Abstract
In systemic amyloidoses, widespread deposition of protein as amyloid causes severe organ dysfunction. It is necessary to discriminate among the different forms of amyloid to design an appropriate therapeutic strategy. We developed a proteomics methodology utilizing two-dimensional polyacrylamide gel electrophoresis followed by matrix-assisted laser desorption/ionization mass spectrometry and peptide mass fingerprinting to directly characterize amyloid deposits in abdominal subcutaneous fat obtained by fine needle aspiration from patients diagnosed as having amyloidoses typed as immunoglobulin light chain or transthyretin. Striking differences in the two-dimensional gel proteomes of adipose tissue were observed between controls and patients and between the two types of patients with distinct, additional spots present in the patient specimens that could be assigned as the amyloidogenic proteins in full-length and truncated forms. In patients heterozygotic for transthyretin mutations, wild-type peptides and peptides containing amyloidogenic transthyretin variants were isolated in roughly equal amounts from the same protein spots, indicative of incorporation of both species into the deposits. Furthermore novel spots unrelated to the amyloidogenic proteins appeared in patient samples; some of these were identified as isoforms of serum amyloid P and apolipoprotein E, proteins that have been described previously to be associated with amyloid deposits. Finally changes in the normal expression pattern of resident adipose proteins, such as down-regulation of αB-crystallin, peroxiredoxin 6, and aldo-keto reductase I, were observed in apparent association with the presence of amyloid, although their levels did not strictly correlate with the grade of amyloid deposition. This proteomics approach not only provides a way to detect and unambiguously type the deposits in abdominal subcutaneous fat aspirates from patients with amyloidoses but it may also have the capability to generate new insights into the mechanism of the diseases by identifying novel proteins or protein post-translational modifications associated with amyloid infiltration.
The amyloidoses constitute a heterogeneous group of diseases whose common pathological hallmark is the presence of extracellular or intracellular amyloid deposits that lead to cellular toxicity, disruption of anatomical architecture, and organ dysfunction (1). In the systemic forms, widespread extracellular amyloid deposition leads to severe dysfunction of vital organs such as the heart, kidney, and liver, resulting in poor prognosis for long term survival. Despite their lack of similarity in amino acid sequence, the amyloidogenic proteins share certain secondary structural similarities (e.g. the tendency to form β-pleated sheet structures) and generate morphologically similar fibrils that show apple green birefringence under polarized light when stained with Congo red. Diagnosis and type classification of amyloid diseases are based on detecting the insoluble amyloid aggregates and determining which protein is the primary constituent (2–5). More than 10 types of systemic amyloidoses are known to occur in humans: these differ from one another in modality of acquisition, clinical course, and treatment (3, 4). The most common systemic forms worldwide are light chain amyloidosis (AL)1 and reactive amyloidosis in which the fibrils are derived, respectively, from monoclonal immunoglobulin light chains and from the acute phase reactant, serum amyloid A protein. However, hereditary amyloidoses, caused by a large and ever expanding list of mutations in genes that code for normally soluble plasma proteins such as transthyretin (TTR), lysozyme, and apolipoproteins A-I and A-II, are not uncommon and have a particularly high incidence in some geographic regions. Little is known of the mechanism underlying the loss of protein secondary and tertiary stability under physiological conditions and the resultant protein deposition as fibrils in target organs. Post-translational modifications of amyloidogenic and/or associated proteins or changes in the local tissue environment, particularly those related to aging and oxidative stress, have been hypothesized to be important factors in disease onset and progression and may also play a role in the variability of clinical presentation (6–12).
Precise definition of the type of amyloidosis in each patient case is a critical step in establishing the disease prognosis and planning the therapeutic strategy. The tissue of choice for biopsy-based demonstration of fibrillar deposits in candidate patients with systemic disease is abdominal subcutaneous fat that is obtained by fine needle aspiration (13–15). Once the presence of fibrils is confirmed, typing is carried out using a variety of histopathologic techniques to characterize the fibril precursor and/or the deposited protein. This process is complex, and currently the chance of misdiagnosis remains significant (16, 17) as traditional immunohistochemistry is not completely reliable for this purpose (5, 18, 19). The conclusive method for typing amyloidoses is the direct biochemical analysis of tissue deposits. Until now, however, direct biochemical analysis of fibrils has been dependent on their isolation, an approach that often requires complex extraction procedures and large amounts of material and that usually is not suitable for routine clinical practice. Recently the demand for sensitive methods to reliably characterize amyloid deposits in small clinical samples has led to the development of immunochemical and LC/MS-based approaches for the analysis of submilligram amounts of either freshly obtained or formalin-fixed tissues (20–22).
Here we applied a 2D PAGE- and MS-based proteomics approach to characterize and identify proteins deposited in abdominal subcutaneous fat tissue aspirates from patients with different types of systemic amyloidoses. We found profound differences between the 2D adipose tissue proteomes of normal and amyloidosis patients that can be exploited as a diagnostic tool for amyloid typing as the approach utilizes tissue that is routinely collected as part of diagnosis. Furthermore we demonstrate the characterization of the deposited amyloidogenic proteins and their isoforms or fragments, the identification of co-deposited proteins that are known to be associated with amyloid fibrils in various tissues, and apparent variations in the regulation of proteins involved in chaperone activity, cellular energetics, and redox regulation in the adipocytes of diseased patients.
EXPERIMENTAL PROCEDURES
Patient Samples—
Abdominal subcutaneous fat tissue samples used as controls were obtained, with institutional review board approval and informed consent, from seven non-amyloid-affected patients (four male and three female; median age, 72 years) undergoing abdominal surgery at Istituto di Ricovero e Cura a Carattere Scientifico Ospedale San Matteo, Pavia, Italy, during the course of the surgical procedure. Patients with neoplastic or other systemic diseases were excluded. Subcutaneous fat samples from patients with systemic amyloidosis were obtained by fine needle aspiration during the routine diagnostic procedure at the Amyloid Treatment and Research Program, Boston University School of Medicine. One sample (ATTR patient 05-152) was surgically obtained at Boston Medical Center with informed consent during inguinal herniectomy. All patients who had received specific treatments for disease were excluded from the study. Samples were obtained from a total of nine amyloidosis patients (six male and three female; seven patients were diagnosed with AL (six λ and one κ), and two were diagnosed with ATTR) with a median age of 65 years. The presence of amyloid fibrils was evaluated histologically with Congo red staining, and the amyloid load was graded with a score from 0 (negative) to 3 (intensely positive). All specimens were immediately frozen and stored at −80 °C until used. Matched blood samples were also obtained contemporaneously with fat tissue acquisition. Serum was separated by centrifugation and then kept frozen until used.
Protein Extraction from Adipose Tissue—
Thawed adipose tissue samples were washed repeatedly with sterile isotonic saline and once with double distilled water. For large samples (in the range of mg to g of tissue), IEF buffer (7 m urea, 2 m thiourea, 4% CHAPS, 65 mm DTT) was added directly to samples at a ratio of 200 μl/100 mg of tissue. Samples were crushed with a disposable pestle and sonicated in ice-water four times (10 pulses of 5 s each time) at intervals of 15 min with gentle shaking at room temperature between intervals. Samples were then centrifuged at 80,000 × g for 1.5 h at 19 °C (23). The central aqueous layer between the top lipid layer and the cell debris pellet was recovered, residual lipids were removed with a second centrifugation at 25,000 × g for 30 min at 4 °C, and the aliquots were stored at −80 °C. Small patient samples (in the range of 10–20 μg of tissue) were washed, resuspended in 100 μl of IEF buffer, sonicated as described above, and then centrifuged for 1 h at 25,000 × g at 4 °C. The central aqueous layer was recovered and then stored at −80 °C. Large and small sample preparation techniques produced identical results when compared with one another using the same samples. Total protein was quantitated relative to standards using the Bio-Rad Protein Assay.
2D PAGE Analysis—
Protein extracts (amounts equivalent to 10–30 μg of protein) were diluted to a final volume of 300 μl with 100 μl of Destreak™ buffer (Amersham Biosciences), IEF buffer, and pI 3–10 ampholytes (Bio-Rad) at a final concentration 0.02%. For serum samples, an aliquot of 6.25 μl of serum was mixed with 10 μl of 10% SDS, 2.3% DTT; heated to 95 °C for 5 min; and diluted to 500 μl with IEF buffer (24). Sixty-five microliters of this solution were then diluted to a final volume of 300 μl using IEF and Destreak buffers and ampholytes as described above for tissue samples. Seventeen-centimeter ReadyStrip™ IPG strips (Bio-Rad) with non-linear gradients of pH 3–10 were subjected to passive rehydration for 1 h and then to active rehydration at 50 V for 8 h. Isoelectric focusing was performed in a Protean™ IEF cell (Bio-Rad) as follows: 120 V for 1 h, 300 V for 30 min, a linear increase up to 3500 V over 3 h, 5000 V for 10 min, and 8000 V steady until a total of 67,000 V-h had elapsed. After IEF, the strips were subjected to standard disulfide reduction with DTT and cysteine alkylation with iodoacetamide followed by second dimension electrophoresis using 9–16% gradient ReadyGels™ (Bio-Rad). Gels were stained with fixative silver stain, PlusOne™ (Amersham Biosciences); the MS-compatible silver stain ProteoSilver™ Plus (Sigma-Aldrich); or GelCode™ colloidal Coomassie Blue (Pierce). All gels were imaged with an EDAS 290 (Eastman Kodak Co.) or a VersaDoc™ 3000 (Bio-Rad) imaging station, and the results were analyzed using PDQuest™ software (Bio-Rad).
