Abstract
To better understand the structural and functional roles of tryptophan at the membrane/water interface in membrane proteins, we examined the structural and functional consequences of Trp → 1-methyl-tryptophan substitutions in membrane-spanning gramicidin A channels. Gramicidin A channels are miniproteins that are anchored to the interface by four Trps near the C terminus of each subunit in a membrane-spanning dimer. We masked the hydrogen bonding ability of individual or multiple Trps by 1-methylation of the indole ring and examined the structural and functional changes using circular dichroism spectroscopy, size exclusion chromatography, solid state 2H NMR spectroscopy, and single channel analysis. N-Methylation causes distinct changes in the subunit conformational preference, channel-forming propensity, single channel conductance and lifetime, and average indole ring orientations within the membrane-spanning channels. The extent of the local ring dynamic wobble does not increase, and may decrease slightly, when the indole NH is replaced by the non-hydrogen-bonding and more bulky and hydrophobic N-CH3 group. The changes in conformational preference, which are associated with a shift in the distribution of the aromatic residues across the bilayer, are similar to those observed previously with Trp → Phe substitutions. We conclude that indole N-H hydrogen bonding is of major importance for the folding of gramicidin channels. The changes in ion permeability, however, are quite different for Trp → Phe and Trp → 1-methyl-tryptophan substitutions, indicating that the indole dipole moment and perhaps also ring size and are important for ion permeation through these channels.
Tryptophan is unusually abundant in membrane proteins, including ion channels (1), where tryptophans (and tyrosines) tend to cluster near the membrane-water interfacial regions of the transmembrane domains of the proteins (2-4). The interfacial clustering of Trp and Tyr may be important for membrane protein structure and function (2, 4-6).
In addition to its hydrogen bonding ability, the Trp indole ring has a permanent dipole moment pointing from N1 in the five-membered ring to C5 in the six-membered ring (Fig. 1A). Furthermore, the larger aromatic ring of Trp, as compared with Phe or Tyr, is more accessible for π-π and/or cation-π interactions (7, 8).
FIGURE 1.
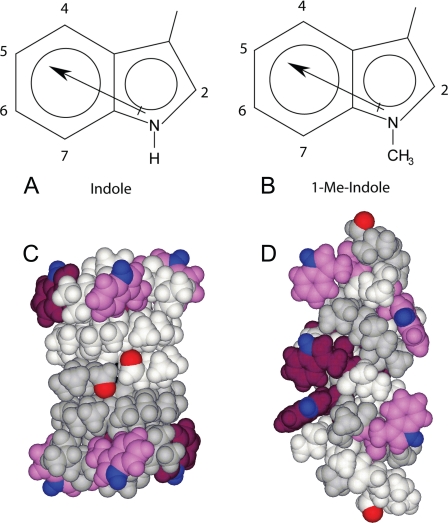
Indole (A) and 1-Me-indole (B) have dipole moments of similar direction and magnitude (∼2.1 Debye for Trp and ∼2.2 Debye for 1-Me-Trp), but 1-Me-indole has lost its hydrogen bonding ability. In the SS transmembrane channel conformation of gA (C), the Trps cluster at the outer subunit interfaces, away from the formyl groups. In DS conformations of gA (D), the Trps are distributed more evenly along the length of the dimer, with Trp9 and Trp11 (maroon) near the molecular center. The different subunits in C and D are shown in gray and white, with formyl oxygens red, Trps either magenta (for Trp13,15) or maroon (for Trp9,11), and indole NH groups blue.
Although these characteristics of the indole side chain make Trp an important component of membrane proteins (2, 9), it is not clear how the characteristics relate to membrane protein structure, function, or lipid interactions. Some insights have been gained by examining free indole analogues in lipid bilayer environments, where NMR investigations led to the suggestion that aromaticity and ring shape are significant factors for the preference of Trp for the polar/nonpolar interface (10, 11). It is unclear, nevertheless, to what extent these and other (12) experiments with small indole analogues, which were not “anchored” by a peptide chain possessing secondary structure, represent the situation in the transmembrane region of a bilayer-spanning protein (4).
To further explore the relative importance of the indole ring dipole moment and hydrogen bonding ability, we used a framework for supporting the indole rings. Previous experiments on gA3 suggested that the indole dipole moment (13-17) has a direct influence on membrane protein function (ion permeation) and that indole NH hydrogen bonding is important for the conformational preference of gA (18, 19). More generally, indole N-H hydrogen bonding appears to be important for protein folding and dynamics (20).
In this study, we make further use of the gA channel framework to position the indole side chain in the bilayer and minimize confounding effects from other (unknown) contributions. gA is a 15-residue peptide with an alternating l- and d-amino acid sequence: formyl-VGALAVVVWLWLWLW-ethanolamide (the d-amino acids are underlined), which forms monovalent cation-permeable transmembrane channels when two 15-residue subunits join from opposite sides of the membrane (5, 21). Each gA subunit has four Trps, which in the single-stranded (SS) β-helical channel structure are localized near the membrane/water interface (Fig. 1C) (22, 23). Changing the membrane environment (24, 25) or the amino acid sequence (18, 19, 26) can alter the gA folding preference, in some cases promoting the folding into double-stranded (DS) dimeric structures (Fig. 1D). The ability to investigate both the conformational equilibrium and ion permeation places gA channels in a class by themselves as a foundation for evaluating the physico-chemical properties and functional importance of Trp residues.
Early studies showed that substitution of any one of the four Trps by Phe decreases the gA single channel conductance (27), with the conductance decrease being more pronounced for the “inner” substitutions, at position 9 or 11 (27). When all four Trps are replaced by Phe, the resulting analogue, designated “gramicidin M,” forms channels with a conductance that is 6-fold less than that of the native gA channels (26, 28). Moreover, the preferred fold of gramicidin M in a lipid environment is one or more DS structures (19, 29), which are not able to conduct ions (18). These results suggest that all four Trps are important for gA structure and function, although the contribution of each Trp may differ. Nevertheless, neither the studies with small indole analogues nor the studies with Trp → Phe substitutions in gA channels could indicate clearly whether the structural and functional changes are due to the loss of the dipole moment, the loss of hydrogen bonding ability, the change in hydrophobicity, or a combination of these or other contributions.
Previously, the importance of the indole ring dipole moment has been studied using Trp → 5-F-Trp substitutions, which enhance the magnitude of the dipole moment yet leave other Trp side-chain properties unchanged (13, 15, 16). In the present study, to focus on the contribution of indole hydrogen bonding, we introduce a methyl group to replace the hydrogen attached to the indole nitrogen (N-H → N-CH3). The resulting 1-methyl-tryptophan (1-Me-Trp) indole ring (Fig. 1B) retains the aromaticity and rigid ring shape, but its ability to be a hydrogen bond donor is blocked by the methyl group. Importantly, the magnitude (2.1 ± 0.1 Debye) and direction of the dipole moments of indole and 1-Me-indole are closely similar (30, 31) to Trp. Using 1-Me-Trp, we synthesized singly substituted gA analogues in which Trp at either position 9, 11, 13 or 15 was replaced. To study the accumulation of effects that may accompany the loss of hydrogen bonding ability, we also made two double substitutions (“inner” pair, 1-Me-Trp9,11; “outer” pair, 1-Me-Trp13,15). In the SS β-helical channel structure, all four Trps are situated close to the bilayer/solution interface (Fig. 1C); in the (misfolded) DS structures (Fig. 1D), the outer Trp13 and Trp15 (colored magenta in Fig. 1) retain positions close to the bilayer/solution interface, whereas the inner Trp9 and Trp11 (colored maroon in Fig. 1) are buried deeper toward the bilayer center than in the SS ion-conducting channels. The properties of the pairwise methylated gA analogues could therefore become very sensitive to the “masking” of the indole-NH hydrogen bond. For completeness, we also prepared the quadruply substituted gA analogue with 1-Me-Trp9,11,13,15.
We examined the 1-Me-Trp gA analogues in lipid environments: structurally by CD spectroscopy, size exclusion chromatography (SEC), and solid state 2H NMR spectroscopy and functionally by single channel experiments, which reveal aspects of both structure and function. Our results indicate that indole hydrogen bonding dominates in determining the conformational preference of the gA channel, whereas the Trp indole ring dipole moment (and bulk) dominate in determining the ion permeability of the channels. In contrast to earlier findings with small indole analogues (10), it is striking how both the ring dipole moment and hydrogen bonding ability assume primary importance when the indole rings are peptide-attached, yet manifest their influence in different ways.
