Abstract
Isothiocyanates (ITCs) found in cruciferous vegetables, including benzyl-ITC (BITC), phenethyl-ITC (PEITC), and sulforaphane (SFN), inhibit carcinogenesis in animal models and induce apoptosis and cell cycle arrest in various cell types. The biochemical mechanisms of cell growth inhibition by ITCs are not fully understood. Our recent study showed that ITC binding to intracellular proteins may be an important initiating event for the induction of apoptosis. However, the specific protein target(s) and molecular mechanisms were not identified. In this study, two-dimensional gel electrophoresis of human lung cancer A549 cells treated with radiolabeled PEITC and SFN revealed that tubulin may be a major in vivo binding target for ITC. We examined whether binding to tubulin by ITCs could lead to cell growth arrest. The proliferation of A549 cells was significantly reduced by ITCs, with relative activities of BITC > PEITC > SFN. All three ITCs also induced mitotic arrest and apoptosis with the same order of activity. We found that ITCs disrupted microtubule polymerization in vitro and in vivo with the same order of potency. Mass spectrometry demonstrated that cysteines in tubulin were covalently modified by ITCs. Ellman assay results indicated that the modification levels follow the same order, BITC > PEITC > SFN. Together, these results support the notion that tubulin is a target of ITCs and that ITC-tubulin interaction can lead to downstream growth inhibition. This is the first study directly linking tubulin-ITC adduct formation to cell growth inhibition.
Isothiocyanates (ITCs)3 are a family of compounds with potential cancer chemopreventive activity. Naturally occurring ITCs found in cruciferous vegetables, including benzyl-ITC (BITC), phenethyl-ITC (PEITC), and sulforaphane (SFN) (Fig. 1A), have demonstrated cancer preventive activity in animals, and increased dietary intake of ITCs has been shown to be associated with a reduced cancer risk in humans (1). It is believed that an important mechanism by which ITCs inhibit tumorigenesis is to suppress proliferation of oncogenic cells by inducing apoptosis and arresting cell cycle progression (1). For example, ITCs have been shown to cause mitochondrial damage, to activate both caspase-dependent and -independent apoptosis (2) and to arrest cell cycle progression, mainly in the G2/M phase; these effects are associated with cyclin B1, cell division cycle 2 and 25C down-regulation, or inhibition (3). However, the upstream biochemical events underlying ITC-induced apoptosis have not been investigated.
FIGURE 1.
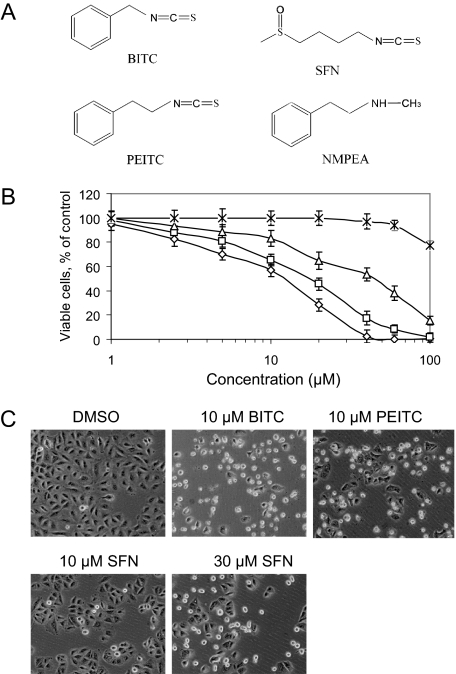
BITC, PEITC, and SFN inhibit A549 cell growth. A, structures of ITCs and NMPEA, an analog of PEITC. B, dose-response growth inhibition after 24-h treatments with BITC (⋄), PEITC (□), SFN (▵), and NMPEA (×); point represents the mean ± S.D. of triplicates. C, phase contrast images of cells after 24-h treatments. Apoptotic cells with shrunken coverage and extensive membrane blebbing appeared after treatment with 10 μm BITC, 10 μm PEITC, and 30 μm SFN. No significant morphological changes were observed in cells treated with vehicle DMSO or 10 μm SFN.