Western Blotting—
After electrophoresis, proteins were transferred to a Millipore™ Q PVDF membrane (Millipore, Billerica, MA) using a TransBlot™ semidry electrophoretic transfer cell (Bio-Rad) and probed with polyclonal rabbit anti-human immunoglobulin light chain or rabbit anti-human transthyretin antibodies (Dako, Glostrup, Denmark), both at a 1:1000 dilution, followed by an alkaline phosphatase-conjugated goat anti-rabbit secondary antibody (Dako) used at a 1:5000 dilution.
In-gel Digestion—
In-gel digestion was conducted as described previously (25). Briefly the protein spots of interest were punch-excised using wide orifice pipette tips. Gel pieces were destained with either Coomassie destain (100 mm NH4CO3, pH 9, 50% acetonitrile) or silver destain (the potassium ferrocyanide-based reagents of the ProteoSilver Plus kit). The gel pieces were washed three times with wash solution 1 (100 mm NH4HCO3, pH 9) and wash solution 2 (100 mm NH4HCO3, pH 9, 50% acetonitrile) followed by wash solution 3 (100% acetonitrile). Gel pieces were swelled in digestion solution (∼10 ng of Trypsin Gold (Promega, Madison, WI) in 50 mm NH4HCO3, pH 9, 5% acetonitrile) and incubated overnight at 37 °C. Peptides were extracted twice with extraction solution 1 (20 mm NH4HCO3, pH 9) and extraction solution 2 (1% trifluoroacetic acid, 50% acetonitrile) followed by extraction solution 3 (100% acetonitrile), each time for 20 min. The entire extraction process was performed in duplicate, and all extraction supernatants were pooled and dried.
MALDI-TOF MS—
Peptides were desalted and prepared for MS using micro-reversed-phase chromatography (ZipTips™, Millipore). Mass spectra were obtained after purified peptides were co-crystallized with the matrix 2,5-dihydroxybenzoic acid onto AnchorChip™ (Bruker Daltonics, Billerica, MA) targets using the dried droplet technique (26, 27) using a Reflex IV™ (Bruker Daltonics) MALDI-TOF MS instrument operated in the positive ion, reflectron mode over the range m/z 400–8000. The intensity of the nitrogen laser irradiation (337 nm, 3-ns pulse duration) was adjusted to ∼3% above threshold value. Signals resulting from 100 to 200 laser shots were summed for each mass spectrum. External calibration of the spectra was achieved using Bruker peptide standards, and internal calibration within 50 ppm was achieved using the masses of known peptide ions as reference values.
MS Data Analysis and Peptide Mass Fingerprinting—
MALDI-TOF mass spectra were deisotoped, and peak lists were generated using the software MoverZ™ from Genomic Solutions (Ann Arbor, MI). Peak lists were submitted to the on-line database search engine Mascot™ at Matrix Science (London, UK) for peptide mass fingerprinting (PMF) analysis against the Swiss-Prot or National Center for Biotechnology Information (NCBI) non-redundant protein databases using the following restrictions: (a) Homo sapiens, (b) trypsin digestion with up to three missed cleavages, (c) ±100-ppm error, and (d) cysteine carbamidomethylation as a fixed modification and methionine oxidation as a variable modification. Identifications were assigned when the score values were returned with p < 0.05 for a false positive and were confirmed by individual inspection of the spectra. Comprehensive peak assignments were accomplished manually using theoretical digest values generated from Mascot-matched sequences (or in the case of immunoglobulin light chain samples, monoclonal light chain sequences established from genetic sequencing of the corresponding patient-derived bone marrow samples) with the on-line software tool MS-Digest (Protein Prospector at University of California, San Francisco, CA) (28).
VL Gene Sequencing and Analysis of VL Gene Usage—
Cells from bone marrow aspirates were treated with NH4Cl to lyse red blood cells, washed, pelleted, and frozen at −80 °C as described previously (29). Total RNA was extracted, and cDNA was synthesized, amplified by multiplex PCR with a set of 5′ primers specific for the framework (FR) 1 region of seven Vλ (VλI, VλII/V, VλIII, VλIVa, VλIVb, and VλVI) and four Vκ (VκI/IV, VκII, and VκIII) families and 3′ λ or κ constant region primers (Integrated DNA Technologies, Coralville, IA) (30), cloned, and sequenced as described previously (31). The clonal sequence was determined on the basis of identity of at least 50% of multiple independently cloned and sequenced products. Sequences were compared using the Jellyfish™ gene analysis software package (Field Scientific, LLC). Once the light chain was identified, additional PCR amplification was performed to correct for minor nucleotide sequence errors introduced by the FR1 primers, and resequencing was carried out. For these experiments, 5′ primers specific for the appropriate VL leader regions and 3′ primers for the appropriate constant region were used. VL genes with the corrected FR1 sequences were evaluated for their homology to the germ line donor sequences using a database of rearranged immunoglobulin genes, V BASE, and the International Immunogenetics Information System, IMGT/V-QUEST (32). The assignation of germ line gene counterpart was done based on maximum homology of the nucleotide sequences. Homology to germ line sequence was determined for complete VL genes with the exception of the codons associated with the variable-joining junction and FR4. All sequences were functional without stop codons, frameshifts, or pseudogenes.
TTR Gene Sequencing—
Genomic DNA was extracted from the peripheral blood leukocytes by standard phenol/chloroform procedure. Exon 4 of the TTR gene was amplified by PCR as described elsewhere (33) The PCR product was subjected to electrophoresis in a 1.5% agarose gel, stained with ethidium bromide, visualized with ultraviolet light, purified by QIAquick purification kit (Qiagen, Valencia, CA), and sequenced in both directions.
RESULTS
The 2D PAGE Proteome of Abdominal Subcutaneous Fat from Non-amyloid (Control) Individuals—
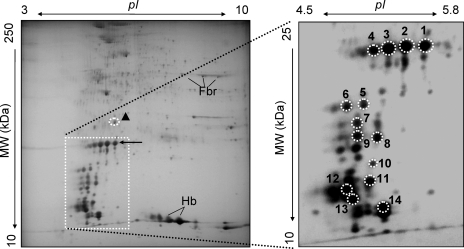
As a basis for comparison with samples from patients with amyloidosis, it was essential to establish a proteomic reference map of non-amyloid adipose tissue. The Congo red-negative abdominal subcutaneous fat samples from individuals (n ≥ 4) without evidence of amyloid disease were analyzed by 2D PAGE followed by Coomassie or silver staining. A representative control 2D map is shown in Fig. 1. Selected landmark spots (Fig. 1, circled spots, spot numbers 1–14), which were relatively abundant and present in roughly equal amounts across all the samples, were identified by in-gel protease digestion, MALDI-TOF MS, and PMF analyses as listed in Table I. Notably although low levels of some blood proteins were detected, the thorough washing of the specimens had minimized contamination by red cell or plasma proteins, and the vast majority of the observed spots were derived from adipose tissue-resident proteins.
Fig. 1.
The 2D PAGE proteome of abdominal subcutaneous fat tissue from control samples. Adipose tissue from normal, non-amyloid patients was subjected to plasma removal, dissolution in IEF buffer, delipidation, reduction and alkylation, and 2D PAGE analysis followed by Coomassie or silver staining as described in the text. Shown is a representative example of a silver-stained gel from a control sample (30 μg); the pI range is 3–10, and the molecular mass range is 10–250 kDa. The identities of landmark spots numbered 1–14 (circled) were assigned on the basis of in-gel digestion, MALDI-TOF MS, and peptide mass fingerprint analyses; the assignments of these spots are indicated in Table I.
Table I.