MATERIALS AND METHODS
Fmoc Addition to 1-Me-l-Trp
Fmoc-1-Me-l-Trp was prepared (32) by reacting N-(9-fluorenylmethoxy-carbonyloxy)succinimide and 1-Me-l-Trp (Sigma) and was separated by filtration and extraction from ethyl acetate/aqueous HCl (0.1 m) solution. The raw product was further purified by recrystallization from ethyl acetate twice. The final yield was about 50%, and the purity of Fmoc-1-Me-l-Trp was confirmed to be >95% by 1H NMR.
Peptide Synthesis and Purification
The 1-Me-Trp gramicidin analogues were synthesized on a PerkinElmer Life Sciences/Applied Biosystems 433A synthesizer (32) using standard Fmoc chemistry and Trp Wang resin (Advanced ChemTech, Louisville, KY) except for three analogues: 1-Me-Trp15, 1-Me-Trp13,15, and 1-Me-Trp9,11,13,15. For these analogues, Fmoc-1-Me-l-Trp was preloaded to HMP resin (Applied Biosystems division of PerkinElmer Life Sciences) prior to peptide synthesis using a method adapted from Ref. 33. The analogues were finished by formylation using p-nitrophenyl formate and cleaved from the resin using 20% ethanolamine in DMF (34). They were purified by Sephadex LH-20 (Amersham Biosciences) gel permeation chromatography (solvent: MeOH), and analyzed by reversed-phase HPLC (Zorbax SB80; 4.6 × 50 mm, 3.5 μm; MacMod Analytical, Chadds Ford, PA). The molecular mass of each analogue was confirmed by electrospray ionization mass spectroscopy (Mass Consortium, San Diego, CA).
Circular Dichroism Spectroscopy and Size Exclusion Chromatography
Dimyristoylphosphatidylcholine (DMPC) and dioleoylphosphatidylcholine (DOPC) were purchased from Avanti Polar Lipids (Alabaster, AL). gA/lipid dispersions (1:30) for CD spectroscopy and SEC were prepared as described in Ref. 35. The final gA concentration in each sample was determined by UV absorbance at 280 nm, using ε = 20,840 m-1 cm-1 (36). (The values of ε and λmax are very similar for Trp and 1-Me-Trp.)
CD spectra were recorded at 22-24 °C using a Jasco J710 CD Spectrometer with a 1-mm path length cell, 1.0-nm bandwidth, 0.2-nm step resolution, and 50-nm/min scan speed. Each spectrum is an average of 12 scans.
SEC was used to estimate the fractions of DS and SS conformers of each gA analogue in lipid suspensions. When gA (analogue)/lipid dispersions are injected into an Ultrastyrogel 1000-Å column (7 μm, 7.8 × 300 mm, mobile phase: 100% tetrohydrofuran at 1.0 ml/min; Waters, Inc., Milford, MA), the head-to-head β6.3 SS conformers are dissociated by the solvent and therefore elute as monomers, along with unfolded or unstructured conformations. In contrast, double-stranded conformers, which are stabilized by greater numbers of hydrogen bonds, do not dissociate and therefore elute as dimers (37).
Solid State 2H NMR
Selective 2H Labeling—A selective 2H labeling procedure at low temperature was adapted from original methods of Bak (38, 39). Trifluoroacetic acid-D (CF3COOD) and D2O were purchased from Cambridge Isotope Laboratories (Andover, MA). Fmoc-1-Me-Trp (1 mmol) was dissolved in 4 ml of CF3COOD and incubated for 3 h at 4°C in darkness, after which the reaction was quenched by adding 12 ml of cold D2O. Fmoc-1-Me-Trp was isolated by centrifugation (13,000 rpm at 4 °C). The amino acid was washed once with D2O and twice with H2Oat 22 °C and dried under vacuum (10-3 mm Hg). The extent of deuterium exchange at each position on the indole ring was estimated from the loss of signal intensity for each individual aromatic hydrogen in the 1H NMR spectrum. When samples were incubated in total darkness, no damage to Fmoc-1-Me-Trp was observed from the 1H NMR spectra. A chirobiotic-T HPLC column (Advanced Separation Technologies Inc., Whippany, NJ) was used to confirm that no racemization had occurred during the exchange reaction. The partially 2H-labeled Fmoc-1-Me-Trp was then used for synthesis of singly substituted analogues.
Preparation of Oriented Samples—Samples for solid state NMR were prepared as described by Van der Wel (40). For each sample, 4 μmol of gA analogue and 80 μmol of DMPC (Avanti Polar Lipids Inc. Alabaster, AL) were used to maintain a 1:20 gA analogue/lipid molar ratio. The samples were incubated at 40 °C for several days to ensure optimal alignment before measuring.
All NMR measurements were done on a Bruker AMX2 300 spectrometer, modified with a high power amplifier to achieve the large spectral width required for solid state deuterium NMR. The proper alignment of the lipid bilayers in each glass plate sample was confirmed by 31P NMR measurements as described in Ref. 40. The 2H experiments were done using a quadrupolar echo pulse sequence (41) with an echo delay of 65 μs, 1.5 million scans, and with 30-ms interpulse time. All measurements were done at 40 °C, when the lipids are in the liquid crystalline phase.
Data Analysis—2H NMR spectra have been used to
examine the orientations of Trp indole rings in gA channels
(16,
42-44).
Because deuterium has a spin number of 1, each deuterium produces two
symmetric peaks in a 2H NMR measurement. Quadrupolar splittings
were converted to orientation angles for the C-D bonds using the following
equation
(44),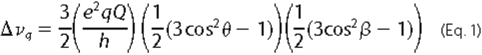
where e2qQ/h is the quadrupolar coupling constant, which has a static value of ∼180 kHz for deuterons on aromatic rings (45), θ is the angle between a particular C-D bond and the gA helix axis, and β is the angle between the helix axis and the direction of the magnetic field, either 0 or 90°. For gA channels, the helix axis is aligned with the membrane normal (44, 46). Assignments of quadrupolar splittings to the C2-D and C5-D bonds for 1-Me-Trps at positions 9 and 11 are based on the corresponding C2 and C5 assignments from partially labeled Trp in gA (44). Assignments for 1-Me-Trp13 and 1-Me-Trp15 were deduced from spectral assignments of Hu et al. (43) for (unmethylated) Trp13 and Trp15 in gA, using the principle of least change. The minimum θ angle difference between a particular C-D bond in 1-Me-Trp and the corresponding Trp was calculated based on the equation above and the observed change in Δνq. Changes in indole ring orientation and dynamics upon 1-methylation were estimated by calculating the backbone-independent ring orientation angles ρ1 and ρ2 and the “effective” quadrupolar coupling constant, as described previously (44). In this method, which does not depend upon knowledge of the local backbone structure, the angle ρ1 represents rotation about an axis normal to the ring plane and the ring bridge, whereas angle ρ2 represents rotation about an axis in the ring plane yet normal to the bridge (see Fig. 1 of Ref. 44).
Electrophysiology
Planar bilayers were formed from n-decane solutions (2.5%, w/v) of diphytanoylphosphatidylcholine (from Avanti) across a hole (∼1.6-mm diameter) in a Teflon® partition that separates two aqueous solutions of unbuffered (pH ∼ 6) salt solution, 1.0 m CsCl or NaCl, which was prepared fresh each day. Single channel experiments were done at 25 ± 1 °C using the bilayer punch method (47), with pipette tip diameter of ∼30 μm and a Dagan 3900 patch clamp amplifier (Dagan Corp., Minneapolis, MN). Unless otherwise stated, the measurements were done at an applied potential of 200 mV.
The gA analogues were added from ethanolic stock solutions to the electrolyte solution on both sides of the bilayer to final concentrations of ∼10-11 m. The final ethanol concentration was less than 0.25%, a concentration that has no effect on channel properties.