It is well known that ITCs can induce cellular oxidative stress by rapidly conjugating and thus depleting cells of glutathione (GSH) (4). As electrophiles, ITCs readily form conjugates with thiols, including the thiols in GSH and cellular proteins; this conjugation reaction is reversible (5). In fact, the facile reaction between ITCs and cellular thiols is a driving force for enriching intracellular ITC levels up to millimolar levels (4). Recently, we have shown, using 14C-PEITC and 14C-SFN, that the initial conjugation predominantly occurs with cellular GSH, but with increasing time, protein binding gradually becomes the major reaction, at least in part because of dissociation of ITC from ITC-GSH conjugates (6). Eventually, proteins are the major binding sites of ITCs inside cells; for example, PEITC-protein conjugates account for 87% of total cellular uptake after 4 h of treatment. Most importantly, the time course of this protein binding correlated well with the inhibition of proliferation and the induction of apoptosis, providing preliminary evidence suggesting that modification of cellular proteins via direct covalent binding to ITCs may be an early event for apoptosis induction. These results prompted us to identify which specific proteins are ITC binding targets in vivo, to determine how these proteins interact with ITCs, and to begin characterizing the functional consequences of these reactions. The studies described here are important because they not only define the chemical basis of ITC-induced cell growth inhibition, they also identify structural information that will enable the rational design of new, more specific and more potent ITC-related compounds for cancer prevention and treatment studies.
Here we report that tubulin was identified by two-dimensional gel electrophoresis as a potential protein target of ITCs. Tubulin-containing microtubules form one of three main cytoskeletons in eukaryotic cells and play a pivotal role in a variety of cellular processes, involving cell division, motility, and intracellular trafficking (7). The dynamics of α- and β-tubulin polymerization and depolymerization, from heterodimers, is required to form mitotic spindles, which are needed to segregate replicated chromosomes to the two daughter cells. Thus, anti-microtubule or anti-mitosis drugs have gained much attention in anti-cancer drug discovery efforts (8). In this study we examined the role of tubulin binding in cell growth arrest effects by BITC, PEITC, and SFN.
MATERIALS AND METHODS
Cells and Chemicals—The human lung cancer cell line A549 (ATCC, Manassas, VA) was grown and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen) at 37 °C in 5% CO2. SFN was provided by Dr. Stephen Hecht (University of Minnesota). 14C-SFN (50 mCi/mmol) was a gift from Dr. Shantu Amin (Pennsylvania State University, Hershey). 14C-PEITC (55 mCi/mmol) was purchased from American Radiolabeled Chemicals (St. Louis, MO). Pure (>99%) porcine tubulin was purchased from Cytoskeleton Inc. (Boulder, CO). Sequencing-grade trypsin was obtained from Promega (Madison, WI). PEITC, BITC, 5,5′-dithiobis(2-nitrobenzoic acid), N-methyl phenethylamine (NMPEA), vinblastine, dimethyl sulfoxide (DMSO), and all other reagents were the highest grade available from Sigma, unless otherwise noted.
Cell Proliferation Assay—Cell growth curves were measured by the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS), assay using the CellTiter 96 Aqueous One Solution cell proliferation assay kit (Promega, Madison, WI). Briefly, cells were plated in 96-well plates at a density of 104 cells/well (200 μl). Twenty four hours after plating, cells were treated with different doses of each ITC for 24 h. Then 20 μl of the MTS solution was added. After 2 h, the absorbance was measured at 490 nm with a Synergy HT fluorescent microplate reader (BIO-TEK Instruments, Inc., Winooski, VT). The rate of growth inhibition was calculated based on the control wells, which were treated with vehicle only (0.1% v/v DMSO) and taken as 100% growth, and wells with cell-free medium were taken as base line. The data are presented as the average of triplicate wells. NMPEA, a PEITC structural analog, was used as an inhibition negative control.
Cell Staining for Cell Cycle Analysis—Cells were treated with 10 μm BITC, 10 μm PEITC, 10 and 30 μm SFN for up to 24 h. After the ITCs were added to the media, aliquots of cells were taken every 4 h for 24 h and fixed with ice-cold 80% ethanol, treated with 500 μg/ml RNase A (Sigma), and subsequently stained with 25 μg/ml propidium iodide (PI) (Sigma).