Assignments of selected protein spots from 2D PAGE, MALDI-TOF MS, and PMF analysis of control fat tissue
Gel spots (as designated in Fig. 1) were excised and subjected to in-gel digestion with trypsin, reduction with DTT and carbamidomethylation, MALDI-TOF MS, and PMF analysis using Mascot as described (stipulating trypsin digestion with up to three missed cleavages, ±100-ppm error, cysteine carbamidomethylation as a fixed modification, and methionine oxidation as a variable modification). For all the database search settings used in the experiments reported, a MOWSE score >53 was determined to yield a p < 0.05 for false positive assignments. ID, identity; SOD, superoxide dismutase; P, phosphate.
| Spot no. (Fig. 1) | Spot ID | NCBInr accession no. | MOWSE score | Number of matching peptides (sequence coverage) |
|---|---|---|---|---|
| 1 | Albumin | gi|55669910 | 342 | 51 (76%) |
| 2 | Actin β | gi|14250401 | 106 | 23 (54%) |
| 3 | Vimentin | gi|62414289 | 350 | 56 (74%) |
| 4 | GP96 | gi|61656607 | 91 | 25 (29%) |
| 5 | Glycerol-3-P dehydrogenase | gi|1708026 | 114 | 16 (48%) |
| 6 | Annexin A2 | gi|18645167 | 424 | 61 (94%) |
| 7 | Aldo-keto reductase I | gi|4503285 | 100 | 16 (53%) |
| 8 | Peroxiredoxin 6 | gi|4758638 | 106 | 16 (68%) |
| 9 | Mn-SOD, mitochondrial | gi|38503339 | 152 | 19 (77%) |
| 10 | αB-Crystallin isoform | gi|4503057 | 120 | 16 (75%) |
| 11 | αB-Crystallin isoform | gi|4503057 | 93 | 14 (64%) |
| 12 | Hemoglobin β | gi|442850 | 121 | 17 (100%) |
| 13 | Adipocyte fatty acid-binding protein | gi|52695841 | 92 | 7 (51%) |
| 14 | 14-3-3 | gi|83754467 | 121 | 16 (40%) |
2D PAGE Proteomics Characterization of Amyloid Deposits in the Adipose Tissue of Patients with Amyloidosis—
Abdominal subcutaneous fat samples were obtained from nine amyloidosis patients, and their amyloid infiltration was graded according to the extent of their Congo red-positive staining. As noted above, all the control samples were negative for amyloid deposition. The results for the patient samples are shown in Table II. For all patients, the type of amyloidosis had been characterized by the demonstration of a plasma cell dyscrasia for AL or by gene typing that resulted in the detection of an amyloidogenic TTR mutation known to be correlated with ATTR. Patient fat samples were subjected to 2D PAGE analysis and staining procedures similar to those used for the control samples (Figs. 2 and 4 show representative 2D gels from AL and ATTR samples, respectively). Pronounced spots were visible in all the AL and ATTR samples in regions of the gels corresponding to the predicted molecular weights and pI values of the circulating amyloidogenic precursors, thus suggesting that these precursors had been deposited as amyloid in the fat tissue. Furthermore ladders and trains of additional pronounced spots (heterogeneous in molecular weight and/or pI) appeared to be closely associated with the previous spots and thus were hypothesized to be fragments, variants, or post-translationally modified isoforms of the amyloidogenic precursors. The total relative intensity of these stained spots correlated qualitatively and semiquantitatively with the grade of amyloid infiltration as evaluated by Congo red staining of the tissue.
Table II.
Characteristics of the fat samples obtained from patients with amyloidoses
The degree of amyloid infiltration in the abdominal subcutaneous fat aspirates was assessed by Congo red staining on a scale from 0 to 3. Positives designate the detection of fibrillar deposits. AL patients were typed by demonstration of a plasma cell dyscrasia followed by sequencing of the monoclonal component as described under “Experimental Procedures.” ATTR patients were typed by direct genomic DNA sequencing of the indicated TTR mutation.
| Patient code | Congo red staining intensity | Amyloid type |
|---|---|---|
| 05-131 | 3+ | AL λ |
| 05-133 | 1+ | AL λ |
| 05-138 | 2+ | AL λ |
| 05-141 | 3+ | AL λ |
| 05-151 | 3+ | AL λ |
| 06-002 | 3+ | AL κ |
| 06-095 | 3+ | AL λ |
| 06-028 | 1+ | ATTR V30M |
| 05-152 | 3+ | ATTR V122I |
Fig. 2.
The 2D PAGE proteome of abdominal subcutaneous fat tissue from AL patient 05-141. Adipose tissue aspirates from patients with Ig light chain amyloidosis were processed and subjected to 2D PAGE analysis and silver staining similarly to control samples. Shown here is a representative example of a gel from a sample (25 μg) from patient 05-141. The pI range is 3–10, and the molecular mass range is 10–250 kDa (left panel). In the enlargement of the boxed region of this gel (pI range, 4.5–5.8; molecular mass range, 10–25 kDa; shown in the right panel) are contained the majority of novel spots relative to controls. The arrow indicates a train of spots at ∼25 kDa. The spots numbered 1–14 were excised and subjected to in-gel digestion, MALDI-TOF MS, and PMF analyses (spectra from spot numbers 1, 8, and 12 are shown in Fig. 3). All numbered spots were determined to have been derived from the patient's amyloidogenic immunoglobulin light chain. The ▴ symbol indicates a spot that was assigned on the basis of MS and PMF analyses as an amyloid-associated protein, apolipoprotein E; this spot also showed increased staining relative to controls. Fbr and Hb indicate the location of fibrin and hemoglobin subunit species, respectively.
Fig. 4.
The 2D PAGE proteome of the abdominal subcutaneous fat tissue from ATTR patient 05-152 with the V122I mutation. Adipose tissue aspirates from patients with ATTR were processed and subjected to 2D PAGE analysis as described in the text. A shows a representative example of a silver-stained gel from a sample (25 μg) from patient 05-152. The pI range is 3–10, and the molecular mass range is 10–250 kDa. The boxed region designates the area of the gel that exhibits pronounced differences from control gels. The spots numbered 1–11 were excised and subjected to in-gel digestion, MALDI-TOF MS, and PMF analyses (the spectrum from spot number 1 is shown in Fig. 5). All spots were determined to be derived from TTR. The additional novel spots, indicated with the ▴ and ▪ symbols, were identified as the amyloid-associated proteins apolipoprotein E and serum amyloid P, respectively. This same sample was analyzed by 2D PAGE followed by Western blotting with polyclonal anti-TTR antisera. B shows the boxed regions of the gel from A with the results of Western blotting with polyclonal anti-human TTR antisera. The pI range is 4.5–5.9, and the molecular mass range is 10–35 kDa. Immunoreactive spots correspond to the numbered spots in A.
Characterization of Ig Light Chains in the 2D Gel Proteome of Fat Tissue Deposits from AL Patients—
The novel spots found in 2D PAGE analysis of proteins isolated from fat tissue biopsy samples from AL patients had clusters (largely in vertical ladders and horizontal trains) of many spots distributed across a wide molecular weight region. In most cases, they were located in the acidic side of the gel (Fig. 2, left panel, boxed region). In all cases, they included spots with an apparent molecular mass of ∼25 kDa, consistent with full-length immunoglobulin light chains. Relatively few spots were present in this region of the 2D gels from the control samples.
The representative 2D PAGE analysis shown in Fig. 2 was derived from the fat aspirate of AL patient 05-141. It displayed a main train of five spots of about 25 kDa (left panel, indicated by an arrow) in the region extending from approximately pI 5.0 to 5.5. These spots were associated with numerous other novel spots in a region with the same approximate pI range but extending down to about 10–11 kDa (left panel, boxed region, and right panel, enlargement). Again contamination with plasma proteins was minimal as indicated by the lack of a predominant albumin spot, although a small amount of intratissue clotting may have occurred as evidenced by the presence of fibrin and hemoglobin spots. Fourteen of the novel spots across the previously described gel area (Fig. 2, spot numbers 1–14) were excised and digested in-gel with trypsin, and the resultant peptides were analyzed by MALDI-TOF MS (Fig. 3, spectra from spot numbers 1, 8, and 12 shown vertically aligned). The spots of higher molecular weight (Fig. 2, spot numbers 1–4) were identified by Mascot PMF analysis as λ light chains (Ig λ C Sut, NCBInr accession number gi|224503, with MOWSE scores ranging from 70 to 135). The spots of lower molecular weight (Fig. 2, spot numbers 5–14), for most of which PMF did not return a significant score, were determined to contain the same (or very similar) light chain sequences on the basis of the presence of multiple peptide ions that were shared with the mass spectra of digests from the spots that were assigned from the PMF analysis. Probably because the residual portions of the truncated light chain proteins were dominated by the variable regions of the Ig light chains, the Mascot scores tended to be relatively low.
Fig. 3.
MALDI-TOF MS characterization of Ig light chain species in the fat tissue 2D proteome of AL patient 05-141. All numbered spots shown in Fig. 2 were subjected to in-gel digestion and MALDI-TOF MS analysis as described. Shown are the aligned spectra of peptides derived from spot numbers 1, 8, and 12 over the range m/z 700–3000. Major peptide ions in the spectrum from spot number 1 are labeled with observed m/z values and the corresponding amino acid intervals (bold) as assigned by PMF analysis and by comparison with theoretical values calculated from the cDNA-derived immunoglobulin light chain sequence obtained from the patient's plasma cell clone (shown above the spectra). Peptide ions displaying oxidation (+Ox), sodium adduction (+Na), and water loss (−H2O) are indicated by arrows extending from the peaks corresponding to their unmodified forms. Total protein coverage represented by these peptide ions is shown underlined in the sequence above. Dotted lines between spectra indicate the stepwise disappearance of C-terminal peptide ions from the spectrum for spot number 1 as compared with the spectrum for spot number 8 and thence to the spectrum of spot number 12, consistent with progressive C-terminal truncation of the protein species through this series of spots. The minimal Ig light chain amino acid sequence suggested to be present in each spot by the detected peptides is indicated in parentheses below the spot number. T, trypsin autolysis peptide; aa, amino acids.