The current signal was filtered at 2 kHz, digitized at 20 kHz, and digitally filtered at 500 Hz. Single channel current transitions were detected on-line and analyzed as described previously (48-50). The reported single current transition amplitudes and lifetime distributions are based on at least four independent determinations, two at each polarity. The lifetime histograms were transformed into survivor distributions, and average channel lifetimes (τ) were determined by fitting a single exponential distribution: N(t)/N(0) = exp(-t/τ), where N(t) denotes the number of channels with a lifetime longer than time t to each histogram (49).
RESULTS
CD Spectroscopy and Size Exclusion Chromatography—When the gA sequence is changed, CD spectra and size exclusion chromatograms allow one to measure the overall secondary structure. Native gA can adopt several different conformations (Fig. 1, C and D), depending on the solvent or lipid environment. In phospholipid vesicles, native gA forms predominantly the head-to-head, right-handed, SS β6.3-helical cation-conducting conformation (Fig. 1C), whose CD spectrum is characterized by positive maxima at ∼218 and ∼235 nm and negative ellipticity below 208 nm (Fig. 2) (35). This spectrum is distinct from the CD spectra of other, nonconducting conformers (24, 51).
FIGURE 2.
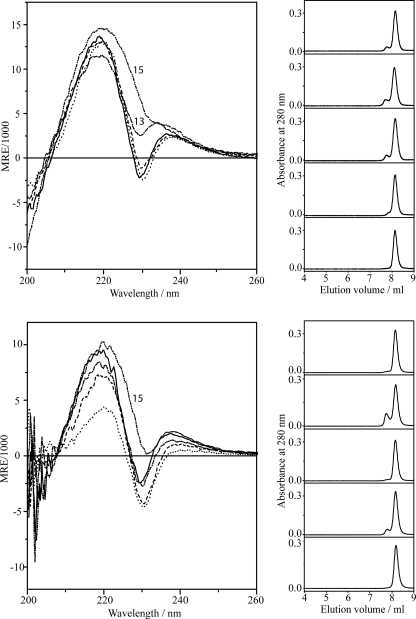
CD spectra and SEC elution profiles for singly substituted 1-Me-Trp analogues in DMPC vesicles (top) and in DOPC vesicles (bottom). The SEC elution profiles were normalized for comparison. From top to bottom, gA (------), 1-MeTrp9 (···), 1-Me-Trp11 (-- -- --), 1-Me-Trp13 (--·--·--; labeled 13), 1-Me- Trp15 (--··--··--; labeled 15).
The population of gA conformations can also be assayed using SEC. When gA/lipid dispersions are injected into an SEC column, with tetrahydrofuran as the mobile phase, the SS channel structure, which is held together by only six intermolecular hydrogen bonds (52), is disrupted by the tetrahydrofuran, such that the peptides elute as monomers, not dimers. By contrast, when a DS dimer (Fig. 1D), which is stabilized by 28 hydrogen bonds (53), is injected, it does not dissociate and elutes as a dimer. SEC thus allows one to estimate the ratio of DS dimers to SS channels for a gramicidin analogue in a particular environment. Consistent with the CD spectra, the SEC profiles show that native gA elutes from DMPC or DOPC mostly as monomers (Fig. 2, right). (Due to the complexity of the conformational mixture, sample-to-sample variations in DS conformer content range from 0 to 10% (54, 55) and are reflected also in the depth of the CD minimum near 230 nm (Figs. 2 and 3); they are not considered significant.)
FIGURE 3.
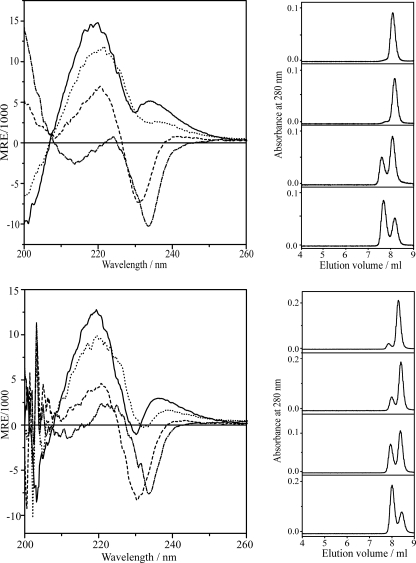
CD spectra and SEC elution profiles for doubly and quadruply substituted 1-Me-Trp analogues in DMPC vesicles (top) and DOPC vesicles (bottom). The SEC elution profiles were normalized for comparison. From top to bottom, gA (------), 1-Me-Trp13,15 (···), 1-MeTrp9,11 (-- -- --), 1-Me-Trp9,11,13,15 (--·--·--).
Singly Substituted 1-Me-Trp Analogues—The CD spectra of singly substituted gramicidins in DMPC vesicles (Fig. 2, top left) have mostly positive ellipticity from ∼208 to ∼260 nm, with two peaks at ∼220 and ∼235 nm, characteristics that are similar to those of native gA in DMPC bilayers. Similar CD spectra are also observed in DOPC vesicles (Fig. 2, bottom left). These results indicate that the singly substituted 1-Me-Trp analogues adopt predominantly the SS β6.3-helical channel conformation, in accord with the size exclusion chromatograms (Fig. 2).
Except for [1-Me-Trp9]gA in DOPC (which shows about 85% monomer), in both DMPC and DOPC vesicles, more than 90% of each singly substituted 1-Me-Trp analogue elutes from the SEC column as monomers (Fig. 2, right), indicating that the single 1-Me-Trp analogues retain the SS β6.3-helical channel structure in lipid bilayers, consistent with the CD spectra in Fig. 2.
Doubly and Quadruply Substituted 1-Me-Trp Analogues—Fig. 3 (top left) shows CD spectra of doubly and quadruply 1-Me-Trp-substituted gA analogues in DMPC vesicles. Compared with native gA and the singly substituted analogues, the CD spectra of [1-Me-Trp9,11]gA and [1-Me-Trp9,11,13,15]gA have much lower ellipticity between 208 and 260 nm. The spectrum of [1-Me-Trp9,11]gA reaches a minimum at ∼230 nm; [1-Me-Trp9,11,13,15]gA has an even more negative ellipticity at ∼234 nm. These results show that both analogues fold into more than one conformation when incorporated into the lipid bilayer and that some of these conformations are DS conformers in addition to the SS β6.3-helical channel conformer. By contrast, the spectrum of [1-Me-Trp13,15]gA is similar to that of native gA, indicating retention of the SS channel conformation in DMPC. These results can be understood by noting that Trp9 and Trp11 are buried in bilayer-spanning DS gA structures (Fig. 1D), whereas Trp13 and Trp15 remain rather interfacial in the DS conformers (Fig. 1D). Similar trends as in DMPC were observed when the analogues were incorporated into DOPC vesicles (Fig. 3, bottom left). Together, the CD spectra of the doubly and quadruply substituted gAs show that N-methylation of Trp9 and Trp11 has large and significant effects on the conformational preference in both DMPC and DOPC lipid bilayers. Conversely, N-methylation at positions 13 and 15 has only minor influence on folding.
Again, the SEC elution profiles for the doubly and quadruply substituted gramicidins in DMPC and DOPC vesicles (Fig. 3, right) conform to predictions based on the CD results. When the “outer” pair of Trps is methylated, the SEC profile of [1-Me-Trp13,15]gA shows, similarly to native gA, that the analogue elutes mostly as monomers. By contrast, significant amounts of DS dimer are observed in the SEC profiles for both [1-Me-Trp9,11]gA and [1-Me-Trp9,11,13,15]gA. Indeed, the majority of [1-Me-Trp9,11,13,15]gA (whether in DMPC or DOPC) elutes as DS dimers. The loss of hydrogen bonds (and increased hydrophobicity) due to N-methylation of Trp9,11 permits these indole rings to bury more deeply in the bilayer (cf. Fig. 1D), thereby shifting the gA structure from the SS channel conformation toward the DS dimer conformation.
Solid State 2H NMR—The orientations of the N-methyl indole rings in the singly substituted gA analogues were examined in 2H NMR experiments.
