Abstract
The eukaryotic translation initiation factor 5A (eIF5A) is the only cellular protein that contains the unique polyamine-derived amino acid, hypusine [Nε-(4-amino-2-hydroxybutyl) lysine]. Hypusine is formed in eIF5A by a novel post-translational modification reaction that involves two enzymatic steps. In the first step, deoxyhypusine synthase catalyzes the cleavage of the polyamine spermidine and transfer of its 4-aminobutyl moiety to the ε-amino group of one specific lysine residue of the eIF5A precursor to form a deoxyhypusine intermediate. In the second step, deoxyhypusine hydroxylase converts the deoxyhypusine-containing intermediate to the hypusine-containing mature eIF5A. The structure and mechanism of deoxyhypusine synthase have been extensively characterized. Deoxyhypusine hydroxylase is a HEAT-repeat protein with a symmetrical superhelical structure consisting of 8 helical hairpins (HEAT motifs). It is a novel metalloenzyme containing tightly bound iron at the active sites. Four strictly conserved His-Glu pairs were identified as iron coordination sites. The structural fold of deoxyhypusine hydroxylase is entirely different from those of the other known protein hydroxylases such as prolyl 4-hydroxylase and lysylhydroxylases. The eIF5A protein and deoxyhypusine/hypusine modification are essential for eukaryotic cell proliferation. Thus, hypusine synthesis represents the most specific protein modification known to date, and presents a novel target for intervention in mammalian cell proliferation.
Keywords: deoxyhypusine, deoxyhypusine hydroxylase, deoxyhypusine synthase, eIF5A, hypusine, post-translational modification, spermidine
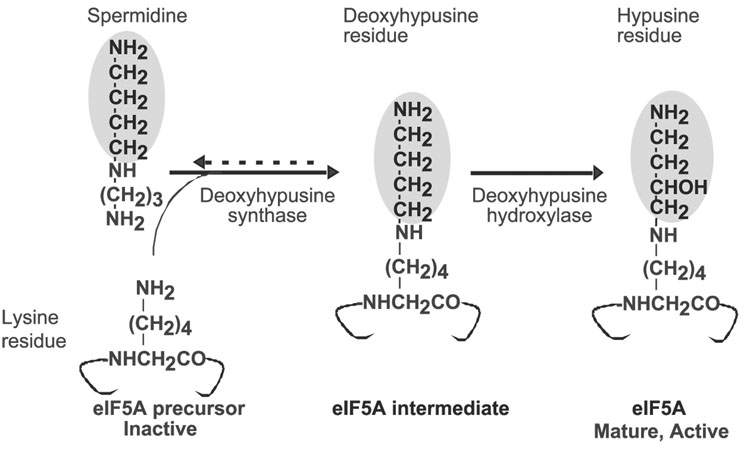
Polyamines can be covalently incorporated into proteins through the transglutaminase reaction and by hypusine biosynthesis. In the latter, the polyamine, spermidine, is utilized for the post-translational modification of a single cellular protein, the precursor of eukaryotic translation initiation factor 5A (eIF5A), to form a unique amino acid, hypusine [Nε-(4-amino-2-hydroxybutyl)lysine] (1–4). Hypusine was originally discovered and identified by Shiba et al. and was named “hypusine” based on its structure, which consists of two moieties, hydroxyputrescine and lysine (5). In the course of a study aimed at the identification of a cellular protein substrate of trans-glutaminase, we discovered that hypusine is, in fact, formed from spermidine in two enzymatic steps (6) (Fig. 1): in the first step, deoxyhypusine synthase [EC 2.5.1.46] catalyzes the transfer of the 4-amino butyl moiety of spermidine to the ε-amino group of one specific lysine residue (Lys50 for the human protein) of the eIF5A precursor to form a deoxyhypusine residue (7–9) This intermediate is subsequently hydroxylated by deoxyhypusine hydroxylase [EC 1.14.99.29] to complete hypusine synthesis and eIF5A maturation (10). The biosynthesis of hypusine defines a specific and critical function of polyamines in cell proliferation (2, 3, 11–13).
Fig. 1. Scheme of hypusine biosynthesis in eIF5A.
The 4-aminobutyl moiety transferred from spermidine to the eIF5A precursor is shaded.
eIF5A is a small, acidic protein that is highly conserved throughout eukaryotes. Sequence conservation is especially high around the hypusine residue, underscoring the importance of this unusual protein modification throughout eukaryotic evolution. The early observation of a correlation between hypusine synthesis and cell growth suggested an important role for hypusine in cell proliferation (14–16). Inhibitors of deoxyhypusine synthase and deoxyhypusine hydroxylase cause growth arrest in various mammalian cells (17, 18). Furthermore, the cytostasis observed in spermidine-deprived cells after treatment with an inhibitor of S-adenosylmethionine decarboxylase was attributed to depletion of hypusine-containing eIF5A (11). Direct evidence for the essential nature of eIF5A and its modification derives from gene inactivation studies in Saccharomyces cerevisiae. Disruption of the eIF5A genes (TIF51A and TIF51B) (19, 20), or of the deoxyhypusine synthase gene (21, 22), produces a lethal phenotype, indicating the importance of the deoxyhypusine modification in the viability of eukaryotic cells. The second step, deoxyhypusine hydroxylation, was also presumed to be essential, based on cell growth inhibition by inhibitors of this enzyme. Recent cloning and identification of the deoxyhypusine hydroxylase gene (23) provide new insights into the evolutionary progression of eIF5A protein and its modification enzymes.
Although eIF5A is intimately involved in eukaryotic cell proliferation, the true physiological function of this essential factor has yet to be elucidated. eIF5A stimulates methionyl-puromycin synthesis (24), in a model assay for the first peptide bond formation. eIF5A may be a bimodular protein interacting with both RNA and proteins, and is presumed to have an important role in the translation machinery. However, a rapid depletion of genetically engineered unstable eIF5A in S. cerevisiae resulted in only a modest decrease in total protein synthesis (25). This finding led to a proposal that eIF5A is an initiation factor specific for a subset of mRNA’s (25, 26). Additional proposed functions of eIF5A include: a cellular cofactor of HIV-1 REV (27), a factor involved in nuclear export (28) and mRNA turnover (29, 30). Recent studies with temperature-sensitive mutants and their suppressors point to a role of eIF5A in cell wall integrity, and actin polarity (31). It is not clear whether the various observed phenotypes are direct or indirect consequence of eIF5A depletion or dysfunction and how the various effects are interrelated. It is possible that eIF5A is a multifunctional protein involved in several critical cellular processes.