Concurrently with this proteomics analysis, the cDNA encoding this patient's amyloidogenic light chain was cloned and sequenced from the plasma cell clone isolated from the bone marrow. The cDNA-deduced amino acid sequence was found to belong to the λ6 subfamily (Fig. 3, above the spectra). It had 217 residues with a theoretical molecular mass of 23,278 Da and a pI of 5.35, values that were very close to those observed by 2D PAGE for the intact light chain species (Fig. 2, spot number 1). The full-length species detected in the serum of the same patient are shown in Fig. 7A. Moreover the observed m/z values of the peptide ions observed in the mass spectra of the tryptic digests from spot numbers 1–14 matched the theoretical values of the tryptic and semitryptic peptides calculated on the basis of the λ6 protein sequence deduced from the plasma cell clone. This result confirmed that the circulating precursor was the same light chain protein that was deposited in fat tissue. In particular, it is important to note that the tryptic peptides derived from the spots at ∼25 kDa covered both the N and the C termini of the full-length amino acid sequence (Fig. 3, spectrum for spot number 1 and as underlined in the sequence above). In contrast, as the apparent molecular weights of the analyzed spots decreased, C-terminal peptide ions progressively disappeared from the mass spectra (e.g. in Fig. 3, spectra from spot numbers 8 and 12 exhibit loss of the C-terminal peptide ions, whose calculated positions are indicated by dotted lines extending from spectrum to spectrum). Therefore, the lower spots on the 2D gel (Fig. 2, spot numbers 5–14) were determined to originate from the variable and joining regions of the light chain (residues 1–112) and to retain variable-length portions of the constant region. Thus, both the full-length precursor and a heterogeneous population of its N-terminal fragments were found together in the deposits. All 112 amino acids comprising the light chain variable and joining regions and at least the first three residues of the constant region were retained in all the Ig light chain-derived proteins present in the analyzed spots.
Fig. 7.
2D PAGE analysis of the sera from patients with AL and TTR amyloidoses demonstrates the presence of the full-length, but not truncated, isoforms of the amyloidogenic precursors. Approximately 70 μl of matched serum samples were processed and subjected to 2D PAGE analysis as described. A shows a representative example of a silver-stained gel from AL patient 05-141, and B shows the gel from ATTR patient 05-152. The boxed areas in A and B correspond to the boxed regions of the gels displayed in Figs. 2 and 4, respectively. Circled spots were assigned on the basis of in-gel digestion followed by MALDI-TOF MS and PMF analyses of the proteolytic peptides. The data were compared with publicly available human plasma 2D reference maps (available from ExPASy) and proteomics databases. The protein in the spot indicated by the arrow in A was assigned on the basis of MS and PMF analyses as the patient's full-length Ig light chain. The spot aligned precisely with the spot at 25 kDa in the fat tissue analysis from the same patient that is indicated by an arrow in Fig. 2. Spots indicated by the arrows in B aligned with the two most basic spots in the train of spots at 13–14 kDa in the gel from this patient's fat tissue (indicated in Fig. 4 as spots number 1 and 2). 1, apolipoprotein A-I; 2, haptoglobin; 3, transthyretin. A sample equivalent to that in B was subjected 2D PAGE followed by Western analysis using polyclonal anti-human TTR antisera. C shows the region of this 2D Western blot corresponding to the boxed region in B. Arrows in C indicate immunoreactive spots corresponding to the silver-stained spots marked with arrows in B. The ▾ symbol indicates a weakly immunoreactive spot at ∼35 kDa.
In addition to truncated species, products resulting from other post-translational modifications and/or ex vivo degradations were observed in the MALDI-TOF mass spectra of the analyzed light chain as indicated in Fig. 3. In all spots, methionine-containing peptides 1–16 (m/z 1771.84) and 1–24 (m/z 2690.24) were found to be extensively oxidized. Additionally an ion (at m/z 1968.05), corresponding to a loss of 18 Da from the peptide 116–134 ([M + H]+ m/z 1986.02), was always present in the mass spectra of digests from the spots that contained this region of the amino acid sequence and suggested the facile loss of water within this peptide.
Characterization of Transthyretin in the Fat Tissue 2D Proteome of ATTR Patients—
In the 2D PAGE analysis of Congo red-positive fat samples from patients with ATTR, novel, strongly stained spots were located in a region of the gels corresponding to a pI of ∼5.5 and a molecular mass of 13–14 kDa, consistent with the migration of full-length, monomeric TTR. Fig. 4A shows an example of a 2D PAGE analysis of adipose tissue from an ATTR patient (05-152) who was heterozygotic for the variant in which there is substitution of isoleucine for valine at TTR residue 122 (V122I), resulting in the cardiomyopathic species. A pronounced cluster of spots was located in the above mentioned pI and molecular weight regions (Fig. 4, A (silver-stained gel) and B (anti-TTR Western analysis of the region boxed in A)). This cluster consisted of two major trains, each containing three spots (Fig. 4A, spot numbers 1–3 and 4–6), which had nearly equal staining intensity but were shifted in molecular weight and pI relative to one another. The migration of the upper train of spots (numbers 1–3) was consistent with full-length monomeric TTR. Fig. 7, B and C, show the apparently full-length TTR species detected in the serum of the same patient. The position of the lower train of spots (spot numbers 4–6) was consistent with TTR species that had lost a charged fragment. Additional novel spots of lesser intensity were present in numerous trains of spots having higher molecular weights (Fig. 4A, spot numbers 7–11). These may consist of dimers or multimers of intact or truncated TTR that remained associated during sample preparation and electrophoresis despite the strongly denaturing conditions. Western blotting with polyclonal anti-human TTR antibodies demonstrated strong immunoreactivity against most of the novel protein spots (Fig. 4B).
All the spots corresponding to the major immunoreactive species were subjected to in-gel digestion, MALDI-TOF MS, and PMF analyses. Fig. 5 shows a representative spectrum derived from the spot designated number 1 in Fig. 4. All the spots at 13–14 kDa and at 35 kDa (Fig. 4A, spot numbers 7–11) contained both wild-type TTR and the V122I variant (e.g. for spot number 1, NCBI accession number gi|1827569 with a MOWSE score of 185). For most spots assigned as TTR-related proteins, the detected peptide ions yielded almost complete sequence coverage (Fig. 5, sequence coverage is underlined). The N-terminal TTR peptide 1–15 was not detected in the MALDI-TOF mass spectra of any spots; however, this observation alone is not sufficient to indicate N-terminal truncation because this peptide is known to be difficult to detect by MALDI MS (7). Peptide 16–21 (at m/z 672.42) was detected only in spot numbers 1–3 and 7–11 and was not observed in the spectra of spot numbers 4–6, whereas the TTR peptide corresponding to residues 22–34 (at m/z 1366.74) was detected in the mass spectra of all spots analyzed. These overall results are consistent with the hypothesis that the lower train of spots contained N-terminally truncated forms of TTR.
Fig. 5.
MALDI-TOF MS characterization of wild-type and V122I variant TTR in the 2D SDS-PAGE proteome of an ATTR patient. All spots numbered on the 2D gel shown in Fig. 4 (2D PAGE analysis of a reduced and alkylated sample from patient 05-152) were subjected to in-gel digestion with trypsin and MALDI-TOF MS analysis as described. Shown is the mass spectrum of tryptic peptides derived from spot number 1 over the range m/z 500–3200. Major peptide ions are labeled with observed m/z values and the corresponding amino acid intervals (bold) as assigned by PMF analysis and by comparison with predicted values calculated from the genomic DNA-derived sequences for wild-type TTR and its V122I variant. Peptide ions displaying single (+Ox) and double (+2Ox) oxidation are indicated by arrows extending from the peaks assigned to their unmodified forms. Total protein coverage represented by these peptide ions is shown underlined in the TTR sequence above the spectrum. The wild-type and variant residues at position 122 are both indicated in bold. The inset shows an expansion of the spectrum over the range m/z 2480–2540. Wild-type and V122I variant peptide ion pairs, corresponding to TTR peptides 105–127 and 104–126 as indicated, are separated by a mutation-derived mass shift of +14 Da. Two additional pairs, corresponding to TTR peptides 105–126 and 104–127, are also designated. T, trypsin autolysis peptide; wt, wild type.
The set of ions corresponding to peptides containing TTR residue 122 occurred in pairs that each consisted of both the wild-type valine-containing peptide ion and the variant isoleucine-containing peptide ion, separated by the calculated mass shift of 14 Da. Four such peptide ion pairs are visible in the spectrum shown in Fig. 5; the region including two of these is enlarged in the figure inset. The fact that peptides from both the amyloidogenic variant and the wild-type TTR were found in all these spots indicates that the two TTR species were co-deposited into fibrils and were identically processed to produce the array of TTR-related products that are heterogeneous in molecular weight and pI. Given that the valine-to-isoleucine substitution is unlikely to have a significant impact on the MALDI ionization efficiencies of the peptides, the ratios of the signal intensities of the ions corresponding to the wild-type and variant peptides can be used to estimate the relative abundances of the two protein species. Thus, it can be concluded that the patient had essentially equimolar deposition of wild-type and V122I variant TTR species into the fat tissue fibrils.