Selective Deuterium Labeling—Because no 2H-labeled 1-Me-Trp is commercially available, we prepared Fmoc-1-Me-Trp partially labeled with 2H. The N-methyl group speeds the kinetics of the isotope exchange on the indole ring. Previously, we observed, based on integrals from 1H NMR spectra, about 70% exchange at C-2 and 20% exchange at C-5 in the indole ring of Fmoc-l-Trp, during a 3-h exchange at 4 °C, with no significant exchange at other positions (44). In the case of Fmoc-1-Me-Trp, the exchange rates are faster, with more than 90% of the hydrogen at C-2 and ∼70% at C-5 exchanging during 3 h at 4 °C. Four singly substituted gA analogues were synthesized, using the partially 2H-labeled Fmoc-1-Me-Trp and examined by solid state 2H NMR in hydrated DMPC bilayers.
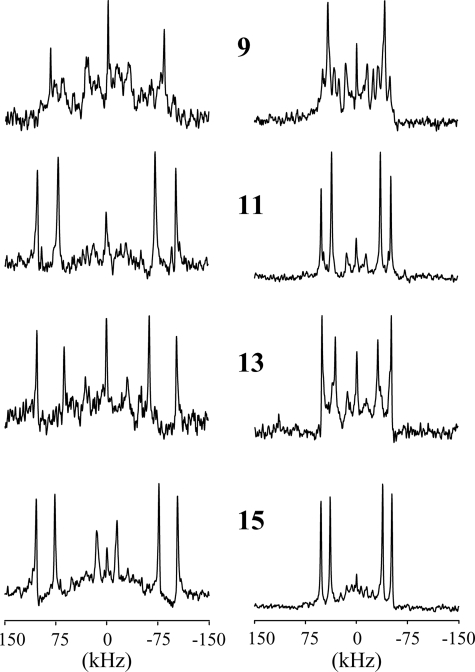
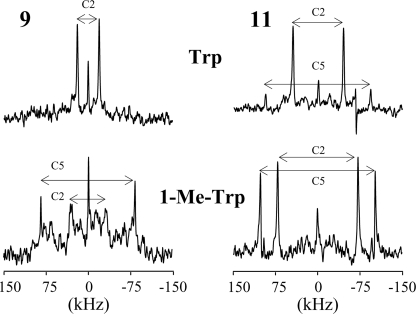
Spectral Assignments—Fig. 4 shows the 2H NMR spectra of partially labeled 1-Me-Trp analogues, at position 9, 11, 13, or 15, with samples oriented at β = 0 or β = 90°. Two pairs of major peaks are observed in each spectrum, corresponding to the 2H labels on carbons 2 and 5 of the 1-Me-Trp indole ring. For each sample, the quadrupolar splittings at β = 90° are about half the values observed at β = 0, indicating rapid axial rotation of the SS β-helix relative to the membrane normal (56).
FIGURE 4.
2H NMR spectra of singly substituted and 1-Me-Trp partially labeled gA analogues in DMPC (from top to bottom): 1-Me-Trp9, 1-Me-Trp11, 1-Me-Trp13, and 1-Me-Trp15. Left, β = 0°; right, β = 90° sample orientation; temperature, 40 °C.
The resonances at C2 and C5 in partially 2H-labeled Trp9 and Trp11 in gA have been assigned (Fig. 5) (see also Ref. 44). In each case, the deuteron at C2 has the smaller quadrupolar splitting (∼43 kHz in Trp9 and ∼96 kHz in Trp11), whereas the C5 deuteron exhibits a larger splitting (∼151 kHz in Trp9, and ∼188 kHz in Trp11). To assign the spectra for 1-Me-Trp, we assumed that the change of quadrupolar splitting for each ring C-D bond in response to 1-methylation would be relatively small (principle of “least change”). In this way, the quadrupolar splittings in the 1-Me-Trp9 and 1-Me-Trp11 spectra (Figs. 4 and 5) were assigned based on their relative closeness to corresponding peaks in the Trp9 and Trp11 spectra. Also for 1-Me-Trp13 and 1-Me-Trp15, we assigned the C5 deuteron a larger quadrupolar splitting than the C2 deuteron (Table 1). Δνq for the C2 deuterons increases substantially upon methylation, whereas Δνq increases more modestly for C5 deuterons (Table 1).
FIGURE 5.
Comparison of 2H NMR spectra of partially labeled Trp (top) and 1-Me-Trp (bottom) at positions 9 (left) and 11 (right), respectively, in gA channels, hydrated in DMPC bilayers. β = 0° sample orientation; temperature, 40 °C.
TABLE 1.
Comparison of quadrupolar splittings of 1-Me-Trp versus Trp in gA
See “Results” for assignment information. All samples were oriented at β = 0°. A quadrupole coupling constant of 180 kHz (static value) was used to estimate the ΔΘ values.
|
Trp position
|
C2 deuteron Δνq
|
ΔΘ
|
||
|---|---|---|---|---|
| Trpa | 1-Me-Trp | |||
| kHz | degrees | |||
| 9 | 43 | 62 | 2.7 | |
| 11 | 96 | 143 | 6.9 | |
| 13 | 106 | 126 | 2.9 | |
| 15 | 125 | 153 | 4.3 | |
|
Trp position
|
C5 deuteron Δνq
|
ΔΘ
|
||
|---|---|---|---|---|
| Trpa | 1-Me-Trp | |||
| kHz | degrees | |||
| 9 | 151 | 168 | 2.7 | |
| 11 | 188 | 204 | 2.9 | |
| 13 | 205 | 206 | 0.2 | |
| 15 | 205 | 207 | 0.4 | |
Data from Ref. 44.
Data Analysis—Table 1 shows the minimum θ angle differences for C2-2H and C5-2H between 1-Me-Trp and Trp at each position (assuming comparable spectral assignments (see above) and using the static QCC value of 180 kHz). The Δθ values show that position 11 exhibits the largest difference for both C2 and C5, whereas θ for C5 of Trp13 and Trp15 changes little after N-methylation. The θ angles calculated from 2H NMR splittings are time-averaged values with respect to the membrane normal, and the small differences in θ between 1-Me-Trp and Trp suggest that N-methylation, within the context of the SS β-helical channel conformation, causes only modest changes in the indole ring orientations or dynamics. The large quadrupolar splittings observed for 2H of C5 in three of the 1-Me-Trp rings suggest that rings 11, 13, and 15 retain their high principal order parameters (Szz in a range of ∼0.86-0.92) (44, 46) with minimal independent ring motions beyond the global motions of the gA transmembrane helix. As a corollary, the static QCC values for these rings are reduced only slightly when multiplied by Szz (46).
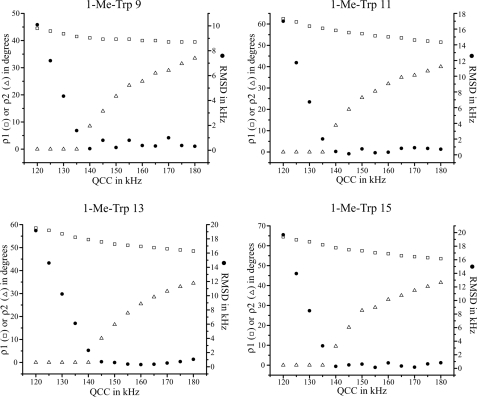
Trp indole ring orientations can be studied independently of the peptide backbone conformation by modeling the orientation of an equivalent yet unconstrained indole ring with respect to the membrane normal and the direction of the magnetic field (44). (Because spectral data were available only for the C2 and C5 deuterons of 1-Me-Trp, we did not consider the relatively small asymmetry parameters for the C-D tensors but note that higher values of Szz and QCC should be used (46).) As a check on the ring dynamics, the ρ1 and ρ2 angles (44) required to “best fit” the C2 and C5 splittings (lowest root mean square deviation) of each 1-Me-Trp were examined for each “apparent” QCC value from 120 to 180 kHz (Fig. 6). (“Apparent” QCC is the product of Szz and static QCC (44).) Similar to the results for unmethylated Trps (44), ρ1 does not vary much as QCC varies, whereas ρ2 varies considerably as the apparent QCC is increased from 140 to 180 kHz in the calculation. The root mean square deviation value for each position falls into a range between 0.5 to 1.5 kHz, when the apparent QCC is above 140 kHz, which is within the experimental error range of 2H NMR splittings. The high root mean square deviation values at apparent QCC values below 135 kHz suggest (again) that N-methylation does not induce additional overall ring motion (not even for Trp9).
FIGURE 6.