Evolution of eIF5A, deoxyhypusine synthase and deoxyhypusine hydroxylase
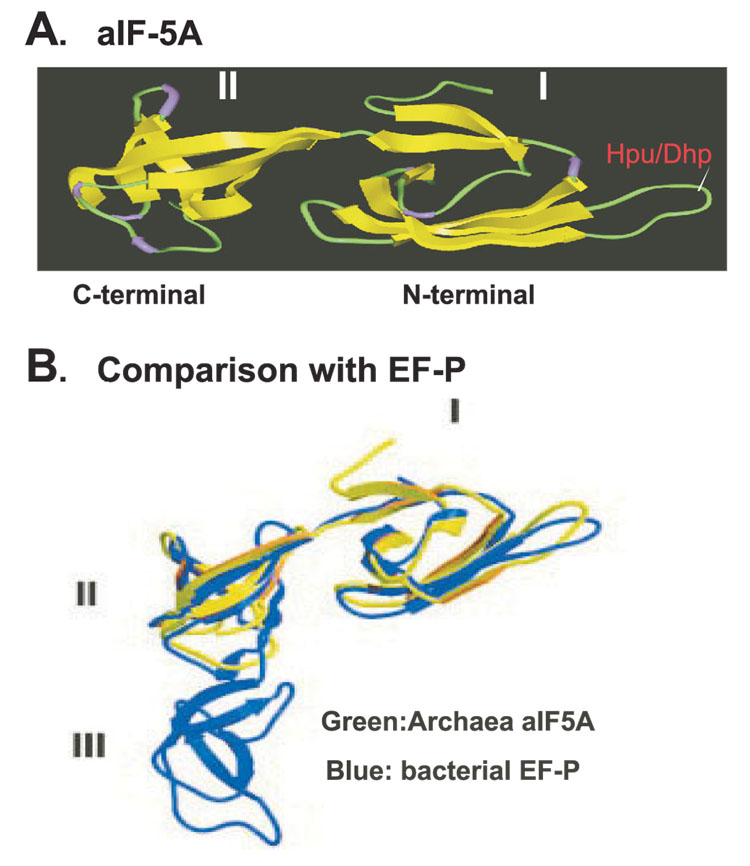
Hypusine is found in all eukaryotes, and in certain archaea, but has not thus far been detected in eubacteria (2, 3, 32). On this ground, eIF5A and hypusine synthesis had been considered a eukaryote-specific event. However, bacteria do contain a distant homolog of eIF5A, elongation factor P (EF-P), with significant sequence and structural similarities (33). EF-P is an essential protein in bacteria that stimulates the peptidyl transferase activity of ribosomes (34). Like eIF5A, it stimulates methionyl-puromycin synthesis in vitro, suggesting even a functional conservation between bacterial EF-P and eIF5A. The structure of eIF5A is predicted to be similar to those of archaeal IF5A (aIF5A)(Fig.2).X-ray structures of aIF5A (35) reveal that this protein consists of two well-defined domains: the N-terminal domain, which contains the hypusine/deoxyhypusine modification site in an exposed loop, and the C-terminal domain, which is similar to the oligonucleotide-binding domain found in several RNA-binding proteins (Fig. 2). EF-P consists of three domains, N-terminal domain I and two repetitive C-terminal domains II and III, and the overall structure resembles tRNA (33). The structures of domains I and II of EF-P are super imposable on the N- and C- terminal domains, respectively, of the aIF5A (Fig. 2). Thus, it seems highly likely that eIF5A evolved from a bacterial elongation factor, EF-P.
Fig. 2. Comparison of crystal structures of aIF5A and EF-P.
A: X-ray structure of archaeal IF5A precursor containing two domains: N-terminal domain (I) and C-terminal domain (II), The location of the lysine residue (in domain I) that undergoes modification to deoxyhypusine or hypusine is indicated. B: Structure of bacterial elongation factor P (EF-P) containing three domains (I–III) in blue, aIF5A in green is superimposed on domains I and II of EF-P. Abbreviations: Hpu, hypusine; Dhp, deoxyhypusine. Panel B reproduced with permission from Ref. 33. Copyright (2004) National Academy of Sciences, USA.
Two or more eIF5A genes have been identified in many eukaryotic organisms, including fungi, plants, vertebrates, and mammals (3, 36). In the yeast S. cerevisiae, the two EIF5A genes, TIF51A and TIF51B, encode two isoforms of 92% identity. Each gene has evolved to become differentially expressed, such that TIF51A (aerobic gene) and TIF51B (anaerobic gene) are reciprocally regulated by oxygen (37). In certain vertebrates, including fish, amphibians and chicken, the two EIF5A genes seem to be co-expressed. In contrast, most mammalian cells and tissues normally express predominantly the EIF5A1 gene and only one isoform protein eIF5A1. eIF5A-2 mRNA is found only in certain tissues, such as testis and brain, suggesting a tissue-specific expression of the EIF5A2 gene. That two protein isoforms have been maintained in mammals, while the respective mRNA sequences have diverged, suggests a differentiated function of eIF5A2 in mammals. A high amplification of the EIF5A2 gene was reported in human ovarian cancer tissues and cells (38), and eIF5A2 isoform proteins are detected in human cancer cell lines, an ovarian cancer line, UACC1598, and a colorectal cancer line, SW-480 (36). The EIF5A2 gene was proposed as a candidate oncogene in ovarian cancer, based on a common amplification of this gene (3q26 locus) and on transformation of the human hepatoma cell line LO2 by over-expression of eIF5A2 (39). The potential role of eIF5A2 in human cancer and the mechanism of regulation of its expression need further investigation.
The emergence of the eIF5A precursor and the hypusine biosynthetic enzymes shows an interesting evolutionary progression. eIF5A (hypusine form) occurs in all eukaryotes examined, consistent with the presence of genes encoding eIF5A (two or more genes), deoxyhypusine synthase (one gene), and deoxyhypusine hydroxylase (one gene). It has been proposed that in certain plants, a homospermidine synthase evolved from deoxyhypusine synthase by gene duplication (40). eIF5A and DHS are seen in all archaeal proteomes suggesting that the first step of modification (lysine residue to deoxyhypusine residue) is likely essential in archaea. However, no orthologs of the eukaryotic DOHH exist in the available archaeal proteomes. Various archaea were reported to contain either deoxyhypusine or hypusine, or a mixture of both, depending on the branch (32). If hypusine indeed occurs in certain branches of archaea, it is not clear how it was produced, given that no DOHH homolog is found in archaea. Neither hypusine, nor deoxyhypusine has been detected in bacteria. Consistent with this, most commonly studied bacteria lack both the DHS and DOHH genes. However, several cyanobacteria, myxobacteria, Thermus, Chlorobium, Rhodopirellula, Bdellovibrio, Zymomonas, Caulobacter and Cytophaga contain a DHS cognate that appears to have been laterally transferred from the archaea. It is not known if EF-P can be modified through a DHS cognate–mediated reaction in these bacteria.