Oxidized forms of the tryptophan-containing peptides 35–48 and 36–48 were also detected in the spectra (Fig. 5). Their mass shifts, relative to the unmodified peptides, exhibited a characteristic oxidation pattern that indicated conversions of the native tryptophan to hydroxytryptophan (+16 Da), N-formylkynurenine (+32 Da), and kynurenine (+4 Da) (34). We have observed previously that Trp-41 in TTR is particularly susceptible to oxidation (8, 33). Within the trains of TTR-related spots on the gels analyzed here, we found that the extent of oxidation at position 41 (Trp-41) increased with a decrease in the observed pI (data not shown).
Identification of Non-fibrillar Proteins Known to Associate with Amyloid Deposits—
Numerous non-fibrillar proteins are known to associate with amyloid deposits in various tissues (35), and therefore, we anticipated that some amyloid-associated proteins might be identified among the novel spots appearing in the 2D PAGE gel separations of proteins from patient samples. MALDI-TOF MS and PMF analyses confirmed this hypothesis through the assignments of trains of spots to isoforms of serum amyloid P and apolipoprotein E (e.g. Figs. 2 and 4, isoforms of serum amyloid P and apolipoprotein E designated by the symbols ▴ and ▪, respectively). Notably the quantities of these amyloid-associated proteins varied among the samples from different patients and showed only weak correlation with the Congo red staining-based estimates of each patient's grade of amyloid fibril load.
Disease-related Changes in the Expression of Adipose Tissue-resident Proteins in Patients with Amyloidosis—
In addition to our characterization of the deposition of amyloidogenic and amyloid-associated proteins in adipose tissue, we were interested to determine whether there were any other disease-related changes in the adipose tissue proteome. Therefore, we examined whether the presence of fibril deposits was associated with any consistent perturbations in the intensity of spots that could be assigned to adipose tissue-resident proteins. The results for samples from a representative control and four of these patients are shown in Fig. 6. In the fat tissue proteomes of nearly all patients with amyloidosis, we observed alteration in the expression of the small heat shock protein αB-crystallin. Two different isoforms of this protein were expressed in normal fat tissue. The more abundant of these had a pI of ∼6.7 and an apparent molecular mass of 22 kDa, whereas the less abundant form had a pI close to 6.3 and molecular mass slightly less than 22 kDa (Fig. 6A, CONTROL panel). Marked decreases in the intensities of the spots for both isoforms were observed in the 2D gel images of the proteins from the amyloid deposits of six patients diagnosed with AL and/or ATTR amyloidoses (Fig. 6A, PATIENTS panels). A marked increase in both isoforms of αB-crystallin, particularly in the more acidic and slightly smaller isoform, was visible in samples from two more cases (one patient with AL and one with ATTR) (data not shown). In the 2D gels of samples from patients with amyloidoses, additional protein spots exhibited staining intensities that were reduced relative to controls. These spots were identified as peroxiredoxin 6 (e.g. 2D gel panels in Fig. 6B) and aldo-keto reductase I; the intensities of both were reduced on the 2D gels of five patients (e.g. 2D gel panels in Fig. 6C). The intensities of the spots attributed to peroxiredoxin were reduced in samples from all six patients, and the intensities of those assigned to aldo-keto reductase I were reduced in samples from five of the six patients.
Fig. 6.
Disease-associated decreases observed in the expression of spots assigned to adipocyte proteins in patients with AL and ATTR amyloidoses. Details of regions of the 2D PAGE proteomes of abdominal subcutaneous fat tissue aspirates (as numerically coded in the upper left corner of each image) are shown in comparison with those from normal controls. After staining of the gel spots, image exposure levels were matched to the total non-amyloidogenic protein load and to the intensity of the non-amyloid-associated landmark spots. Circled spots, which demonstrated pronounced decreases in the staining intensity in the samples from most patients, were identified by in-gel digestion, MALDI-TOF MS, and PMF analyses. A, isoforms of αB-crystallin (αB-CRY) with approximate molecular mass of 22 kDa. B, peroxiredoxin 6 with approximate molecular mass of 25 kDa. C, aldo-keto reductase with approximate molecular mass of 37 kDa. LC, an amyloidogenic light chain spot from one patient (06-095) is visible in the detail.
Identification of the Circulating Amyloidogenic Precursor in Full-length, but Not Truncated, Isoforms in the Sera of AL and ATTR Patients—
To determine the extent of similarity between the circulating forms of the amyloidogenic proteins and their counterparts deposited as amyloid fibrils in fat tissue, serum samples and fat aspirates were acquired from patients during the same clinical sessions, and the serum samples were also analyzed by 2D PAGE followed by silver staining. Fig. 7 shows 2D gels of representative samples from patients diagnosed with AL and ATTR. In both AL and ATTR serum samples, full-length circulating amyloidogenic proteins were readily detected as strongly staining spots, similar to those detected in the fat tissue analysis. However, the heterogeneous populations of amyloidogenic protein spots of various pI values and molecular weights that characterized the 2D gels of the fat deposits were not observed in the analyses of serum samples: no spots that corresponded to the spots assigned as truncated light chain or TTR spots in fat samples were observed in the 2D gel separations of the serum samples from the same patients (e.g. the boxed areas in Fig. 7, A and B, correspond to the boxed areas in Figs. 2 and 4). For example, a single spot identified as a λ light chain by immunoblotting and MS (Fig. 7A, arrow), shown in the 2D gel of the serum from AL patient 05-141, aligned exactly with the most basic protein species in the train of spots at 25 kDa on the 2D gel of the matching fat tissue (Fig. 2, arrow). The peptide ions detected in the mass spectrum acquired from this serum spot corresponded to those present in the spectra from the spots assigned to Ig light chain from the fat tissue and covered both the N and the C termini of the predicted amyloidogenic light chain sequence, thus confirming that this serum spot indeed represented the full-length form of circulating precursor. The additional spots corresponding to the charge and molecular weight isoforms present in the patient's adipose tissue sample (Fig. 2, right panel, spot numbers 5–14) were not detected in the same region of the gel of the serum sample. Similarly the amyloidogenic protein region of the 2D gel of the serum from ATTR patient 05-152 (Fig. 7B) showed only a pair of prominent spots that had approximately equal intensities, and both were identified as TTR by immunoblotting as shown in Fig. 7C. These immunoreactive spots corresponded to the two most basic spots in the train at 13–14 kDa on the 2D gel of the matching fat tissue (Fig. 4, spot numbers 1 and 2). Additional serum TTR species were detected by immunoblotting as a faint spot (Fig. 7C) and corresponded to a TTR spot at 35 kDa on the gel from the fat tissue (Fig. 4B). Thus, within the range of detection by silver staining, the abundant forms of amyloidogenic proteins detected by 2D PAGE analysis in serum were significantly less heterogeneous in pI and molecular weight than those detected in the amyloid deposits in fat tissue.
DISCUSSION
The investigation of the proteome of amyloid-infiltrated tissues constitutes a novel approach to the study of systemic amyloid diseases. Here we report the first application of the well established 2D PAGE differential display technique, combined with MALDI-TOF MS peptide mass fingerprinting, to the analysis of subcutaneous adipose tissue obtained by fine needle aspiration from patients with systemic amyloidoses. We developed a rapid and simple procedure that efficiently solubilizes amyloid deposits from small amounts of subcutaneous fat aspirate material and prepares the samples for high resolution 2D PAGE analysis. The method provides a way to characterize protein deposits and detect changes in protein expression in the adipose tissue from patients with different types of systemic amyloidoses. We consider this approach to be a valuable complement to traditional diagnostic procedures used for patients with systemic amyloidoses.
In the 2D PAGE gels of all the patient fat aspirate samples we analyzed, we found novel, disease-associated spots. The 2D gel migration of these spots proved to be consistent with the positions we established for amyloidogenic proteins and their fragments; the amyloid-related spots were easily located by comparison with 2D PAGE maps of control samples. Furthermore the 2D PAGE maps of the samples from different types of amyloidosis were distinct from one another. Unequivocal identification of the novel spots was accomplished by mass spectrometry and peptide mass fingerprint analyses and by immunoblotting. The combination of these approaches precisely assigned the amyloidogenic protein present in each of the deposits and also defined co-localized proteins. Additionally the presence of specific amino acid substitutions that result from genetic mutations that lead to the hereditary forms of amyloidosis could be directly detected by MS of the deposited species on the basis of characteristic mass shifts in proteolytic peptides. Because fat aspirates are routinely acquired for histological studies during clinical evaluation for systemic amyloid disease, the described approach is practical and feasible for virtually all patients.
For the purpose of the biochemical characterization of the protein constituents of tissue amyloid deposits, separation on a 2D gel is suitable for resolving the complexity of the species deposited in fat without laborious and time-consuming extraction and purification of the amyloid fibrils. However, it is important to emphasize the critical role of careful tissue washing in this analytical procedure because circulating amyloidogenic precursors present in contaminating plasma could bias the results if they are not efficiently removed from the sample obtained by biopsy. It should be noted that repeated cycles of washing with isotonic saline solution efficiently eliminated the common plasma-related precursors that were not part of the amyloid deposits, including immunoglobulin light chains or transthyretin, so that they were not visible in the 2D gels of our control samples.