Graphs to show the variation of the best fit ρ1 and ρ2, and the minimum value of root mean square deviation, as functions of apparent QCC [(static QCC) × Szz] for 1-Me-Trp indole ring at positions 9, 11, 13, and 15, respectively. See also Ref. 44.
In fact, the larger quadrupolar splittings at both C2 and C5 (Table 1) suggest that N-methylation may restrict the ring motions. This finding is contrary to a simple notion that N-methylation, by blocking the hydrogen bonding ability of the Trp indole ring, could allow for increased ring motion. Perhaps the loss of the ability to form hydrogen bonds (and the increased bulk and hydrophobicity associated with the added-CH3 group) may restrict the conformational space available to the side chain. With only two C-D bond orientations known for 1-Me-Trp, however, the results are insufficient to determine the “best” QCC value. Assuming that the best fit QCC values are about the same as for Trp at each position (∼155 kHz for positions 9 and 11, ∼165 kHz for positions 13 and 15, corresponding to Szz values of 0.86 and 0.92, respectively (46)), the ρ1 and ρ2 angles were calculated using these particular QCC values; they are listed in Table 2. Compared with the corresponding Trp indole rings, within the context of the SS channel conformation of gA, the ring orientations of 1-Me-Trps are only slightly changed. The largest observed change is only 8.5° in ρ1 for Trp11.
TABLE 2.
Influence of 1-methylation upon ρ1 and ρ2 for the Trps in gramicidin channels
It is assumed in this analysis that the ring QCC values remain unchanged on methylation (see “Materials and Methods” for details). The values for the unmethylated Trps are from Ref. 46.
|
Trp
|
ρ1
|
ρ2
|
QCC
|
Me-Trp
|
|
|---|---|---|---|---|---|
| Δρ1 | Δρ2 | ||||
| degrees | degrees | kHz | degrees | degrees | |
| 9 | 37.0 | 23.0 | 155 | +3.5 | +0.5 |
| 11 | 46.0 | 23.5 | 155 | +8.5 | +4.5 |
| 13 | 46.5 | 27.0 | 165 | +3.5 | +1 |
| 15 | 50.5 | 30.5 | 165 | +4.5 | +4.5 |
Single Channel Experiments—Single channel experiments were conducted for [1-Me-Trp9]gA and [1-Me-Trp15]gA in both 1 m NaCl and 1 m CsCl. Other 1-Me-Trp gA analogues were characterized only in 1 m CsCl.
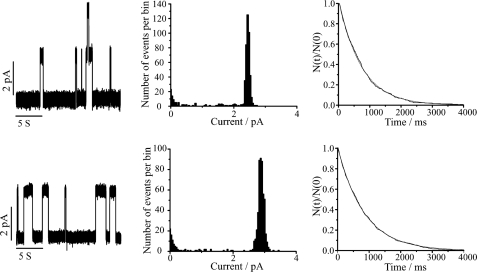
Single Channel Characteristics in NaCl—Fig. 7 shows current traces, amplitude histograms, and lifetime histograms of the singly substituted 1-Me-Trp9 and 1-Me-Trp15 analogues in 1 m NaCl and diphytanoylphosphatidylcholine. For both analogues, the single channel conductances and lifetimes are close to those of native gA channels (Table 3) (27, 57, 58).
FIGURE 7.
Single channel current traces, current transition amplitude histograms, and lifetime histograms (from left to right) for 1-Me-Trp9[gA] (top) and 1-Me-Trp15[gA] (bottom). Only a single predominant channel type is observed in the current transition amplitude and lifetime histograms. Conditions were as follows: Diphytanoylphosphatidylcholine, 1 m NaCl, 200 mV, 25 °C.
TABLE 3.
Summary of single channel conductances (g) and mean channel lifetimes (τ) for 1-Me-Trp gA analogues
Conditions were as follows: diphytanoylphosphatidylcholine, 1 m salt, 200 mV, 25 °C.
| Analogue | ga | τ | Salt |
|---|---|---|---|
| picosiemens | ms | ||
| gA | 47 | 680 | CsCl |
| [1-Me-Trp9]gA | 45 | 770 | CsCl |
| [1-Me-Trp11]gA | 43 | 1200 | CsCl |
| [1-Me-Trp13]gA | 50 | 790 | CsCl |
| [1-Me-Trp15]gA | 51 | 870 | CsCl |
| [1-Me-Trp9,11]gA | 39 | 1300 | CsCl |
| [1-Me-Trp13,15]gA | 47 | 940 | CsCl |
| [1-Me-Trp9,11,13,15]gA | 36 | 2200 | CsCl |
| gA | 15.0b | 840b | NaCl |
| [1-Me-Trp9]gA | 12.0 | 730 | NaCl |
| [1-Me-Trp15]gA | 14.5 | 830 | NaCl |
| [Phe9]gA | 6.0b | 1000b | NaCl |
| [Phe15]gA | 10.9b | 790b | Nacl |
S.E. of measurement: ± 0.5 - 1.0 picosiemens.
Data from Ref. 27 are shown for comparison.
Single Channel Characteristics in CsCl—Similar to the observations with NaCl, the channels formed by the singly substituted 1-Me-Trp analogues retain similar Cs+ conductances as that of the native gA channel (Table 3). The similarities in the single channel conductances for both Na+ and Cs+ are consistent with the finding from 2H NMR that the indole ring and ring dipole orientations change little upon 1-methylation of each Trp in the gA channel.
Single substitutions at position 9, 13, or 15 have modest effects on the single channel lifetimes, but the substitution at position 11 gives rise to channels with a significantly longer lifetime (∼1200 ms) than was observed with gA. The patterns in the lifetime results are similar to those with Trp → Phe substitutions (27), wherein the substitution of Trp11 also increased the single channel lifetime. The relative characteristics of the [1-Me-Trp9]gA and [1-Me-Trp15]gA channels in 1 m CsCl, compared with gA channels, are similar to the observations in 1 m NaCl.
When both Trp9 and Trp11 are replaced by 1-Me-Trp, the Cs+ conductance decreases, and the single channel lifetime increases to about twice that of native gA channels (presumably reflecting the substitution at position 11). In contrast, the conductance and lifetime of channels with 1-Me-Trp at positions 13 and 15 are comparable with those of native gA. When all four Trps are substituted by 1-Me-Trp, the channel lifetime increases to more than 3-fold that of native gA, whereas the conductance remains similar to that of [1-Me-Trp9,11]gA channels. Throughout the series of 1-Me-Trp substitutions considered here, the single channel lifetime is more sensitive to the Trp methylation than is the single channel conductance.
Comparison of Trp → 1-Me-Trp-substituted and Trp → Phesubstituted gA Channels—Table 4 shows a comparison of the single channel lifetimes of channels with single and quadruple Trp → 1-Me-Trp and Trp → Phe substitutions (in both subunits), normalized by the lifetimes of native gA channels. For single Trp → 1-Me-Trp or Trp → Phe substitutions, the relative lifetimes are close to 1, except at position 11, where the lifetime ratio is 1.8 for 1-Me-Trp and 2.7 for Phe. For the quadruple substitutions, however, the relative lifetime increases to 3.2 for 1-Me-Trp but decreases to ∼0.4 for Phe. This divergence between the quadruple 1-Me-Trp and Phe substitutions suggests that the Trp indole ring shape and dipole moment are important for gA channel function (in addition to any effect of hydrogen bond formation).
TABLE 4.
Comparison of relative single channel lifetimes of gramicidin channels having Trp → 1-Me-Trp and Trp → Phe substitutions
DISCUSSION
Disrupting the hydrogen bond forming ability of Trp indole rings by N-methylation of the indole ring of one or more Trps in the gA sequence, together with structural and functional characterization of the resulting analogue channels, provides insight into the more general issue of the role of Trp hydrogen bond formation for integral membrane proteins. Besides removing the hydrogen bond, the N-methyl group also increases the hydrophobicity of the indole ring. Here we discuss the consequences of this N-methylation for gA folding preference, channel function, and indole ring orientation.
Folding Preference of 1-Me-Trp gA Analogues—Gramicidin A can adopt different structural conformations depending on different solvent environments (51, 59). There are two major groups of conformers with different folding patterns. One consists of a single structure, the head-to-head, right-handed, and SS β6.3-helix (Fig. 1C), which forms a monovalent cation-permeable channel. The other group of conformers is the family of interconverting DS helices with varying pitch and number of residues per turn (51); an example is shown in Fig. 1D (53, 60). The overall conformational equilibrium can be described as SS channel ↔ monomers ↔ DS dimer.