Gene disruption studies demonstrated the essential nature of the eIF5A and DHS genes in S. cerevisiae and in higher eukaryotes (19–22). However, the single DOHH gene, YJR070C, does not appear to be essential in S. cerevisiae, since the DOHH null strain grows only slightly more slowly than its parent strain (23). DOHH seems to be functionally more significant in the fission yeast, S. pombe. A E66K mutation in Mmd1p, the S. pombe DOHH homolog, caused a temperature-sensitive growth and abnormal distribution and morphology of mitochondria at non-permissive temperature (41). These phenotypes were attributed to mmd1 mutant protein defective in promoting microtubule assembly at the non-permissive temperature. These findings suggest a role for DOHH or eIF5A (the hypusine-containing form) in microtubule stability and function in S. pombe. In higher organisms, i.e. C. elegans (42) or Drosophila melanogaster (43), inactivation of the DOHH gene is recessively lethal. Thus, the essentiality of the final step of hypusine modification has probably evolved only in multicellular eukaryotes.
Mechanism of dseoxyhypusine synthase (DHS) reaction
Deoxyhypusine synthase catalyzes a complex sequence of reactions, involving two substrates, spermidine and eIF5A(Lys), and a cofactor, NAD, to convert one specific lysine residue of the eIF5A precursor to a deoxyhypusine residue. This enzyme exhibits an absolute specificity toward its protein substrate, eIF5A(Lys), and also a very narrow specificity toward spermidine. Studies with eIF5A-derived polypeptides with stepwise truncation, from either the N- or the C-terminus, revealed that a large eIF5A polypeptide (larger than aa30-aa80) is required for its substrate function (44). Of the various spermidine analogs tested, only the closely related compounds, homospermidine, aminopropyl cadaverine, cis- and trans-unsaturated spermidine, and N8-methyl- and N8-ethyl spermidines act as donor substrates for DHS, but caldine or N1-methyl spermidine do not (45).
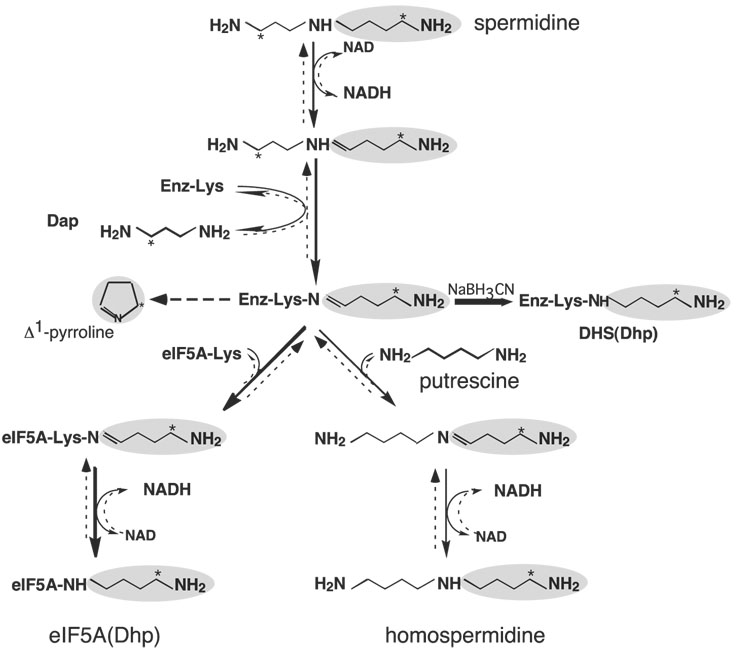
The deoxyhypusine synthesis occurs in four steps [Fig. 3, steps I–IV, solid arrows from Spd to eIF5A(Dhp)] (46): (i) NAD-dependent dehydrogenation of spermidine to dehydrospermidine, (ii) cleavage and transfer of the 4-aminobutyl moiety from dehydrospermidine to the ε-amino group of an active site lysine residue (Lys329 for the human enzyme) to form a covalent enzyme-imine intermediate, (iii) transfer of the 4-aminobutyl moiety from the enzyme intermediate to the ε-amino group of a specific lysine residue of eIF5A precursor (Lys 50 for the human protein) to form an eIF5A-imine intermediate, and (iv) reduction of this intermediate by enzyme-bound NADH to form a deoxyhypusine residue. The involvement of a transient enzyme-imine intermediate was demonstrated by trapping it into a stable enzyme-substrate adduct after NaBH3CN reduction of the mixture containing the enzyme, [1,8-3H]spermidine and NAD, but no eIF5A(Lys). The labeled component of the enzyme-adduct was identified as deoxyhypusine, indicating that the acceptor of the labeled 4-butyl amine moiety was a lysine residue of the enzyme. The site of this enzyme-imine intermediate formation was identified as Lys329 for the human DHS by amino acid sequencing of the labeled tryptic peptide. The critical role of this residue in catalysis was confirmed by site-directed mutagenesis studies (47). DHS mutant enzymes with Lys329 substituted with either Arg or Ala were totally inactive, although they were capable of binding eIF5A precursor protein. Furthermore, the S. cerevisiae DHS mutant enzyme with the active site Lys350 substituted with Arg could not support the growth of S. cerevisiae DHS null strain (22).
Fig. 3. Mechanism of the deoxyhypusine synthase reaction.
The pathways from spermidine leading to deoxyhypusine synthesis and to homospermidine synthesis are indicated by solid arrows. The reversal pathways from eIF5A(Dhp) back to spermidine and from homospermidine to spermidine are indicated by broken arrows. The 4-aminobutyl moiety is shaded and the position of [3H] derived from [1,8-8H]spermidine is indicated by *. In the absence of acceptor, the 4-aminobutyl moiety of the enzyme-imine intermediate is cyclized to form Δ1-pyrroline. The reduction of the transient enzymeimine intermediate to a stable adduct by NaCNBH3 is indicated by a bold arrow. Abbreviations are: Dap, 1,3-diaminopropane; DHS(Dhp): deoxyhypusine-containing deoxyhypusine synthase. Modified from Ref. 45.