The amyloid deposits in fat that we examined were not homogeneous in their deposited protein species but rather were composed of a population of species derived from the amyloidogenic precursor that was unexpectedly heterogeneous in terms of pI and molecular weight in contrast to the relatively narrow molecular weight distribution we observed for the circulating forms of these proteins. The complexity of the deposited amyloid proteins was particularly pronounced in AL where multiple, different, highly abundant species were simultaneously detected. In the 2D gel separations of samples from all the AL patients, the full-length light chains could be clearly distinguished from the fragments and constituted a significant fraction of the deposits. Additionally we detected numerous fragments that retained the N terminus but exhibited various degrees of truncation within the C-terminal constant region. These findings were consistent with previously reported results obtained by other techniques (36–38). Both the earlier reports and our results suggest that the variable region, rather than the constant region, makes the greater contribution to amyloid formation. However, the presence of a large amount of full-length light chain in the deposits indicates that truncation is presumably not a necessary step for protein unfolding and deposition into fibrils.
The considerable charge heterogeneity of the deposited light chains, as resolved by isoelectric focusing, provides evidence for multiple post-translational modifications of the light chains that apparently affect a large fraction of the deposited species. The observed ladder patterns of spots on the gels were consistent with progressive protein truncation, whereas the observed train patterns may have resulted from oxidation, phosphorylation, deamidation, or loss of singly charged residues from the light chain species. Protein deamidation, in particular, has been demonstrated to correlate with reduced amyloidogenic protein stability and seems to favor aggregation (39). Interestingly it seems likely that the observed light chain modifications occurred during, or subsequent to, the process of deposition of these proteins in tissue because light chain fragments or charge isoforms were not detected in the corresponding sera. Changes in the surface-exposed residues of the fibril protein constituents, occurring during their slow turnover in fibrils, could account for some or all of the described modifications. Truncation could be due to the activity of tissue proteases (40) on regions of the light chains that were not tightly packed into the fibril core. Alternatively modifications could occur prior to deposition if minute amounts of modified circulating forms have a high propensity to unfold, thus increasing their exposure to proteases, oxidative modifications, and non-enzymatic hydrolysis and thereby shortening their half-life in the blood and/or promoting rapid deposition in tissues and increasing the relative abundances of these forms in the fat tissue as compared with the serum.
Similar findings, in terms of fragmentation and charge heterogeneity, were observed for the deposited transthyretin protein in the ATTR samples. In these cases, a strong tendency of the protein to associate in multimers was also evidenced by the presence of high molecular weight TTR species that could not be dissociated even under strongly denaturing and reducing conditions. This is the first report of the extensive biochemical characterization of the amyloid deposits in ATTR caused by a cardiomyopathic TTR variant, V122I, which has higher prevalence in the African-American population (41). The presence of truncated TTR (typically lacking the first 46–51 residues from the N terminus) in the amyloid deposits has been described both in familial ATTR and in senile systemic amyloidosis (9, 42, 43). In the V122I amyloid sample we analyzed for this study, the truncated species constituted approximately half of the deposited TTR as evidenced by staining intensity. According to the MS results, a major truncation point occurred before position 22 as a result of cleavage significantly closer to the N terminus than was reported for other variants (44). Additionally it is interesting that wild-type and variant species were found by MS to co-migrate with approximately equal abundance in all the TTR spots. These results implied a pathogenic (or at least cooperative) role for wild-type TTR in the context of the mutant TTR species, and they also indicated that the modifications of both forms of TTR were equal in mode and extent. Wild-type and variant TTR were both truncated in the same manner in the region before residue 22. This previously unreported finding suggested that the presence of this particular variant species influenced the processing of the wild-type protein before or after deposition. The fact that fragments of the precursor were not detectable in serum by 2D PAGE analysis was consistent with the hypothesis that truncation may occur on exposed regions of the sequence after the co-deposition of the two species into fibrils in an approximately equimolar ratio. A progressive increase in the extent of oxidation of TTR tryptophan 41 was observed as spots increased in acidity within the same trains. This result suggested that the modification might have occurred, at least in part, in vivo and was not solely an artifact resulting from sample handling between the SDS-PAGE and MS analyses, consistent with our previous findings.2 This modification may have biological significance in that it may be the consequence of oxidative stress that occurred in the tissue after amyloid deposition. Alternatively tryptophan oxidation could have been promoted by oxidative stress in the circulation prior to amyloid deposition. Such conditions may lead to a loss of stability and to protein aggregation. Further studies are needed to establish the precise natures of each of the TTR and light chain modifications and their exact role(s) in the process of amyloid formation.
The identity of all the species that were recognized by the anti-TTR antibodies on Western blot was confirmed by MALDI MS and PMF analysis of the corresponding spots in the gel. The presence of TTR species with an apparent molecular mass higher than the monomer (about 35 kDa) in 2D PAGE gels under reducing conditions has been reported previously in normal plasma (see the human plasma map on the Swiss-2DPAGE Website). The hypothesis that the presence of proteins whose binding to TTR could have been responsible for its higher molecular weight migration was considered. However, in all the TTR spots (numbered in the patient sample in Fig. 4), TTR was the only protein identified. The consistent detection of altered expression of certain resident proteins in the majority of amyloid-infiltrated adipose samples examined in this study is intriguing and suggests the possibility that these proteins contribute to the development of amyloidosis. The amount of amyloid in affected tissue differs considerably among patients (Congo red intensity scores ranging from 1 to 4), and this changes the proportion of non-amyloid-derived proteins on the total protein load. Direct comparison between raw maps from patients and controls could thus lead to underestimation of the expression of resident proteins in the former. To overcome this issue, before comparison, the exposure levels in images from patient samples were matched to the total estimated non-amyloid protein load, and the intensity of non-amyloid-associated landmark spots, which were not expected to change as a consequence of amyloid deposition (such as β-actin, spot number 2 in Fig. 1), was used for spot intensity normalization. After this correction, most proteins did not appear to decrease significantly in patients’ samples, whereas the reduction of the intensity of the αB-crystallin, peroxiredoxin 6, and aldo-keto reductase 1 spots was confirmed. However, a detailed analysis of all dysregulated adipocyte proteins was beyond the scope of the present study given the apparent diversity of the fat tissue proteomes obtained from even this small set of patients. Larger scale studies will be required before concrete conclusions can be made regarding the full suite of amyloid deposit-associated proteins and their roles.
Alteration in the expression of αB-crystallin by adipocytes, observed in both AL and ATTR fat samples, was particularly striking. αB-Crystallin, a small intracellular chaperone with antiapoptotic activity (45), has been extensively studied in vitro for its potential to inhibit amyloid fibril formation, specifically the formation of fibrils from the Aβ protein, β2-microglobulin, and α-synuclein (46–48). However, αB-crystallin has never been reported to inhibit fibrillogenesis in AL or TTR amyloidosis. The expression of αB-crystallin is known to be dysregulated in the brain in many neurodegenerative disorders (49). In particular, it has been shown to be increased in the astroglia of patients with Alzheimer disease, but immunohistochemistry did not show that it co-localized with amyloid plaques in these patients (50). In fact, an increase in αB-crystallin has been linked to reductive stress (51). In the samples examined here, the expression level of αB-crystallin was drastically reduced in the majority of our adipose tissue samples obtained from patients with systemic amyloidoses. Considering that adipocytes are not involved in the synthesis of the amyloidogenic precursors, the significance of this finding in the setting of systemic amyloidoses is unclear, but it does suggest that the extracellular amyloid deposits can influence, or can be influenced by, the expression of intracellular proteins involved in protein folding and cell survival. Furthermore the reduction in expression of αB-crystallin relative to control samples may not be confined to adipocytes but may also occur in other tissues. In particular, deficient chaperone activity of αB-crystallin in tissues that produce amyloidogenic precursors could underlie the secretion of these pathogenic unfolded or misfolded proteins.
The expression of two other adipocyte-resident proteins, peroxiredoxin 6 and aldo-keto reductase I, was also reduced in most of the fat samples from amyloid patients that we examined in this study. These two proteins are involved in the regulation of cellular energetics and redox status. These findings suggested the contribution of oxidative stress, which has been hypothesized to mediate amyloid-associated damage in other cell types (52, 53), to the pathologic perturbations in adipose tissue in systemic amyloidoses. The aberrant expression of proteins involved in the antioxidant response, in particular peroxiredoxins, has been demonstrated previously in neurodegenerative disorders (49, 54).
Extension of this pilot study to a large cohort of patients can take advantage of the easy accessibility of this appropriate patient tissue, the subcutaneous fat aspirate, and should enable deeper and potentially highly fruitful investigations into the causes and consequences of amyloid fibril deposition in systemic amyloidoses. Further studies are needed to determine whether the altered and varying expression for adipocyte-resident proteins that we report here may enhance amyloid fibril deposition or occur in response to fibril deposition and to investigate how these proteins may participate in pathogenesis of amyloid diseases.
Supplementary Material
Acknowledgments
We kindly thank Prof. Paolo Dionigi and staff, Drs. Curtis Barry, Michael Rosenzweig, Giovanni Palladini, and all the patients who provided the fat tissue samples.