Structural (22, 52) and functional studies (5) show that the predominant conformation of gA in lipid bilayers is the SS β6.3-helical channel structure. The four Trps at the C-terminal cluster at the membrane-water interface (Fig. 1C) and are important for the SS conformational preference of gA in lipid bilayers (18, 19). The DS structures (Fig. 1D) are disfavored, probably because of the energetic cost to bury the four Trps within the lipid bilayer hydrophobic core (18). This means, for native gA in a lipid environment, that the equilibrium shifts from the DS conformers that dominate in organic solvents toward the SS channel structure. The CD and SEC results (Figs. 2 and 3) show that this preference for the SS structure holds true when only one Trp is N-methylated and for the “outer” pair of substitutions in [1-Me-Trp13,15]gA. In contrast, methylation of the “inner” pair of indole rings allows Trp9 and Trp11 to become more buried within the lipid acyl chains (Fig. 1D) and shifts the conformational preference. Based on the SEC elution profiles, more than 30% of [1-Me-Trp9,11]gA and about 60% of [1-Me-Trp9,11,13,15]gA form DS dimers in both DMPC and DOPC vesicles, indicating that the DS conformation becomes increasingly favored when the indole nitrogens of the “inner” pair of Trp9 and Trp11 are methylated.
In contrast to what would be expected from studies on small molecules (10) or our results with single 1-Me-Trp substitutions (Fig. 2), the double substitution with 1-Me-Trp9,11 clearly allows the “inner” pair of Trps to bury more deeply within the lipid acyl chain region, thereby increasingly favoring a DS conformation. The results with [1-Me-Trp9,11]gA and [1-Me-Trp9,11,13,15]gA show that the Trp hydrogen bonding is important for the indole interfacial localization and demonstrates the increased sensitivity afforded by the double substitutions.
Our conclusion, that indole NH hydrogen bond formation is important for anchoring Trp residues to the interface, is further strengthened by the results in Table 5, where we compare the conformational preferences (as deduced from SEC) of gA analogues with Trp → Phe and Trp → 1-Me-Trp substitutions. The parallels among the two sets of analogues are striking. The greater increase in the proportion of DS conformers upon modification of the “inner” Trp9,11 (compared with the “outer” Trp13,15) is observed not only with Trp → 1-Me-Trp but also with Trp → Phe substitutions (19). The common feature of Trp → Phe and Trp → 1-Me-Trp is the removal of the ability of the indole NH to form hydrogen bonds. Thus, when the hydrogen bonding ability is lost, the more hydrophobic 1-Me-indole ring becomes able to bury deeper within the membrane than the indole ring. In DS conformers, Trp9,11 are buried deeper than Trp13,15 (Fig. 1), and Trp9,11 are expected to be more important than Trp13,15 in determining the gA fold, as is observed. The energetic consequences of N-methylation are position-dependent, and (somewhat) additive.
TABLE 5.
Comparison of conformational preferences of gramicidin analogues having Trp → 1-Me-Trp and Trp → Phe substitutions
The areas of peaks for SS and DS conformers in size exclusion chromatograms are compared and reported as the percentage of subunits in the single-stranded conformation.
Single Channel Lifetimes—A gA channel forms when two subunits associate to form the SS β-helical channel; the channel disappears when the subunits dissociate. The subunit/subunit interface formed by the first five residues of each subunit near the bilayer center is important for the single channel lifetime (57, 61, 62). The lifetime also depends on hydrophobic matching between the channel exterior and the host lipid bilayer (25). Replacing amino acids other than those at the subunit interface, such as Trp → Phe (27) or Trp → Gly substitutions (63, 64), or changing the d-Leu “spacer” residues between the Trps (65) also alters the single channel lifetime (66), presumably by altering the orientations and/or lipid interactions of the anchoring Trp side chains.
Analogues having 1-Me-Trp retain the same subunit/subunit interface and the same length as native gA, but the Trp interactions with lipids or with neighboring side chains may be altered. A maximum lifetime is observed when all four Trps are methylated (Table 4). In contrast to 4-fold methylation, 4-fold substitution of all gA Trps by Phe causes a net reduction in lifetime (Table 4). Although both [Phe9,11,13,15]gA and [1-Me-Trp9,11,13,15]gA prefer DS conformations in lipid bilayers, once the SS channels form, the [1-Me-Trp9,11,13,15]gA channels are more stable than the [Phe9,11,13,15]gA channels. The larger hydrophobic aromatic ring systems of the 1-Me-Trp residues stabilize the SS channel more than the smaller Phe ring systems, demonstrating that factors other than hydrogen bond formation are important.
Among the single substitutions, [1-Me-Trp11]gA channels have the highest lifetime, ∼1200 ms (Table 3). Similarly, a Trp → Phe substitution at position 11 causes a much longer single channel lifetime than substitution of Phe at position 9, 13, or 15 (Table 4), although a Trp → Gly substitution at position 11 causes no change in lifetime (64). Removing the Trp11 hydrogen bond (only), whether by methylation (Table 4) or by conversion to Phe (27), significantly increases the channel lifetime. The destabilizing effect of a hydrogen bond at Trp11 is seen also in [Phe9,13,15]gA, where the channel lifetime is reduced to only 5 ms (27). The similar results obtained with the 1-Me-Trp and Phe substitutions indicate that Trp11 among the four Trps is uniquely situated, a notion that is supported also by results with doubly substituted channels. Methylation of the inner pair in [1-Me-Trp9,11]gA yields a longer channel lifetime than methylation of the outer pair in [1-Me-Trp13,15]gA (Table 3). Maybe the Trp11 indole ring is separated from the membrane/water interface by a particularly inopportune distance.
Ion Permeation—Changing the Trp indole ring dipole moment has significant effects on the ion permeability of gA channels (13, 15, 16). N-Methylation of the Trp indole ring has little effect on the direction or magnitude of the indole dipole moment (30), and the methyl substitutions would be expected to cause only modest alterations in the electrostatic interactions between the indole dipoles and the permeating ions (as long as the average side chain orientations relative to the channel pore are preserved). Not surprisingly, therefore, single 1-Me-Trp substitutions have only small effects on the single channel conductances (Table 3 and Fig. 7). Only the inner pair, doubly substituted and the quadruply substituted analogues form channels that have obviously lower single channel conductances than the native gA channels.
When single channel conductances in 1.0 m NaCl are compared (Fig. 7 and Table 3), the [1-Me-Trp9] and [1-Me-Trp15]gA channels have higher conductances than the [Phe9]gA and [Phe15]gA channels (27). Moreover, the [Phe9,11,13,15]gA channels have a Cs+ conductance that is 6-fold less than that of gA channels (at 200 mV) (26), whereas the conductance of [1-Me-Trp9,11,13,15]gA channels is reduced by only 25%. This comparison of 1-Me-Trp versus Phe further demonstrates the importance of the Trp dipole moment for the ion permeability of the gA channels.
1-Me-Trp Indole Ring Orientation—The singly substituted 1-Me-Trp analogues retain the SS channel conformation in lipid bilayers (Table 5). Within this context, the 1-Me-Trp ring orientations remain similar to those of the Trps in native gA; the changes in the average indole ring orientations are modest (and with little effect on the single channel conductances) but clearly measurable. For all Trps, the positive Δρ1 and Δρ2 values indicate that the 1-Me-indole rings all rotate in the same direction upon methylation (Table 2). Because the observed changes somehow result from the loss of hydrogen-bonding ability, it is not surprising that position 11 has the largest Δρ1 (∼9°).
Implication for Membrane Proteins—Although Trp often is considered to be hydrophobic, Trp residues have long been described as “anchoring” residues in membrane proteins (2, 4, 5), and as early as 1984, it was found, using [Trp1]gA channels, that there is a significant penalty associated with burying Trp residues in the bilayer core (62). More recently, it was shown that Trps near the C terminus of a membrane-spanning α-helix display a distinctly different behavior from those near the N terminus, reflected in large differences in the allowed side chain torsion angles and ring motions, highlighting the directionality of an α-helix relative to the bilayer/solution interface (67).