Reversibility of the DHS reaction and homospermidine synthesis activity of DHS
Like the plant deoxyhypusine synthase from tobacco and Senecio vernalis (48), human DHS can accommodate putrescine as an alternative butylamine acceptor instead of eIF5A(Lys) resulting in the formation of homospermidine from spermidine (Fig. 3, solid arrows from Spd to homospermidine) (45). However, the Km for putrescine (1.12 mM) is much higher than that for eIF5A(Lys) (1.5 µM) (values for human DHS), indicating that deoxyhypusine synthesis is the preferred pathway of the DHS reaction. Furthermore, all the DHS-catalyzed reactions are reversible and DHS can facilitate interconversion of spermidine, eIF5A(Dhp) and homospermidine by way of a common enzyme-imine intermediate (Fig. 3) (45). When deoxyhypusine-containing eIF5A (with a radiolabel in the 4-aminobutyl moiety of deoxyhypusine residue) was incubated with DHS, NAD and 1,3-diaminopropqne, radioactive spermidine was formed by an efficient transfer of the 4-aminobutyl moiety from the deoxyhypusine residue back to 1,3-diaminopropane by way of the same enzyme-imine intermediate [Fig. 3, broken arrows from eIF5A (Dhp) to Spd]. When putrescine was added in place of 1,3-diaminopropane, radiolabeled homospermidine was generated [from eIF5A(Dhp) to homospermidine]. Thus, the aminobutyl moiety of the enzyme-imine intermediate can be transferred to any one of the three acceptors, eIF5A(Lys), putrescine or 1,3-diaminopropane leading to the synthesis of deoxyhypusine, homospermidine or spermidine, respectively. In contrast to the deoxyhypusine-containing protein, no reversal was observed with hypusine-containing eIF5A, suggesting that hydroxylation at the 4-aminobutyl side chain of the deoxyhypusine residue prevents DHS-mediated reversal of the modification. Whereas the first step of hypusine synthesis is reversible, the second step, DOHH-mediated hydroxylation, locks eIF5A into an active hypusine form, thereby making the overall reaction an irreversible protein modification.
X-ray structure of human recombinant DHS and DHS inhibitor studies
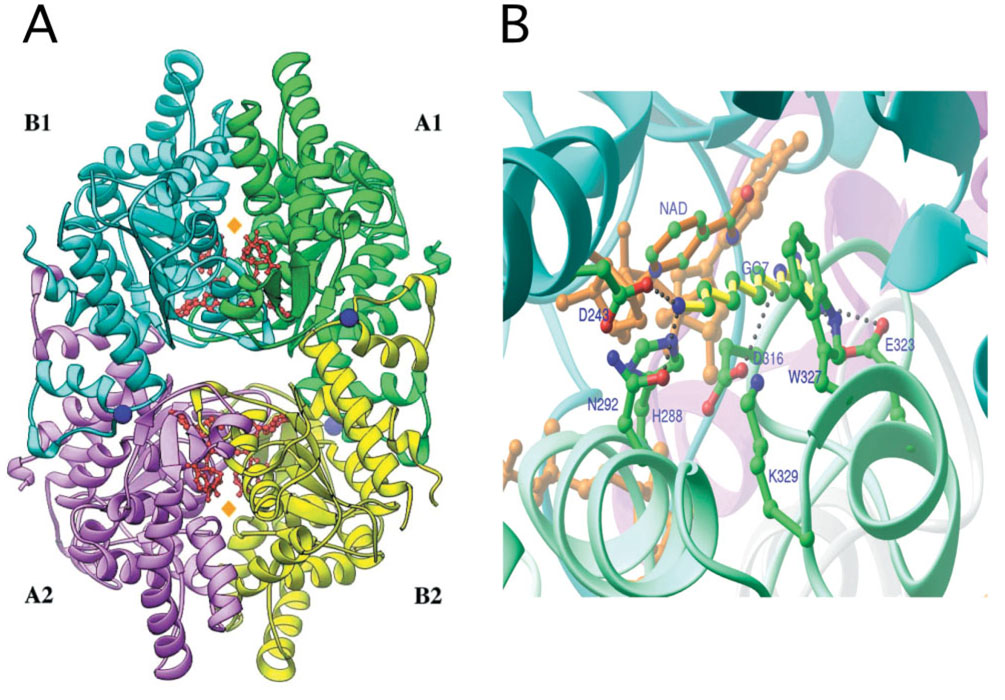
Crystal structures of human recombinant deoxyhypusine synthase have been determined for its complex with the cofactor NAD (Form I) (49), and its ternary complex with NAD and an inhibitor, GC7, (Form II) (50) (Fig. 4). Human DHS is a tetrameric enzyme composed of four identical subunits of 40 kDa. Each subunit contains a nucleotide-binding (Rossmann) fold. The tetramer is comprised of two tightly associated dimers and contains four spermidine-binding sites, two in each dimer interface. In the Form I crystals, which were obtained under conditions of acidic pH and high ionic strength, the entrance to the substrate-binding active sites was blocked by the N-terminal two-turn α helix (ball-and-chain motif), suggesting an inactive form of the enzyme. The structure of the Form II crystal grown under physiological conditions (at low ionic strength and at close to neutral pH) probably represents an active enzyme. In this form, the ball-and-chain motif could not be detected in the electron density, suggesting that it swings freely and no longer blocks the active site entrance. The competitive inhibitor, GC7, was bound in the deep tunnel of the spermidine-binding site. The topology of the active site provides a basis for the development of structure-based inhibitors.
Fig. 4. Tetrameric structure of DHS in complex with NAD (A) and active site of DHS with NAD and GC7 bound (B).
Modified from Ref. 50.
Prior to the determination of the DHS structure, the spermidine-binding site of DHS was probed with a series of diamine and polyamine analogs to identify strong inhibitors of DHS (51). These studies revealed a requirement for two basic moieties separated by 7–8 methylenes for effective binding to the active site of the enzyme. Derivatives with bulky groups in the methylene backbone or secondary amino group were ineffective, indicating a narrow groove for the spermidine-binding pocket. Of numerous spermidine analogs synthesized and tested, N1-guanyl-1,7-diaminoheptane (GC7) and its methyl derivative were the most potent inhibitors of deoxyhypusine synthase in vitro and in cultured cells (17, 52). GC7 caused arrest of proliferation in various mammalian cells, including a panel of human cancer cell lines. Although GC7 does not seem to interfere with other enzymes of polyamine biosynthesis and metabolism and DHS is the only known target for GC7, the possibility that GC7 exerts other deleterious effects on cell growth cannot be excluded.