Footnotes
Published, MCP Papers in Press, May 12, 2008, DOI 10.1074/mcp.M700545-MCP200
The abbreviations used are: AL, light chain amyloidosis; ATTR, transthyretin amyloidosis; 2D, two-dimensional; PMF, peptide mass fingerprinting; TTR, transthyretin; VL, variable region, light chain; FR, framework region; MOWSE, molecular weight search.
M. E. McComb, A. Lim, E. A. Berg, L. H. Connors, M. Skinner, and C. E. Costello, unpublished results.
This work was supported, in whole or in part, by National Institutes of Health Grants P41 RR10888 and S10 RR15942 and Contract N01 HV28178 (to C. E. C.) and Grant P01 HL 68705 (to D. C. S.). This work was also supported by contributions from the Gerry Foundation, the Young Family Amyloid Research Fund, and the David Jamieson Amyloid Research Fund (to M. S.) and by grants from the Italian Ministry of Health, the Cariplo Foundation “Network Operativo per la Biomedica di Eccellenza in Lombaria (NOBEL) project,” the Foundation IRCCS Policlinico San Matteo, and Ghislieri College Pavia (to G. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Merlini, G., and Bellotti, V. ( 2003) Molecular mechanisms of amyloidosis. N. Engl. J. Med. 349, 583–596 [DOI] [PubMed] [Google Scholar]
- 2.Westermark, P., Benson, M. D., Buxbaum, J. N., Cohen, A. S., Frangione, B., Ikeda, S. I., Masters, C. L., Merlini, G., Saraiva, M. J., and Sipe, J. D. ( 2005) Amyloid: toward terminology clarification—report from the Nomenclature Committee of the International Society of Amyloidosis. Amyloid 12, 1–4 [DOI] [PubMed] [Google Scholar]
- 3.Obici, L., Perfetti, V., Palladini, G., Moratti, R., and Merlini, G. ( 2005) Clinical aspects of systemic amyloid diseases. Biochim. Biophys. Acta 1753, 11–22 [DOI] [PubMed] [Google Scholar]
- 4.Falk, R. H., Comenzo, R. L., and Skinner, M. ( 1997) Medical progress—the systemic amyloidoses. N. Engl. J. Med. 337, 898–909 [DOI] [PubMed] [Google Scholar]
- 5.Picken, M. M. ( 2007) New insights into systemic amyloidosis: the importance of diagnosis of specific type. Curr. Opin. Nephrol. Hypertens. 16, 196–203 [DOI] [PubMed] [Google Scholar]
- 6.Saraiva, M. J. ( 2001)Transthyretin amyloidosis: a tale of weak interactions. FEBS Lett. 498, 201–203 [DOI] [PubMed] [Google Scholar]
- 7.Lim, A., Prokaeva, T., McComb, M. E., O'Connor, P. B., Theberge, R., Connors, L. H., Skinner, M., and Costello, C. E. ( 2002) Characterization of transthyretin variants in familial transthyretin amyloidosis by mass spectrometric peptide mapping and DNA sequence analysis. Anal. Chem. 74, 741–751 [DOI] [PubMed] [Google Scholar]
- 8.McComb, M. E., Lim, A., Théberge, R., Prokaeva, T., Connors, L. H., Skinner, M., and Costello, C. E. ( 2004) Clinical diagnosis of familial transthyretin amyloidosis by quadrupole orthogonal time-of-flight mass spectrometry, in Proceedings of the 50th ASMS Conference on Mass Spectrometry and Allied Topics, Orlando, FL, 2002; Abstract WOCam-11:15, American Society for Mass Spectrometry, Santa Fe, NM
- 9.Kingsbury, J. S., Théberge, R., Karbassi, J. A., Lim, A., Costello, C. E., and Connors, L. H. ( 2007) Detailed structural analysis of amyloidogenic wild type transthyretin using a novel purification strategy and mass spectrometry. Anal. Chem. 79, 1990–1998 [DOI] [PubMed] [Google Scholar]
- 10.Sekijima, Y., Wiseman, R. L., Matteson, J., Hammarström, P., Miller S. R., Sawkar, A. R., Balch, W. E., and Kelly, J. W. ( 2005) The biological and chemical basis for tissue-selective amyloid disease. Cell 121, 73–85 [DOI] [PubMed] [Google Scholar]
- 11.Zhang, Q., and Kelly, J. W. ( 2005) Cys-10 mixed disulfide modifications exacerbate transthyretin familial variant amyloidogenicity: a likely explanation for variable clinical expression of amyloidosis and the lack of pathology in C10S/V30M transgenic mice? Biochemistry 44, 9079–9085 [DOI] [PubMed] [Google Scholar]
- 12.Connors, L. H., Jiang, Y., Budnik, M., Théberge, R., Prokaeva, T., Bodi, K. L., Seldin, D. C., Costello, C. E., and Skinner, M. ( 2007) Heterogeneity in primary structure, post-translational modifications and germline gene usage of nine full length amyloidogenic κ1 immunoglobulin light chains. Biochemistry 46, 14259–14271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duston, M. A., Skinner, M., Meenan, R. F., and Cohen, A. S. ( 1989) Sensitivity, specificity, and predictive value of abdominal fat aspiration for the diagnosis of amyloidosis. Arthritis Rheum. 32, 82–85 [DOI] [PubMed] [Google Scholar]
- 14.van Gameren, I. I., Hazenberg, B. P., Bijzet, J., and van Rijswijk, M. H. ( 2005) Diagnostic accuracy of routine versus dedicated studied subcutaneous abdominal fat tissue for detection of systemic amyloidosis. Arthritis Rheum. 52, S247–S247 [DOI] [PubMed] [Google Scholar]
- 15.Westermark, P., Davey, E., Lindbom, K., and Enqvist, S. ( 2006) Subcutaneous fat tissue for diagnosis and studies of systemic amyloidosis. Acta Histochem. 108, 209–213 [DOI] [PubMed] [Google Scholar]
- 16.Anesi, E., Palladini, G., Perfetti, V., Arbustini, E., Obici, L., and Merlini, G. ( 2001) Therapeutic advances demand accurate typing of amyloid deposits. Am. J. Med. 111, 243–244 [DOI] [PubMed] [Google Scholar]
- 17.Lachmann, H. J., Booth, D. R., Booth, S. E., Bybee, A., Gilbertson, J. A., Gillmore, J. D., Pepys, M. B., and Hawkins, P. N. ( 2002) Misdiagnosis of hereditary amyloidosis as AL (primary) amyloidosis. N. Engl. J. Med. 346, 1786–1791 [DOI] [PubMed] [Google Scholar]
- 18.Linke, R. P., Oos, R., Wiegel, N. M., and Nathrath, W. B. J. ( 2006) Classification of amyloidosis: misdiagnosing by way of incomplete immunohistochemistry and how to prevent it. Acta Histochem. 108, 197–208 [DOI] [PubMed] [Google Scholar]
- 19.Kebbel, A., and Rocken, C. ( 2006) Immunohistochemical classification of amyloid in surgical pathology revisited. Am. J. Surg. Pathol. 30, 673–683 [DOI] [PubMed] [Google Scholar]
- 20.Murphy, C. L., Wang, S., Williams, T., Weiss, D. T., and Solomon, A. ( 2006) Characterization of systemic amyloid deposits by mass spectrometry. Methods Enzymol. 412, 48–62 [DOI] [PubMed] [Google Scholar]
- 21.Kaplan, B., Vidal, R., Kumar, A., Ghiso, J., and Gallo, G. ( 1999) Immunochemical microanalysis of amyloid proteins in fine-needle aspirates of abdominal fat. Am. J. Clin. Pathol. 112, 403–407 [DOI] [PubMed] [Google Scholar]
- 22.Kaplan, B., Martin, B. M., Livneh, A., Pras, M., and Gallo, G. R. ( 2004) Biochemical subtyping of amyloid in formalin-fixed tissue samples confirms and supplements immunohistologic data. Am. J. Clin. Pathol. 121, 794–800 [DOI] [PubMed] [Google Scholar]
- 23.Corton, M., Villuendas, G., Botella, J. I., San Millan, J. L., Escobar-Morreale, H. F., and Peral, B. ( 2004) Improved resolution of the human adipose tissue proteome at alkaline and wide range pH by the addition of hydroxyethyl disulfide. Proteomics 4, 438–441 [DOI] [PubMed] [Google Scholar]
- 24.Hughes, G. J., Frutiger, S., Paquet, N., Ravier, F., Pasquali, C., Sanchez, J. C., James, R., Tissot, J. D., Bjellqvist, B., and Hochstrasser, D. F. ( 1992) Plasma protein map—an update by microsequencing. Electrophoresis 13, 707–714 [DOI] [PubMed] [Google Scholar]
- 25.Perlman, D. H., Berg, E. A., O'Connor, P. B., Costello, C. E., and Hu, J. ( 2005) Reverse transcription-associated dephosphorylation of hepadnavirus nucleocapsids. Proc. Natl. Acad. Sci. U. S. A. 102, 9020–9025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karas, M., Bachmann, D., Bahr, U., and Hillenkamp, F. ( 1987) Matrix-assisted ultraviolet-laser desorption of nonvolatile compounds. Int. J. Mass Spectrom. 78, 53–68 [Google Scholar]
- 27.Karas, M., and Hillenkamp, F. ( 1988) Laser desorption ionization of proteins with molecular masses exceeding 10000 daltons. Anal. Chem. 60, 2299–2301 [DOI] [PubMed] [Google Scholar]