These positional (and directional) effects of Trp urge caution when interpreting the results of Trp scanning mutagenesis along transmembrane segments (68, 69), because a Trp-induced change in membrane protein insertion or function could result not only from steric effects but also from the energetic consequences of inserting a Trp at the protein/bilayer boundary.
Multiple Trp → 1-Me-Trp substitutions in gA may cause considerable changes in the distribution between SS and DS conformers, with only modest changes in single channel conductance. Because the changes in conformational preference parallel those for Trp → Phe substitutions (Table 5), the preference is determined, at least in part, by the energetic cost of burying Trp residues in the bilayer hydrophobic core. Because the single channel conductances are changed much less by Trp → 1-Me-Trp substitutions than by the corresponding Trp → Phe substitutions (70), with little (13, 16) change in the average indole ring orientation, the indole ring dipole moment is a determinant of the ion permeability of the channels.
Our conclusion, that indole hydrogen bonding is important for the interfacial preference of Trp in membrane-spanning proteins, seems to disagree with the conclusions from studies using small, freely mobile, Trp side chain analogues (10-12). To resolve the apparent conflict, we note that the small molecule experiments probe only the most likely (minimum energy) position, whereas our experiments using gA channels probe the consequences of “forcing” the Trp side chain into the bilayer core. Molecular dynamics simulations (71-73) suggest that there may be a significant barrier for burying indole side chains in the bilayer core (and even a small barrier for the Phe side chain analogue, benzene). The small molecule measurements appear unable to detect modest differences (on the order of a kcal/mol or so (74)) in the energetic cost for burying indole or N-Me-indole in the bilayer hydrophobic core, whereas the gA experiments are able to detect such differences. The present results demonstrate the importance of examining the structural (and functional) importance of amino acid residues using a framework wherein the side chain position(s) with respect to the bilayer is “fixed” by a transmembrane (channel) structure.
Conclusions—The conformational changes observed for gA analogues with Trp → 1-Me-Trp substitutions are similar to those observed previously with Trp → Phe substitutions, indicating that indole hydrogen bonding is important for the interfacial preference of Trp in membrane-spanning proteins. When comparing the present results with those observed previously for small molecule Trp side chain analogues, however, we note that the “interfacial preference” of Trp actually is an “aversion to being buried” in the bilayer hydrophobic core.
The structure and function of the conducting channels formed by gA analogues with Trp → 1-Me-Trp substitutions are similar to those of native gA channels. The changes in Trp indole ring orientation upon 1-methylation are less than 5° (except for position 11, where Δρ1 = 8.5°). Single substitutions of Trp → 1-Me-Trp have little effect on the single channel conductance or lifetime except for [1-Me-Trp11]gA channels, which, like [Phe11]gA channels, have a 2-fold longer lifetime, an effect due to the destabilizing influence of an indole hydrogen bond at this particular location (depth) with respect to the membrane/water interface.
We conclude that the hydrogen bonding ability of the Trp indole ring is important for the interfacial preference of Trp (and thus for the conformational preference of gA and the formation of gA channels). The Trp side chain dipole moment is important for the ion permeability of the channels.
Acknowledgments
We thank Patrick van der Wel for help with calculating the orientations of the 1-Me-Trp indole rings.
This work was supported, in whole or in part, by National Institutes of Health Grants RR15569 and GM70971. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: gA, gramicidin A; DMPC, 1,2-dimyristoylphosphatidylcholine; DOPC, 1,2-dioleoylphosphatidylcholine; DS, double-stranded; QCC, quadrupolar coupling constant; SEC, size exclusion chromatography; SS, single-stranded; Fmoc, N-(9-fluorenyl)methoxycarbonyl; 1-Me-Trp, 1-methyl-tryptophan.
References
- 1.Wallace, B. A., and Janes, R. W. (1999) Adv. Exp. Med. Biol. 467 789-799 [DOI] [PubMed] [Google Scholar]
- 2.Schiffer, M., Chang, C. H., and Stevens, F. J. (1992) Protein Engr. 5 213-214 [DOI] [PubMed] [Google Scholar]
- 3.Cowan, S. W., Schirmer, T., Rummel, G., Steiert, M., and Ghosh, R. (1992) Nature 358 727-733 [DOI] [PubMed] [Google Scholar]
- 4.Killian, J. A., and von Heijne, G. (2000) Trends Biochem. Sci. 25 429-434 [DOI] [PubMed] [Google Scholar]
- 5.O'Connell, A. M., Koeppe, R. E., II, and Andersen, O. S. (1990) Science 250 1256-1259 [DOI] [PubMed] [Google Scholar]
- 6.Koeppe, R. E., II (2007) J. Gen. Physiol. 130 223-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burley, S., and Petsko, G. (1988) Adv. Protein Chem. 39 125-189 [DOI] [PubMed] [Google Scholar]
- 8.Dougherty, D. A. (1996) Science 271 163-168 [DOI] [PubMed] [Google Scholar]
- 9.Killian, J. A. (1998) Biochim. Biophys. Acta 1376 401-416 [DOI] [PubMed] [Google Scholar]
- 10.Yau, W.-M., Wimley, W. C., Gawrisch, K., and White, S. H. (1998) Biochemistry 37 14713-14718 [DOI] [PubMed] [Google Scholar]
- 11.Persson, S., Killian, J. A., and Lindblom, G. (1998) Biophys. J. 75 1365-1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kachel, K., Asuncion-Punzalan, E., and London, E. (1995) Biochemistry 34 15475-15479 [DOI] [PubMed] [Google Scholar]
- 13.Andersen, O. S., Greathouse, D. V., Providence, L. L., Becker, M. D., and Koeppe, R. E., II (1998) J. Am. Chem. Soc. 120 5142-5146 [Google Scholar]
- 14.Hu, W., and Cross, T. A. (1995) Biochemistry 34 14147-14155 [DOI] [PubMed] [Google Scholar]
- 15.Busath, D. D., Thulin, C. D., Hendershot, R. W., Phillips, L. R., Maughan, P., Cole, C. D., Bingham, N. C., Morrison, S., Baird, L. C., Hendershot, R. J., Cotten, M., and Cross, T. A. (1998) Biophys. J. 75 2830-2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotten, M., Tian, C., Busath, D. D., Shirts, R. B., and Cross, T. A. (1999) Biochemistry 38 9185-9197 [DOI] [PubMed] [Google Scholar]
- 17.Cole, C. D., Frost, A. S., Thompson, N., Cotten, M., Cross, T. A., and Busath, D. D. (2002) Biophys. J. 83 1974-1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durkin, J. T., Providence, L. L., Koeppe, R. E., II, and Andersen, O. S. (1992) Biophys. J. 62 145-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salom, D., Pérez-Payá, E., Pascal, J., and Abad, C. (1998) Biochemistry 37 14279-14291 [DOI] [PubMed] [Google Scholar]
- 20.Samanta, U., Pal, D., and Chakrabarti, P. (2000) Proteins 38 288-300 [PubMed] [Google Scholar]
- 21.Andersen, O. S., Koeppe, R. E., II, and Roux, B. (2007) in Biological Membrane Ion Channels (Chung, S. H., Andersen, O. S., and Krishnamurthy, V., eds) pp. 33-80, Springer, New York
- 22.Ketchem, R. R., Hu, W., and Cross, T. A. (1993) Science 261 1457-1460 [DOI] [PubMed] [Google Scholar]