Cloning and expression of deoxyhypusine hydroxylase (DOHH)
We recently cloned the DOHH gene (23) by screening a S. cerevisiae GST-ORF library (constructed in the laboratory of Dr. Eric M. Phizicky, University of Rochester) containing a collection of 6144 strains (representing the whole S. cerevisiae genome), each strain expressing one gene as a GST-ORF fusion protein (53). By screening this library for expression of DOHH activity as a GST-fusion protein, we identified YJR070C as the gene for S. cerevisiae DOHH. Previously, YJR070C had been identified as a gene encoding an eIF5A-binding protein LIA1 (Ligand of eIF5A), of then unknown function (54). YJR070C is the only gene encoding DOHH activity in S. cerevisiae, since the YJR070C null strain contains only deoxyhypusine and lacks hypusine.
Homolog genes of YJR070C are found in all eukaryotes from fungi to human, with only one DOHH homolog gene in each species (23, 41). Like eIF5A and DHS, DOHH is also highly conserved in the eukaryotic kingdom. The sequence of human DOHH, encoded by the homolog gene HLRC1, is 48% identical and 61% similar to that of S. cerevisiae DOHH (Fig. 5A). The yeast YJR070C encodes a protein of 325 amino acids (36 kDa) and the human gene, HLRC1, a protein of 302 amino acids (32 kDa). Both yeast and human recombinant enzymes displayed comparable activities in converting the deoxyhypusine-containing eIF5A to the hypusine-containing eIF5A. The cloned human DOHH gene was also functional upon transfection into human and other mammalian cells (23). When 293T cells were transfected with eIF5A-1 vector alone, a high expression of eIF5A precursor protein was detected. However, the overexpressed eIF5A protein was not modified and was largely in the precursor form, presumably due to a limiting level of endogenous DHS activity in these cells. When 293T cells were cotransfected with both eIF5A and DHS expression vectors, the deoxyhypusine-containing intermediate eIF5A(Dhp), but not mature eIF5A, accumulated. Only upon co-transfection with three vectors encoding eIF5A, DHS and DOHH, respectively, was increased production of mature eIF5A (hypusine form) observed. These findings demonstrate that the cloned human DOHH gene expresses functional DOHH activity in mammalian cells and that co-expression of all three proteins, eIF5A, DHS and DOHH, is required for overproduction of fully modified eIF5A.
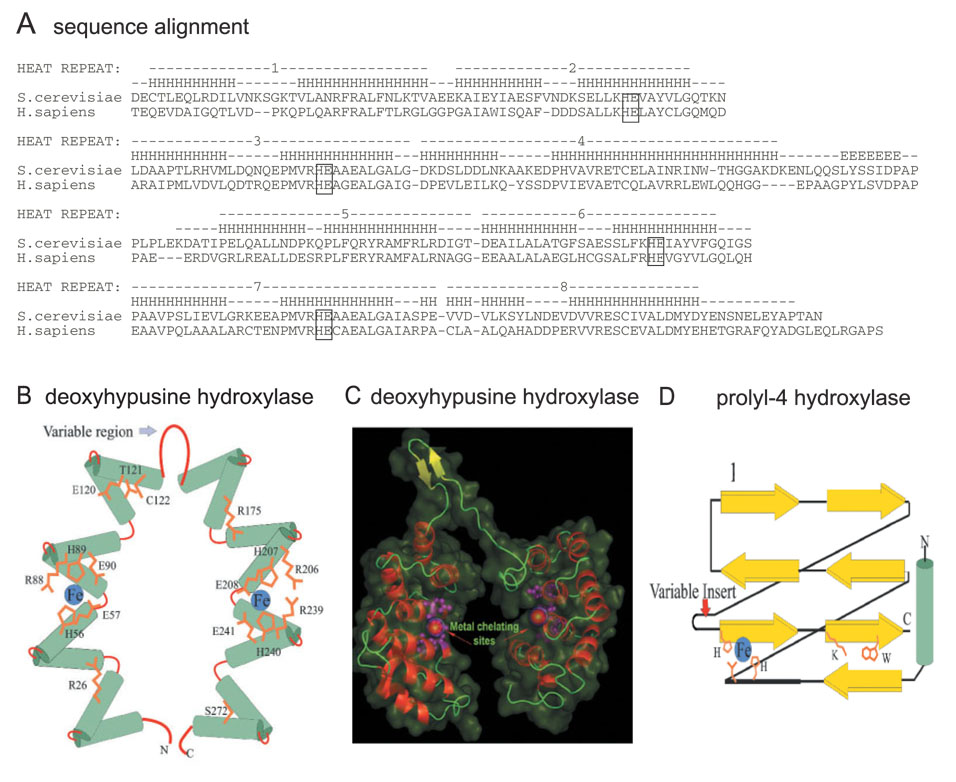
Fig. 5. Sequence and predicted structure of DOHH.
A: Sequence alignment of S. cerevisiae and human DOHH protein. A more complete comparison of DOHH sequences is given in Ref. 23. The conserved HE (HisGlu) pairs are boxed. Schematic diagram (B) and 3D model (C) of DOHH in comparison with prolyl 4-hydroxylase (D) Modified from Ref. 23.
A structure model of DOHH: DOHH is a HEAT-repeat–containing metalloenzyme
Examination of the DOHH sequence alignment (Fig. 5A) reveals that this protein belongs to a family of HEAT-repeat–containing proteins, [named for human huntingtin (H), elongation factor 3 (E), a subunit of protein phosphatase 2A (A) and the target of rapamycin (TOR)] which commonly mediate protein–protein interactions (55). In a variety of bacterial and eukaryotic proteins, the HEAT motif, an α-helical hairpin (a pair of α-helices) of roughly 50 amino acids is tandemly repeated to form super-helical structures. Computer modeling of DOHH predicts a structure consisting of eight HEAT-repeats in a symmetrical dyad of four HEAT motifs connected by a variable region (Fig. 5A). Circular dichroism spectral analysis of pure recombinant human DOHH revealed a high content of alpha-helical structure (77 ± 2.1%) consistent with the value calculated from the predicted model (76–78%). Four highly characteristic and strictly conserved HE (histidine-glutamate) motifs are present, one pair in each dyad. A three-dimensional homology model of DOHH was constructed using the pre-existing crystal structure of a HEAT-repeat protein, the E. coli protein YibA (PDB: 1OYZ) as a template. This model shows that the C-terminal helices of the HEAT repeats line the “inner” circumference of the curved toroid formed by the HEAT-repeats and that the predicted metal-chelating sites lie in the interior of the concave structure (Fig. 5B and C). The variable region between the two symmetric halves is likely to form a “hinge-like” structure that might allow flexibility to accommodate the substrate protein in the interior cavity. However, such homology-based structural models only provide an approximate structure for the protein and its possible metal interactions.