Cell Staining for Mitosis Marker—Cells were fixed and stained according to a modified protocol (9). Briefly, after being fixed in 80% ice-cold ethanol for 2 h at -20 °C, cells were washed once with 1% BSA in PBS, permeated in 1 ml of PBS containing 1% BSA, 0.25% Triton X-100 for 5 min on ice. After blocking with PBS containing 1% BSA, 10% normal goat serum on ice for 1 h, 5 μg of anti-phosphorylated histone H3 antibody (rabbit polyclonal IgG, Upstate Biotechnology) was added to each sample. The cells were incubated overnight at 4 °C, then washed twice with 5 ml of 1% BSA in PBS and resuspended in 180 μl of PBS containing 1% BSA, 10% normal goat serum. Fluorescein-conjugated goat anti-rabbit (Invitrogen) and PI were added (7 and 2 μg per sample, respectively). Cells were incubated for 1 h at 4 °C. After washed three times with 1% BSA in PBS, cells were centrifuged and treated with 500 μg/ml RNase A for 30 min.
Flow Cytometry—Cell cycle distributions were measured by flow cytometry. Flow cytometry was done on a FACSCalibur (BD Biosciences). Fluorescein isothiocyanate and PI were excited with a 488 nm laser, and data were collected through 530- and 630-nm bandpass filters, respectively.
Caspase-3 Activity—The assay was performed according to assay kit manufacturer's protocol. Briefly, cells were treated with 10 μm BITC, 10 μm PEITC, 10 and 30 μm SFN for 24 h. Cells were harvested at 4-h intervals by scraping, and lysed by freezing and thawing three times in 100 μl of assay buffer (50 mm HEPES, 100 mm NaCl, 10 mm dithiothreitol, 1 mm EDTA, 0.1% CHAPS, pH 7.4). The lysates were centrifuged at 10,000 × g for 15 min. Ten μl of cell lysate supernatant was mixed with 70 μl of assay buffer and 20 μl of 50 μm Ac-DEVD-rhodamine 110 (R110) substrate (Roche Applied Science). The mixture was incubated at 37 °C in the dark for 1 h. The release of rhodamine from the substrate was monitored with excitation at 485 nm and emission at 528 nm every 10 min with a Synergy HT fluorescent microplate reader. The unit of activity for caspases-3 is expressed as nanomoles of R110 released per min per mg of protein.
Two-dimensional Gel Electrophoresis—The procedures used were performed according to established protocols (10). Cells at 50% confluency were treated with 30 μm 14C-PEITC or 14C-SFN for 1 h. Cells were then lysed in 7 m urea, 2 m thiourea, 2% CHAPS, 1% ASB-14, 0.2% carrier ampholytes 4–7, 40 mm Tris, 0.0002% bromphenol blue, 1× protease inhibitor mix (GE Healthcare) on ice for 30 min. The cell lysate was centrifuged at 13,000 × g for 5 min to remove the insoluble matter. The supernatant, concentration adjusted to 0.5 mg of protein in 450 μl, was applied to an IPG drystrip of 24 cm at pH 4–7 (Bio-Rad). After 12 h of in-gel rehydration at 50 V, isoelectric focusing was performed in isoelectric focusing cells (Amersham Biosciences) with the following voltage gradients: (i) 0–500-V gradient in 1 h; (ii) 500–10,000-V gradient in 8 h; and (iii) 10,000-V constant until 80,000 V-h. IPG strips were equilibrated in 50 mm BisTris-Cl, 6 m urea, 30% glycerol, 2% SDS, 1% dithiothreitol, 0.005% bromphenol blue, pH 6.5. Each strip was loaded on top of a 5–20% gradient BisTris SDS-polyacrylamide gel slab (20 × 26 cm, 1 mm thick), and electrophoresis was performed in 50 mm MOPS, 50 mm BisTris, 0.1% SDS, 1 mm EDTA, pH 6.8, at a constant power of 30 watts for 30 min and 100 watts for 6 h or until the dye front reached the gel bottom. The proteins were stained with colloidal Coomassie Blue (11), and proteome images were acquired by an Expression 1680 flatbed scanner (Epson). The radioactivity images of gels were obtained by exposing air-dried gels to BioMax MR x-ray film (Eastman Kodak Co.) for up to 2 months at -80 °C.