- 28.Clauser, K. R., Baker, P., and Burlingame, A. L. ( 1999) Role of accurate mass measurement (±10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 71, 2871–2882 [DOI] [PubMed] [Google Scholar]
- 29.Comenzo, R. L., Michelle, D., LeBlanc, M., Wally, J., Zhang, Y., Kica, G., Karandish, S., Arkin, C. F., Wright, D. G., Skinner, M., and McMannis, J. ( 1998) Mobilized CD34+ cells selected as autografts in patients with primary light-chain amyloidosis: rationale and application. Transfusion 38, 60–69 [DOI] [PubMed] [Google Scholar]
- 30.Welschof, M., Terness, P., Kolbinger, F., Zewe, M., Dubel, S., Dorsam, H., Hain, C., Finger, M., Jung, M., Moldenhauer, G., Hayashi, N., Little, M., and Opelz, G. ( 1995) Amino acid sequence based PCR primers for amplification of rearranged human heavy and light chain immunoglobulin variable region genes. J. Immunol. Methods 179, 203–214 [DOI] [PubMed] [Google Scholar]
- 31.Weichman, K., Dember, L. M., Prokaeva, T., Wright, D. G., Quillen, K., Rosenzweig, M., Skinner, M., Seldin, D. C., and Sanchorawala, V. ( 2006) Clinical and molecular characteristics of patients with non-amyloid light chain deposition disorders, and outcome following treatment with high-dose melphalan and autologous stem cell transplantation. Bone Marrow Transplant. 38, 339–343 [DOI] [PubMed] [Google Scholar]
- 32.Giudicelli, V., Chaume, D., and Lefranc, M. P. ( 2004) IMGT/V-QUEST, an integrated software program for immunoglobulin and T cell receptor V-J and V-D-J rearrangement analysis. Nucleic Acids Res. 32, W435–W440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim, A., Prokaeva, T., Connors, L. H., Falk, R. H., Skinner, M., and Costello, C. E. ( 2002) Identification of a novel transthyretin Thr59Lys/Arg104His. A case of compound heterozygosity in a Chinese patient diagnosed with familial transthyretin amyloidosis. Amyloid 9, 134–140 [PubMed] [Google Scholar]
- 34.Finley, E. L., Dillon, J., Crouch, R. K., and Schey, K. L. ( 1998) Identification of tryptophan oxidation products in bovine α-crystallin. Protein Sci. 7, 2391–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kisilevsky, R. ( 2000) The relation of proteoglycans, serum amyloid P and apo E to amyloidosis current status, 2000. Amyloid 7, 23–25 [DOI] [PubMed] [Google Scholar]
- 36.Olsen, K. E., Sletten, K., and Westermark, P. ( 1998) Extended analysis of AL-amyloid protein from abdominal wall subcutaneous fat biopsy: Kappa IV immunoglobulin light chain. Biochem. Biophys. Res. Commun. 245, 713–716 [DOI] [PubMed] [Google Scholar]
- 37.Olsen, K. E., Sletten, K., and Westermark, P. ( 1998) Fragments of the constant region of immunoglobulin light chains are constituents of AL-amyloid proteins. Biochem. Biophys. Res. Commun. 251, 642–647 [DOI] [PubMed] [Google Scholar]
- 38.Westermark, P., Benson, L., Juul, J., and Sletten, K. ( 1989) Use of subcutaneous abdominal fat biopsy specimen for detailed typing of amyloid fibril protein-AL by amino acid sequence analysis. J. Clin. Pathol. 42, 817–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nilsson, M. R., Driscoll, M., and Raleigh, D. P. ( 2002) Low levels of asparagine deamidation can have a dramatic effect on aggregation of amyloidogenic peptides: implications for the study of amyloid formation. Protein Sci. 11, 342–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bohne, S., Sletten, K., Menard, R., Buhling, F., Vockler, S., Wrenger, E., Roessner, A., and Rocken, C. ( 2004) Cleavage of AL amyloid proteins and AL amyloid deposits by cathepsins B, K, and L. J. Pathol. 203, 528–537 [DOI] [PubMed] [Google Scholar]
- 41.Yamashita, T., Asl, K. H., Yazaki, M., and Benson, M. D. ( 2005) A prospective evaluation of the transthyretin Ile122 allele frequency in an African-American population. Amyloid 12, 127–130 [DOI] [PubMed] [Google Scholar]
- 42.Westermark, P., and Westermark, G. T. ( 2005) Purification of transthyretin and transthyretin fragments from amyloid-rich human tissues. Methods Mol. Biol. 299, 255–260 [DOI] [PubMed] [Google Scholar]
- 43.Bergstrom, J., Gustavsson, A., Hellman, U., Sletten, K., Murphy, C. L., Weiss, D. T., Solomon, A., Olofsson, B. O., and Westermark, P. ( 2005) Amyloid deposits in transthyretin-derived amyloidosis: cleaved transthyretin is associated with distinct amyloid morphology. J. Pathol. 206, 224–232 [DOI] [PubMed] [Google Scholar]
- 44.Schormann, N., Murrell, J. R., and Benson, M. D. ( 1998) Tertiary structures of amyloidogenic and non-amyloidogenic transthyretin variants: new model for amyloid fibril formation. Amyloid 5, 175–187 [DOI] [PubMed] [Google Scholar]
- 45.Kamradt, M. C., Lu, M., Werner, M. E., Kwan, T., Chen, F., Strohecker, A., Oshita, S., Wilkinson, J. C., Yu, C., Oliver, P. G., Duckett, C. S., Buchsbaum, D. J., LoBuglio, A. F., Jordan, V. C., and Cryns, V. L. ( 2005) The small heat shock protein αB-crystallin is a novel inhibitor of TRAIL-induced apoptosis that suppresses the activation of caspase-3. J. Biol. Chem. 280, 11059–11066 [DOI] [PubMed] [Google Scholar]
- 46.Rekas, A., Adda, C. G., Andrew Aquilina, J., Barnham, K. J., Sunde, M., Galatis, D., Williamson, N. A., Masters, C. L., Anders, R. F ., Robinson, C. V., Cappai, R., and Carver, J. A. ( 2004) Interaction of the molecular chaperone αB-crystallin with α-synuclein: effects on amyloid fibril formation and chaperone activity. J. Mol. Biol. 340, 1167–1183 [DOI] [PubMed] [Google Scholar]
- 47.Raman, B., Ban, T., Sakai, M., Pasta, S. Y., Ramakrishna, T., Naiki, H., Goto, Y., and Rao, C. M. ( 2005) αB-crystallin, a small heat-shock protein, prevents the amyloid fibril growth of an amyloid β-peptide and β2-microglobulin. Biochem. J. 392, 573–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee, S., Carson, K., Rice-Ficht, A., and Good, T. ( 2006) Small heat shock proteins differentially affect Aβ aggregation and toxicity. Biochem. Biophys. Res. Commun. 347, 527–533 [DOI] [PubMed] [Google Scholar]
- 49.Zabel, C., Sagi, D., Kaindl, A. M., Steireif, N., Klare, Y., Mao, L., Peters, H., Wacker, M. A., Kleene, R., and Klose, J. ( 2006) Comparative proteomics in neurodegenerative and non-neurodegenerative diseases suggest nodal point proteins in regulatory networking. J. Proteome Res. 5, 1948–1958 [DOI] [PubMed] [Google Scholar]
- 50.Wilhelmus, M. M. M., Otte-Holler, I., Wesseling, P., de Waal, R. M. W., Boelens, W. C., and Verbeek, M. M. ( 2006) Specific association of small heat shock proteins with the pathological hallmarks of Alzheimer's disease brains. Neuropathol. Appl. Neurobiol. 32, 119–130 [DOI] [PubMed] [Google Scholar]
- 51.Rajasekaran, N. S., Connell, P., Christians, E. S., Yan, L.-J., Taylor, R. P., Orosz, A., Zhang, X. G., Stevenson, T. J., Peshock, R. M., Leopold, J. A., Barry, W. H., Loscalzo, J., Odelberg, S. J., and Benjamin, I. J. ( 2007) Human αB crystallin mutation causes oxido-reductive stress and protein aggregation cardiomyopathy in mice. Cell 130, 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brenner, D. A., Jain, M., Pimentel, D. R., Wang, B., Connors, L. H., Skinner, M., Apstein, C. S., and Liao, R. L. ( 2004) Human amyloidogenic light chains directly impair cardiomyocyte function through an increase in cellular oxidant stress. Circ. Res. 94, 1008–1010 [DOI] [PubMed] [Google Scholar]
- 53.Saraiva, M. J. M. ( 2003) Cellular consequences of transthyretin deposition. Amyloid 10, 13–16 [PubMed] [Google Scholar]
- 54.Krapfenbauer, K., Engidawork, E., Cairns, N., Fountoulakis, M., and Lubec, G. ( 2003) Aberrant expression of peroxiredoxin subtypes in neurodegenerative disorders. Brain Res. 967, 152–160 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.