. 2003;62:241–323. doi: 10.1016/s0065-3527(03)62005-1. [DOI] [PubMed] [Google Scholar]
- 43.Thompson K, Stewart AJA. The measurement and meaning of reproductive effort in plants. Am Nat. 1981;117:205–211. [Google Scholar]
- 44.Stearns SC. Oxford: Oxford University Press; 1992. The evolution of life histories. p. 262. [Google Scholar]
- 45.Stanton ML, Roy BA, Thiede DA. Evolution in stressful environments. I. Phenotypic variability, phenotypic selection, and response to selection in five distinct environmental stresses. Evolution. 2000;54:93–111. doi: 10.1111/j.0014-3820.2000.tb00011.x. [DOI] [PubMed] [Google Scholar]
- 46.Jeger M, Seal S, van der Bosch F. Evolutionary epidemiology of plant virus diseases. Adv Virus Res. 2006;67:163–203. doi: 10.1016/S0065-3527(06)67005-X. [DOI] [PubMed] [Google Scholar]
- 47.Mitchell-Olds T. Pleiotropy causes long-term genetic constraints on life-history evolution in Brassica rapa. Evolution. 1996;50:1849–1858. doi: 10.1111/j.1558-5646.1996.tb03571.x. [DOI] [PubMed] [Google Scholar]
- 48.Pigliucci M, Schlichting CD. Reaction norms of Arabidopsis. V. Flowering time controls phenotypic architecture in response to nutrient stress. J Evol Biol. 1998;11:285–301. [Google Scholar]
- 49.Baker AM, Burd M, Climie KM. Flowering phenology and sexual allocation in single-mutation lineages of Arabidopsis thaliana . Evolution. 2005;59:970–978. [PubMed] [Google Scholar]
- 50.Strickland EH. Some parasites of Simulium larvae and their effects on the development of the host. Biol Bull. 1911;21:302–338. [Google Scholar]
- 51.Mitchell MJ, Cali A. Vairimorpha necatrix (Microsporida: Burenellidae) affects growth and development of Heliothis zea (Lepidoptera: Noctuidae) raised at various temperatures. J Econ Entomol. 1994;87:933–940. [Google Scholar]
- 52.Blaser M, Schmid-Hempel P. Determinants of virulence for the parasite Nosema whitei in its host Tribolium castaneum. J Invertebr Pathol. 2005;89:251–257. doi: 10.1016/j.jip.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 53.Ballabeni P. Parasite-induced gigantism in a snail: A host adaptation? Funct Ecol. 1995;9:887–893. [Google Scholar]
- 54.Dunoyer P, Lecellier CH, Abreu Parizotto E, Himber C, Voinnet O. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell. 2004;16:1235–1250. doi: 10.1105/tpc.020719. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Fagoaga C, López C, Moreno P, Navarro L, Flores R, et al. Viral-like symptoms induced by the ectopic expression of the p23 gene of citrus tristeza virus are citrus specific and do not correlate with the pathogenicity of the virus strain. Mol Plant Microbe Int. 2005;18:435–445. doi: 10.1094/MPMI-18-0435. [DOI] [PubMed] [Google Scholar]
- 56.Geri C, Love AJ, Cecchini E, Barrett SJ, Laird J, et al. Arabidopsis mutants that suppress the phenotype induced by transgene-mediated expression of cauliflower mosaic virus (CaMV) gene VI are less susceptible to CaMV-infection and show reduced ethylene sensitivity. Plant Mol Biol. 2004;56:111–124. doi: 10.1007/s11103-004-2649-x. [DOI] [PubMed] [Google Scholar]
- 57.Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, et al. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev Cell. 2003;4:205–217. doi: 10.1016/s1534-5807(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 58.Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, et al. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Gene Dev. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richner H, Christe P, Oppliger A. Paternal investment affects prevalence of malaria. Proc Natl Acad Sci U S A. 1995;92:1192–1194. doi: 10.1073/pnas.92.4.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hurtrez-Boussès S, Blondel J, Perret P, Fabreguettes J, Renaud F. Chick parasitism by blowflies affects feeding rates in a Mediterranean population of blue tits. Ecol Lett. 1998;1:17–20. [Google Scholar]
- 61.Minchella DJ, Loverde PT. A cost of increased early reproductive effort in the snail Biomphalaria glabrata. Am Nat. 1981;118:876–881. [Google Scholar]