Tubulin in Vitro Polymerization Assay—Purified tubulin was dissolved in the general tubulin buffer (80 mm PIPES, 1 mm MgCl2, 1 mm EGTA, 5% glycerol, and 1 mm GTP, pH 6.9). The tubulin solution (3.0 mg/ml) was then incubated with 3, 10, 30, 100, and 300 μm BITC, PEITC, and SFN for 10 min on ice. The polymerization was initiated, and the measurement of the absorbance at 340 nm began when the 96-well plate was put into a pre-warmed (37 °C) Synergy HT fluorescent microplate reader. The measurements took 1 h. Tubulin of the same concentration was treated with vehicle (DMSO) and 1 μm vinblastine as negative and positive controls for the polymerization inhibition assay. NMPEA was also used as a negative control.
Tubulin in Vivo Polymerization Assay by Indirect Immunofluorescence Microscopy—The assay was performed according to a published method (12). Briefly, 104 cells were cultured on microscope cover glass (12 mm diameter; Fisher) placed in individual wells of a 24-well tissue culture plate (BD Biosciences) for 24 h before being treated with 5 or 10 μm BITC, PEITC, and SFN for 30 min and 1, 2, 4, and 24 h at 37 °C. The cells were then fixed sequentially with 3% paraformaldehyde in Dulbecco's modified Eagle's medium (10 min) and 3% paraformaldehyde in PBS (20 min), quenched with 50 mm NH4Cl in PBS, and permeabilized for 30 min with 1% BSA, 0.075% saponin in PBS. Cells were incubated overnight at 4 °C with mouse monoclonal anti-human tubulin IgG1 (1:400; Sigma, clone DM1A), rinsed three times with 0.1% BSA, 0.075% saponin in PBS, and then incubated with Alexa Fluor 546-conjugated goat anti-mouse IgG (1:800; Molecular Probes) for 1 h at room temperature. Once the labeling was complete, the cover glasses were rinsed twice with PBS and mounted onto microscope slides with a small drop of ProLong AntiFade reagent containing 4′,6-diamidino-2-phenylindole (Molecular Probes). The samples were viewed, and images were taken with a Fluoview-FV300 Laser Scanning Confocal System (Olympus, Tokyo, Japan).
Detection of in Vitro Adduct between Tubulin and ITCs—Purified porcine tubulin (Cytoskeleton Inc.) was dissolved in PBS buffer at 0.5 mg/ml. The solution was incubated at room temperature for 60 min in the presence of 100 μm BITC, PEITC, and SFN, or with same volume of carrier solvent DMSO (untreated). The protein was purified by SDS-PAGE after the reactions. Gel slices from the ∼50-kDa migration position were sent for mass spectrometry analysis.
Detection of in Vivo Adduct between Tubulin and BITC—A549 cells were treated with 20 μm BITC for 1 h. Treated cells were lysed in RIPA buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 0.25% sodium deoxycholate, 1 mm phenylmethylsulfonyl fluoride) on ice for 20 min before centrifugation at 15,000 × g for 10 min. The pellet was washed first by RIPA buffer (once) and then by water (twice) and finally resuspended in 200 μl of water. The resuspension mixture was incubated with DNase I (0.5 μg, Sigma) for 30 min at room temperature before being centrifuged at 300 × g through a 20% sucrose cushion. The recovered pellet was dissolved in 2% SDS and resolved by SDS-PAGE. The gel slices at ∼50 kDa were sent for mass spectrometry analysis.
Mass Spectrometry Analysis—Protein spots of interest from the two-dimensional gel electrophoresis experiments were analyzed by in-gel digestion and nanoflow reversed-phase liquid chromatography (nanoRPLC)-tandem mass spectrometry (MS/MS) as described (13).
Analysis of the products formed from the ITC reaction with tubulin was carried out as follows. An aliquot of tubulin was mixed with an equal volume of 100 mm NH4HCO3, digested with 2 μg of trypsin at 37 °C for 2 h, acidified with trifluoroacetic acid, and desalted by C-18 Zip-Tip microcolumns (Millipore, Billerica, MA). Samples were lyophilized, reconstituted in 0.1% trifluoroacetic acid, and analyzed by matrix-assisted laser desorption/ionization-time-of-flight/time-of-flight (MALDI-TOF/TOF) MS and nanoRPLC-MS/MS as described previously (4, 13, 14). Tandem MS spectra from the nanoRPLC-MS/MS analysis were compared with entries in the UniProt porcine proteomic data base from the European Bioinformatics Institute using SEQUEST software (ThermoElectron) with dynamic modifications of 149, 163, and 177 on cysteine.