- 23.Townsley, L. E., Tucker, W. A., Sham, S., and Hinton, J. F. (2001) Biochemistry 40 11676-11686 [DOI] [PubMed] [Google Scholar]
- 24.Wallace, B. A., Veatch, W. R., and Blout, E. R. (1981) Biochemistry 20 5754-5760 [DOI] [PubMed] [Google Scholar]
- 25.Mobashery, N., Nielsen, C., and Andersen, O. S. (1997) FEBS Lett. 412 15-20 [DOI] [PubMed] [Google Scholar]
- 26.Fonseca, V., Daumas, P., Ranjalahy Rasoloarijao, L., Heitz, F., Lazaro, R., Trudelle, Y., and Andersen, O. S. (1992) Biochemistry 31 5340-5350 [DOI] [PubMed] [Google Scholar]
- 27.Becker, M. D., Greathouse, D. V., Koeppe, R. E., II, and Andersen, O. S. (1991) Biochemistry 30 8830-8839 [DOI] [PubMed] [Google Scholar]
- 28.Heitz, F., Gavach, C., Spach, G., and Trudelle, Y. (1986) Biophys. Chem. 24 143-148 [DOI] [PubMed] [Google Scholar]
- 29.Cotten, M., Xu, F., and Cross, T. A. (1997) Biophys. J. 73 614-623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McClellan, A. L. (1963) Tables of Experimental Dipole Moments, pp. 280-325, W.H. Freeman, San Francisco
- 31.Abribat, G., and Viallet, P. (1970) C. R. Acad. Sci. (Paris) 271 1029-1032 [Google Scholar]
- 32.Greathouse, D. V., Koeppe, R. E., II, Providence, L. L., Shobana, S., and Andersen, O. S. (1999) Methods Enzymol. 294 525-550 [DOI] [PubMed] [Google Scholar]
- 33.Sieber, P. (1987) Tetrahedron Lett. 28 6147-6150 [Google Scholar]
- 34.Greathouse, D. V., Goforth, R. L., Crawford, T., van der Wel, P. C. A., and Killian, J. A. (2001) J. Peptide Res. 57 519-527 [DOI] [PubMed] [Google Scholar]
- 35.Greathouse, D. V., Hinton, J. F., Kim, K. S., and Koeppe, R. E., II (1994) Biochemistry 33 4291-4299 [DOI] [PubMed] [Google Scholar]
- 36.Turner, G. L., Hinton, J. F., Koeppe, R. E., II, Parli, J. A., and Millett, F. S. (1983) Biochim. Biophys. Acta 756 133-137 [Google Scholar]
- 37.Bañó, M. C., Braco, L., and Abad, C. (1988) J. Chromatogr. 458 105-116 [DOI] [PubMed] [Google Scholar]
- 38.Bak, B., Dambmann, C., and Nicolaisen, F. (1967) Acta Chem. Scand. 21 1674-1675 [DOI] [PubMed] [Google Scholar]
- 39.Bak, B., Led, J. J., and Pedersen, E. J. (1969) Acta Chem. Scand. 23 3051-3054 [DOI] [PubMed] [Google Scholar]
- 40.Van der Wel, P. C. A., Strandberg, E., Killian, J. A., and Koeppe, R. E., II (2002) Biophys. J. 83 1479-1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis, J. H., Jeffrey, K. R., Valic, M. I., Bloom, M., and Higgs, T. P. (1976) Chem. Phys. Lett. 42 390-394 [Google Scholar]
- 42.Koeppe, R. E., II, Killian, J. A., and Greathouse, D. V. (1994) Biophys. J. 66 14-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu, W., Lazo, N. D., and Cross, T. A. (1995) Biochemistry 34 14138-14146 [DOI] [PubMed] [Google Scholar]
- 44.Koeppe, R. E., II, Sun, H., van der Wel, P. C. A., Scherer, E. M., Pulay, P., and Greathouse, D. V. (2003) J. Am. Chem. Soc. 125 12268-12276 [DOI] [PubMed] [Google Scholar]
- 45.Gall, C. M., DiVerdi, J. A., and Opella, S. J. (1981) J. Am. Chem. Soc. 103 5039-5043 [Google Scholar]
- 46.Pulay, P., Scherer, E. M., van der Wel, P. C. A., and Koeppe, R. E., II (2005) J. Am. Chem. Soc. 127 17488-17493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andersen, O. S. (1983) Biophys. J. 41 119-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sawyer, D. B., Koeppe, R. E., II, and Andersen, O. S. (1989) Biochemistry 28 6571-6583 [DOI] [PubMed] [Google Scholar]
- 49.Durkin, J. T., Koeppe, R. E., II, and Andersen, O. S. (1990) J. Mol. Biol. 211 221-234 [DOI] [PubMed] [Google Scholar]
- 50.Andersen, O. S., Bruno, M. J., Sun, H., and Koeppe, R. E., II (2007) Methods Mol. Biol. 400 541-568 [DOI] [PubMed] [Google Scholar]
- 51.Veatch, W. R., Fossel, E. T., and Blout, E. R. (1974) Biochemistry 13 5249-5256 [DOI] [PubMed] [Google Scholar]
- 52.Arseniev, A. S., Lomize, A. L., Barsukov, I. L., and Bystrov, V. F. (1986) Biol. Membr. 3 1077-1104 [Google Scholar]
- 53.Langs, D. A. (1988) Science 241 188-191 [DOI] [PubMed] [Google Scholar]
- 54.Killian, J. A., Prasad, K. U., Hains, D., and Urry, D. W. (1988) Biochemistry 27 4848-4855 [DOI] [PubMed] [Google Scholar]
- 55.LoGrasso, P. V., Moll, F. I., and Cross, T. A. (1988) Biophys. J. 54 259-267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Killian, J. A., Taylor, M. J., and Koeppe, R. E., II (1992) Biochemistry 31 11283-11290 [DOI] [PubMed] [Google Scholar]
- 57.Mattice, G. L., Koeppe, R. E., II, Providence, L. L., and Andersen, O. S. (1995) Biochemistry 34 6827-6837 [DOI] [PubMed] [Google Scholar]
- 58.Ashrafuzzaman, M., Lampson, M. A., Greathouse, D. V., Koeppe, R. E., II, and Andersen, O. S. (2006) J. Phys. Condens. Matter 18 S1235-S1255 [Google Scholar]
- 59.Bouchard, M., Benjamin, D. R., Tito, P., Robinson, C. V., and Dobson, C. M. (2000) Biophys. J. 78 1010-1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burkhart, B. M., Li, N., Langs, D. A., Pangborn, W. A., and Duax, W. L. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 12950-12955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Russell, E. W. B., Weiss, L. B., Navetta, F. I., Koeppe, R. E., II, and Andersen, O. S. (1986) Biophys. J. 49 673-686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mazet, J. L., Andersen, O. S., and Koeppe, R. E., II (1984) Biophys. J. 45 263-276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sham, S. S., Fernandez, J. Q., Townsley, L. E., Greathouse, D. V., Andersen, O. S., and Hinton, J. F. (2003) Biochemistry 42 1401-1409 [DOI] [PubMed] [Google Scholar]
- 64.Jordan, J. B., Shobana, S., Andersen, O. S., and Hinton, J. F. (2006) Biochemistry 45 14012-14020 [DOI] [PubMed] [Google Scholar]
- 65.Jude, A. R., Greathouse, D. V., Koeppe, R. E., II, Providence, L. L., and Andersen, O. S. (1999) Biochemistry 38 1030-1039 [DOI] [PubMed] [Google Scholar]
- 66.Koeppe, R. E., II, Hatchett, J., Jude, A. R., Providence, L. L., Andersen, O. S., and Greathouse, D. V. (2000) Biochemistry 39 2235-2242 [DOI] [PubMed] [Google Scholar]
- 67.van der Wel, P. C. A., Reed, N. D., Greathouse, D. V., and Koeppe, R. E., II (2007) Biochemistry 46 7514-7524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hong, K. H., and Miller, C. (2000) J. Gen. Physiol. 115 51-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Subbiah, R. N., Kondo, M., Campbell, T. J., and Vandenberg, J. I. (2005) J. Physiol. 569 367-379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koeppe, R. E., II, Mazet, J.-L., and Andersen, O. S. (1990) Biochemistry 29 512-520 [DOI] [PubMed] [Google Scholar]
- 71.Norman, K. E., and Nymeyer, H. (2006) Biophys. J. 91 2046-2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.MacCallum, J. L., Bennett, W. F. D., and Tieleman, D. P. (2007) J. Gen. Physiol. 129 371-377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.MacCallum, J. L., Bennett, W. F. D., and Tieleman, D. P. (2008) Biophys. J. 94 3393-3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wimley, W. C., and White, S. H. (1993) Biochemistry 32 6307-6312 [DOI] [PubMed] [Google Scholar]
- 75.Koeppe, R. E., II, Providence, L. L., Greathouse, D. V., Heitz, F., Trudelle, Y., Purdie, N., and Andersen, O. S. (1992) Proteins 12 49-62 [DOI] [PubMed] [Google Scholar]