The predicted structure of DOHH is unrelated to those of the majority of previously studied hydroxylases, e.g. Fe(II)-and 2-oxoacid–dependent dioxygenases, such as prolyl 4-hydroxylase. The latter share a common feature in the form of a specific β-jelly roll structure, termed the double stranded beta helix (DSBH) (Fig 5 D) (56). Yet both DOHH and DSBH enzymes seem to contain similar metal binding active sites. Most of the DSBH dioxygenases have a consensus His1-X-D/E-Xn-His2 sequence of appropriately positioned histidine and acidic (E or D) residues that coordinates iron (57). In the case of DOHH, four strictly conserved His-Glu pairs (two pairs on each dyad) were identified as potential metal coordination sites. Indeed, iron was consistently found in purified preparations of the yeast and human enzymes. A single substitution of Ala for any of six specific amino acid residues (H56, H89, E90, H207, H240, E241 of the human enzyme) in the four conserved HE motifs caused a marked reduction in the iron content of the enzyme. Furthermore, alanine substitution of any residue of the four HE motifs completely abolished DOHH activity (Y.S Kim and M.H. Park, unpublished results), indicating the importance of the conserved HE residues in iron binding and catalysis. Although the superhelical structure of DOHH is entirely different from those of DSBH dioxygenases, DOHH enzymes may have convergently evolved a similar irondependent catalytic mechanism.
Concluding remarks
With the recent cloning of the DOHH gene, identification of the enzymes in the hypusine biosynthetic pathway has been completed. Co-expression of all three proteins, eIF5A, DHS and DOHH will permit overproduction of hypusine-containing eIF5A for functional and structural analysis. The mechanism and structure of the first step enzyme, DHS, have been extensively characterized. Although eIF5A and DOHH have been proposed as potential targets of antitumor (58) and anti-HIV-1 therapy (59), no specific inhibitors of DOHH are currently available. The newly cloned DOHH enzyme (probably the catalytic subunit) is under investigation to determine its X-ray structure and catalytic mechanism and to develop specific inhibitors. DHS is absolutely specific for eIF5A substrate and DOHH is expected to elicit a similar specificity. The basis for the strict specificity of molecular interactions between eIF5A substrates and their modifying enzymes is being pursued in an effort to determine the structures of complexes of eIF5A(Lys) with DHS and DOHH. The physiological function and mode of action of eIF5A have been perplexing. However, recent genetic analysis of temperature-sensitive mutants of eIF5A in S. cerevisiae provides some insights into its complex function and warrants further investigation.
Acknowledgments
The research in the author’s laboratory was supported by the Intramural Research Program of the NIH (NIDCR)
Abbreviations
- eIF5A
eukaryotic translation initiation factor, (mature form containing hypusine)
- eIF5A(Lys)
eIF5A precursor containing lysine
- eIF5A(Dhp)
eIF5A intermediate containing deoxyhypusine
- aIF5A
archaeal initiation factor 5A
- EF-P
elongation factor P
- DHS
deoxyhypusine synthase
- DOHH
deoxyhypusine hydroxylase
- GC7
N1-guanyl-1,7-diamonoheptane
REFERENCES
- 1.Park MH, Cooper HL, Folk JE. Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc. Natl. Acad. Sci. USA. 1981;78:2869–2873. doi: 10.1073/pnas.78.5.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park MH, Lee YB, Joe YA. Hypusine is essential for eukaryotic cell proliferation. Biol. Signals. 1997;6:115–123. doi: 10.1159/000109117. [DOI] [PubMed] [Google Scholar]
- 3.Chen KY, Liu AY. Biochemistry and function of hypusine formation on eukaryotic initiation factor 5A. Biol. Signals. 1997;6:105–109. doi: 10.1159/000109115. [DOI] [PubMed] [Google Scholar]
- 4.Cooper HL, Park MH, Folk JE, Safer B, Braverman R. Identification of the hypusine-containing protein Hy+ as translation initiation factor eIF-4D. Proc. Natl. Acad. Sci. USA. 1983;80:1854–1857. doi: 10.1073/pnas.80.7.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shiba T, Mizote H, Kaneko T, Nakajima T, Kakimoto Y. Hypusine, a new amino acid occurring in bovine brain. Isolation and structural determination. Biochim. Biophys. Acta. 1971;244:523–531. doi: 10.1016/0304-4165(71)90069-9. [DOI] [PubMed] [Google Scholar]
- 6.Park MH, Cooper HL, Folk JE. The biosynthesis of protein-bound hypusine (Nε-(4-amino-2-hydroxybutyl)-lysine): lysine as the amino acid precursor and the intermediate role of deoxyhypusine (Nε-(4-aminobutyl)lysine) J. Biol. Chem. 1982;257:7217–7222. [PubMed] [Google Scholar]
- 7.Wolff EC, Lee YB, Chung SI, Folk JE, Park MH. Deoxyhypusine synthase from rat testis: purification and characterization. J. Biol. Chem. 1995;270:8660–8666. doi: 10.1074/jbc.270.15.8660. [DOI] [PubMed] [Google Scholar]
- 8.Chen KY, Dou QP. NAD+ stimulated the spermidine-dependent hypusine formation on the 18 kDa protein in cytosolic lysates derived from NB-15 mouse neuroblastoma cells. FEBS Lett. 1988;229:325–328. doi: 10.1016/0014-5793(88)81149-9. [DOI] [PubMed] [Google Scholar]
- 9.Murphey RJ, Gerner EW. Hypusine formation in protein by a two-step process in cell lysates. J. Biol. Chem. 1987;262:15033–15036. [PubMed] [Google Scholar]
- 10.Abbruzzese A, Park MH, Folk JE. Deoxyhypusine hydroxylase from rat testis: partial purification and characterization. J. Biol. Chem. 1986;261:3085–3089. [PubMed] [Google Scholar]