- 62.Stibor H. Predator induced life-history shifts in a fresh-water cladoceran. Oecologia. 1992;92:162–165. doi: 10.1007/BF00317358. [DOI] [PubMed] [Google Scholar]
- 63.Jennions MD, Telford SR. Life-history phenotypes in populations of Brachyrhaphis episcopi (Poeciliidae) with different predator communities. Oecologia. 2002;132:44–50. doi: 10.1007/s00442-002-0942-4. [DOI] [PubMed] [Google Scholar]
- 64.Testa JW. Population dynamics and life-history trade-offs of moose (Alces alces) in south central Alaska. Ecology. 2004;85:1439–1452. [Google Scholar]
- 65.Day TA, Ruhland CT, Grobe CW, Xiong F. Growth and reproduction of Antarctic vascular plants in response to warming and UV radiation reductions in the field. Oecologia. 1999;119:24–35. doi: 10.1007/s004420050757. [DOI] [PubMed] [Google Scholar]
- 66.Becklin KM, Kirkpatrick E. Compensation through rosette formation: the response of scarlet gilia (Ipomopsis aggregata: Polemoniaceae) to mammalian herbivory. Can J Bot. 2006;84:1298–1303. [Google Scholar]
- 67.Stenstrom A, Jonsdottir IS. Effects of simulated climate change on phenology and life history traits in Carex bigelowii. Nord J Bot. 2006;24:355–371. [Google Scholar]
- 68.Brody AK, Price MV, Waser NM. Life-history consequences of vegetative damage in scarlet gilia, a monocarpic plant. Oikos. 2007;116:975–985. [Google Scholar]
- 69.Reznick DA, Endler JA. The impact of predation on life-history evolution in trinidarian guppies (Poecilia reticulata). Evolution. 1982;36:160–177. doi: 10.1111/j.1558-5646.1982.tb05021.x. [DOI] [PubMed] [Google Scholar]
- 70.Relyea RA. Getting out alive: how predators affect the decision to metamorphose. Oecologia. 2007;152:389–400. doi: 10.1007/s00442-007-0675-5. [DOI] [PubMed] [Google Scholar]
- 71.Rizzo TM, Palukaitis P. Construction of full-length cDNA of Cucumber mosaic virus RNAs 1, 2 and 3: generation of infectious RNA transcripts. Mol Gen Genet. 1990;222:249–256. doi: 10.1007/BF00633825. [DOI] [PubMed] [Google Scholar]
- 72.Zhang L, Hanada K, Palukaitis P. Mapping local and systemic symptom determinants of cucumber mosaic cucumovirus in tobacco. J Gen Virol. 1994;75:3185–3191. doi: 10.1099/0022-1317-75-11-3185. [DOI] [PubMed] [Google Scholar]
- 73.Bonnet J, Fraile A, Sacristán S, Malpica JM, García-Arenal F. Role of recombination in the evolution of natural populations of Cucumber mosaic virus, a tripartite RNA plant virus. Virology. 2005;332:359–368. doi: 10.1016/j.virol.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 74.Lot H, Marrou J, Quiot JB, Esvan C. Contribution à l'étude du virus de la mosaïque du concombre (CMV). Méthode de purification rapide du virus. Ann Phytopathol. 1972;4:25–38. [Google Scholar]
- 75.Sharbel TF, Haubold B, Mitchell-Olds T. Genetic isolation by distance in Arabidopsis thaliana: biogeography and postglacial colonization of Europe. Mol Ecol. 2000;9:2109–2118. doi: 10.1046/j.1365-294x.2000.01122.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect of CMV infection on rosette and inflorescence weight of Arabidopsis accessions. (A) Viral effect on rosette weight of plants estimated as RWi/RWm, where i and m denote infected and mock-inoculated plants, respectively. (B) Viral effect on inflorescence weight estimated as described for (A). Data are mean±standard errors of 10 replicates. The effect of infection is shown for LS-CMV (green), Fny-CMV (blue), and De72-CMV (red). Accessions are divided into allometry groups 1 and 2.
(1.82 KB TIF)
Origin of Arabidopsis thaliana accessions analysed in this work.
(35 KB DOC)
Statistical parameters of analysed host life-history traits.
(90 KB DOC)
Two-way ANOVAs of Arabidopsis life-history traits responses to CMV infection, using accession and virus isolate as factors.
(44 KB DOC)
Two-way ANOVAs of Arabidopsis life-history traits responses to CMV infection, using virus isolate and allometry group as factors.
(44 KB DOC)