- 11.Byers TL, Lakanen JR, Coward JK, Pegg AE. The role of hypusine depletion in cytostasis induced by S-adenosyl-L-methionine decarboxylase inhibition: new evidence provided by 1-methylspermidine and 1,12-dimethylspermine. Biochem. J. 1994;303:363–368. doi: 10.1042/bj3030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chattopadhyay MK, Tabor CW, Tabor H. Spermidine but not spermine is essential for hypusine biosynthesis and growth in Saccharomyces cerevisiae: spermineis converted to spermidine in vivo by the FMS1-amine oxidase. Proc. Natl. Acad. Sci. USA. 2003;100:13869–13874. doi: 10.1073/pnas.1835918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishimura K, Murozumi K, Shirahata A, Park MH, Kashiwagi K, Igarashi K. Independent roles of eIF5A and polyamines in cell proliferation. Biochem J. 2005;385:779–785. doi: 10.1042/BJ20041477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper HL, Park MH, Folk JE. Posttranslational formation of hypusine in a single major protein occurs generally in growing cells and is associated with activation of lymphocyte growth. Cell. 1982;29:791–797. doi: 10.1016/0092-8674(82)90441-x. [DOI] [PubMed] [Google Scholar]
- 15.Chen KY. An 18000-dalton protein metabolically labeled by polyamines in various mammalian cell lines. Biochim. Biophys. Acta. 1983;756:395–402. doi: 10.1016/0304-4165(83)90350-1. [DOI] [PubMed] [Google Scholar]
- 16.Gerner EW, Mamont PS, Bernhardt A, Siat M. Post-translational modification of the protein-synthesis initiation factor eIF-4D by spermidine in rat hepatoma cells. Biochem. J. 1986;239:379–386. doi: 10.1042/bj2390379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park MH, Wolff EC, Lee YB, Folk JE. Anti-proliferative effects of inhibitors of deoxyhypusine synthase: inhibition of growth of Chinese hamster ovary cells by guanyl diamines. J. Biol. Chem. 1994;269:27827–27832. [PubMed] [Google Scholar]
- 18.Hanauske-Abel HM, Park MH, Hanauske AR, Popowicz AM, Lalande M, Folk JE. Inhibition of the G1-S transition of the cell cycle by inhibitors of deoxyhypusine hydroxylation. Biochim. Biophys. Acta. 1994;1221:115–124. doi: 10.1016/0167-4889(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 19.Schnier J, Schwelberger HG, Smit-McBride Z, Kang HA, Hershey JW. Translation initiation factor 5A and its hypusine modification are essential for viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1991;11:3105–3114. doi: 10.1128/mcb.11.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wöhl T, Klier H, Ammer H. The HYP2 gene of Saccharomyces cerevisiae is essential for aerobic growth: characterization of different isoforms of the hypusine-containing protein Hyp2p and analysis of gene disruption mutants. Mol. Gen. Genet. 1993;241:305–311. doi: 10.1007/BF00284682. [DOI] [PubMed] [Google Scholar]
- 21.Sasaki K, Abid MR, Miyazaki M. Deoxyhypusine synthase gene is essential for cell viability in the yeast Saccharomyces cerevisiae. FEBS Lett. 1996;384:151–154. doi: 10.1016/0014-5793(96)00310-9. [DOI] [PubMed] [Google Scholar]
- 22.Park MH, Joe YA, Kang KR. Deoxyhypusine synthase activity is essential for cell viability in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:1677–1683. doi: 10.1074/jbc.273.3.1677. [DOI] [PubMed] [Google Scholar]
- 23.Park J-H, Aravind L, Wolff EC, Kaevel J, Kim YS, Park MH. Molecular cloning, expression and structural prediction of deoxyhypusine hydroxylase: a novel HEAT-repeat-containing metalloenzyme. Proc. Nat. Acad. Sci. USA. 2006;103:51–56. doi: 10.1073/pnas.0509348102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benne R, Hershey JWB. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J. Biol. Chem. 1978;253:3078–3087. [PubMed] [Google Scholar]
- 25.Kang HA, Hershey JW. Effect of initiation factor 5A depletion on protein synthesis and of Saccharomyces cerevisiae. J. Biol. Chem. 1994;269:3934–3940. [PubMed] [Google Scholar]
- 26.Xu A, Jao DL, Chen KY. Identification of mRNA that binds to eukaryotic initiation factor 5A by affinity co-purification and differential display. Biochem. J. 2004;384:585–590. doi: 10.1042/BJ20041232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruhl M, Himmelspach M, Bahr GM, Hammerschmid F, Jaksche H, Wolff B, Aschauer H, Farrington GK, Probst H, Bevec D, Hauber J. Eukaryotic initiation factor 5A is a cellular target of the human immunodeficiency virus type 1 Rev activation domain mediating trans-activation. J. Cell. Biol. 1993;123:1309–1320. doi: 10.1083/jcb.123.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lipowsky G, Bischoff FR, Schwarzmaier P, Kraft R, Kostka S, Hartmann E, Kutay U, Görlich D. Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J. 2000;19:4362–4371. doi: 10.1093/emboj/19.16.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuk D, Jacobson A. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 1998;17:2914–2925. doi: 10.1093/emboj/17.10.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valentini SR, Casolari JM, Oliveira CC, Silver PA, McBride AE. Genetic interactions of yeast eukaryotic translation initiation factor 5A (eIF5A) reveal connections to poly(A)-binding protein and protein kinase C signaling. Genetics. 2002;160:393–405. doi: 10.1093/genetics/160.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zanelli CF, Valentini SR. Pkc1 acts through Zds1 and Gic1 to suppress growth and cell polarity defects of a yeast eIF5A mutant. Genetics. 2005;171:1571–1581. doi: 10.1534/genetics.105.048082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartig D, Schümann H, Klink F. The unique posttranslational modification leading to deoxyhypusine or hypusine is a general feature of the archaebacterial kingdom. System. Appl. Microbiol. 1990;13:112–116. [Google Scholar]
- 33.Hanawa-Suetsugu K, Sekine S, Sakai H, Hori-Takemoto C, Terada T, Unzai S, Tame JR, Kuramitsu S, Shirouzu M, Yokoyama S. Crystal structure of elongation factor P from Thermus thermophilus HB8. Proc. Natl. Acad. Sci. USA. 2004;101:9595–9600. doi: 10.1073/pnas.0308667101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glick BR, Ganoza MC. Identification of a soluble protein that stimulates peptide bond synthesis. Proc Natl Acad Sci USA. 1975;72:4257–4260. doi: 10.1073/pnas.72.11.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim KK, Hung LW, Yokota H, Kim R, Kim SH. Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at 1.8 Å resolution. Proc. Natl. Acad. Sci. USA. 1998;95:10419–10424. doi: 10.1073/pnas.95.18.10419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clement PC, Henderson CA, Jenkins ZA, Smit-McBride Z, Wolff EC, Hershey JWB, Park MH, Johansson HE. Identification and characterization of eukaryotic initiation factor 5A-2. Eur. J. Biochem. 2003;147:4254–4263. doi: 10.1046/j.1432-1033.2003.03806.x. [DOI] [PubMed] [Google Scholar]
- 37.Mehta KD, Leung D, Lefebvre L, Smith M. The ANB1 locus of Saccharomyces cerevisiae encodes the protein synthesis initiation factor eIF-4D. J. Biol. Chem. 1990;65:8802–8807. [PubMed] [Google Scholar]
- 38.Guan XY, Sham JS, Tang TC, Fang Y, Huo KK, Yang JM. Isolation of a novel candidate oncogene within a frequently amplified region at 3q26 in ovarian cancer. Cancer Res. 2001;61:3806–3809. [PubMed] [Google Scholar]
- 39.Guan XY, Fung JM, Ma NF, Lau SH, Tai LS, Xie D, Zhang Y, Hu L, Wu QL, Fang Y, Sham JS. Oncogenic role of eIF-5A2 in the development of ovarian cancer. Cancer Res. 2004;64:4197–4200. doi: 10.1158/0008-5472.CAN-03-3747. [DOI] [PubMed] [Google Scholar]
- 40.Ober D, Harms R, Witte L, Hartmann T. Molecular evolution by change of function. Alkaloid-specific homospermidine synthase retained all properties of deoxyhy-pusine synthase except binding the eIF5A precursor protein. J. Biol. Chem. 2003;278:12805–12812. doi: 10.1074/jbc.M207112200. [DOI] [PubMed] [Google Scholar]
- 41.Weir BA, Yaffe MP. Mmd1p, a novel, conserved protein essential for normal mitochondrial morphology and distribution in the fission yeast Schizosaccharomyces pombe. Mol. Biol. Cell. 2004;15:1656–1665. doi: 10.1091/mbc.E03-06-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugimoto A. High-throughput RNAi in Caenorhabditis elegans: genome-wide screens and functional genomics. Differentiation. 2004;72:81–91. doi: 10.1111/j.1432-0436.2004.07202004.x. [DOI] [PubMed] [Google Scholar]
- 43.Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, Misra S, Rubin GM. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joe YA, Park MH. Structural features of the eIF-5A precursor required for posttranslational synthesis of deoxyhypusine. J. Biol. Chem. 1994;269:25916–25921. [PubMed] [Google Scholar]
- 45.Park JH, Wolff EC, Folk JE, Park MH. Reversal of the deoxyhypusine synthesis reaction. Generation of spermidine or homospermidine from deoxyhypusine by deoxyhypusine synthase. J. Biol. Chem. 2003;278:32683–32691. doi: 10.1074/jbc.M304247200. [DOI] [PubMed] [Google Scholar]
- 46.Wolff EC, Folk JE, Park MH. Enzyme-substrate intermediate formation at lysine 329 of human deoxyhypusine synthase. J. Biol. Chem. 1997;272:15865–15871. doi: 10.1074/jbc.272.25.15865. [DOI] [PubMed] [Google Scholar]
- 47.Joe YA, Wolff EC, Lee YB, Park MH. Enzyme-substrate intermediate at a specific lysine residue is required for deoxyhypusine synthesis. The role of Lys329 in human deoxyhypusine synthase. J. Biol. Chem. 1997;272:32679–32685. doi: 10.1074/jbc.272.51.32679. [DOI] [PubMed] [Google Scholar]
- 48.Ober D, Hartmann T. Deoxyhypusine synthase from tobacco. cDNA isolation, characterization, and bacterial expression of an enzyme with extended substrate specificity. J. Biol. Chem. 1999;274:32040–32047. doi: 10.1074/jbc.274.45.32040. [DOI] [PubMed] [Google Scholar]
- 49.Liao DI, Wolff EC, Park MH, Davies DR. Crystal structure of the NAD complex of human deoxyhypusine synthase: an enzyme with a ball-and-chain mechanism for blocking the active site. Structure. 1998;6:23–32. doi: 10.1016/s0969-2126(98)00004-5. [DOI] [PubMed] [Google Scholar]
- 50.Umland TC, Wolff EC, Park MH, Davies DR. A new crystal structure of deoxyhypusine synthase reveals the configuration of the active enzyme and of an enzyme. NAD.inhibitor ternary complex. J. Biol. Chem. 2004;279:28697–28705. doi: 10.1074/jbc.M404095200. [DOI] [PubMed] [Google Scholar]
- 51.Jakus J, Wolff EC, Park MH, Folk JE. Features of the spermidine-binding site of deoxyhypusine synthase as derived from inhibition studies. Effective inhibition by bis- and mono- guanylated diamines and polyamines. J. Biol. Chem. 1993;268:13151–13159. [PubMed] [Google Scholar]
- 52.Lee YB, Folk JE. Branched-chain and unsaturated 1,7-diaminoheptane derivatives as deoxyhypusine synthase inhibitors. Bioorg. Med. Chem. 1998;6:253–270. doi: 10.1016/s0968-0896(97)10030-x. [DOI] [PubMed] [Google Scholar]
- 53.Martzen MR, McCraith SM, Spinelli SL, Torres FM, Fields S, Grayhack EJ, Phizicky EM. A biochemical genomics approach for identifying genes by the activity of their products. Science. 1990;286:1153–1155. doi: 10.1126/science.286.5442.1153. [DOI] [PubMed] [Google Scholar]
- 54.Thompson GM, Cano VS, Valentini SR. Mapping eIF5A binding sites for Dys1 and Lia1: in vivo evidence for regulation of eIF5A hypusination. FEBS Lett. 2003;555:464–468. doi: 10.1016/s0014-5793(03)01305-x. [DOI] [PubMed] [Google Scholar]
- 55.Andrade MA, Petosa C, O’Donoghue SI, Muller CW, Bork P. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- 56.Aravind L, Koonin EV. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-3-research0007. RESEARCH0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hausinger RP. FeII/alpha-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 58.Clement PM, Hanauske-Abel HM, Wolff EC, Kleinman HK, Park MH. The antifungal drug ciclopirox inhibits deoxyhypusine and proline hydroxylation, endothelial cell growth and angiogenesis in vitro. Int. J. Cancer. 2002;100:491–498. doi: 10.1002/ijc.10515. [DOI] [PubMed] [Google Scholar]
- 59.Andrus L, Szabo P, Grady RW, Hanauske AR, Huima-Byron T, Slowinska B, Zagulska S, Hanauske-Abel HM. Antiretroviral effects of deoxyhypusyl hydroxylase inhibitors—A hypusine-dependent host cell mechanism for replication of human immunodeficiency virus type 1 (HIV-1) Biochem. Pharmacol. 1998;55:1807–1818. doi: 10.1016/s0006-2952(98)00053-7. [DOI] [PubMed] [Google Scholar]