Measurement of Free Thiols in Tubulin—The number of free thiols in tubulin was determined by Ellman assay (15). Lyophilized porcine tubulin (Cytoskeleton Inc., Denver, CO) was dissolved in 0.1 m sodium phosphate, pH 7.4. An aliquot of tubulin solution (0.4 mg/ml) was incubated with 80 and 160 μm BITC, PEITC, and SFN (the ratios of compound to tubulin cysteine were 1:1 and 2:1, respectively) at room temperature in the dark for 30 min. After incubation, tubulin samples were diluted in 4 m guanidine HCl and 1 mm 5,5′-dithiobis-2-nitrobenzoic acid, and the absorbance at 412 nm was measured by a UV-1700 UV/visible spectrophotometer (Shimadzu, Columbia, MD). A standard curve was generated using N-acetylcysteine (Sigma). The calculated extinction coefficient for 2-nitro-5-thiobenzoate dianion at 412 nm was 14,300 m-1 cm-1.
Structural Characterization of Tubulin Treated with ITCs—Circular dichroism spectra were recorded on a Jasco J-810 spectropolarimeter (Jasco, Easton, MD) over the wavelength range of 250 to 190 nm using a 10-mm path length cell and then averaged over three scans. Spectra were recorded on samples containing 2 μm tubulin in 20 mm PIPES, 1 mm MgCl2, 1 mm EGTA, pH 6.8, before and after addition of 40 μm BITC (the ratio of BITC to tubulin cysteine was 2:1). The fraction of helices was estimated from ellipticity at 222 nm using an empirical method (16).
All fluorescence measurements were performed on a PTI QuantaMaster fluorescence spectrophotometer (Photon Technology International, Birmingham, NJ) equipped with the peltier temperature control. The change of intrinsic tryptophan fluorescence of 2 μm tubulin in PIPES buffer (80 mm PIPES, 1 mm MgCl2, 1 mm EGTA, pH 6.8) by compounds was monitored with λ(excitation) = 295 nm and λ(emission) = 329 nm. To avoid photo-bleaching of the samples, the shutter was open for 5 s per min for 35 min. The data were analyzed using software Felix32 based on the mono-exponential decay model.
Western Blotting—A549 cells were treated with different concentrations of BITC, PEITC, and SFN for 4 h. Treated cells were lysed in RIPA buffer on ice for 20 min before centrifugation at 15,000 × g for 10 min. The supernatants were collected as the soluble fraction, whereas the pellets were re-dissolved in SDS buffer (65 mm Tris, pH 7.0, 2% SDS, 50 mm dithiothreitol, 10% glycerol, and 0.001% bromphenol blue) and used as the insoluble fraction. Twenty micrograms of protein were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore). Western blot analysis was performed by incubating the membrane with a mouse α-tubulin antibody (1:2000, clone DM1A; Sigma) and a β-tubulin antibody (1:1000, clone TUB2.1; Sigma) followed by a horseradish peroxidase-labeled goat anti-mouse secondary antibody (1:10,000; GE Healthcare). The targeted proteins were detected by chemiluminescence (Pierce). β-Actin, detected by an antibody (1:10,000, clone AC-74; Sigma), was used as a loading control in this experiment.
RESULTS
BITC, PEITC, and SFN Inhibited Cell Proliferation—Incubation of A549 cells with BITC, PEITC, and SFN for 24 h at a concentration range of 1–100 μm caused a dose-dependent inhibition of cell proliferation (Fig. 1B). The IC50 values were 13.5, 18.3, and 43.0 μm for BITC, PEITC, and SFN, respectively. In contrast, NMPEA, a structural analog of PEITC without the ITC functionality, did not significantly inhibit cell growth except at the highest concentration tested. These results indicate that the ITC functional group is essential for inhibiting cell growth, and the alkyl or arylalkyl side chain moiety dictates the inhibitory potency. These findings are consistent with the cell morphology (Fig. 1C). Significant changes in cell morphology were observed after treatment with BITC and PEITC at 10 μm. At this concentration, however, SFN caused little change. Similar effects were only observed with SFN only at 30 μm or greater concentrations. It should be noted that 10 μm of PEITC is a clinically attainable concentration.4
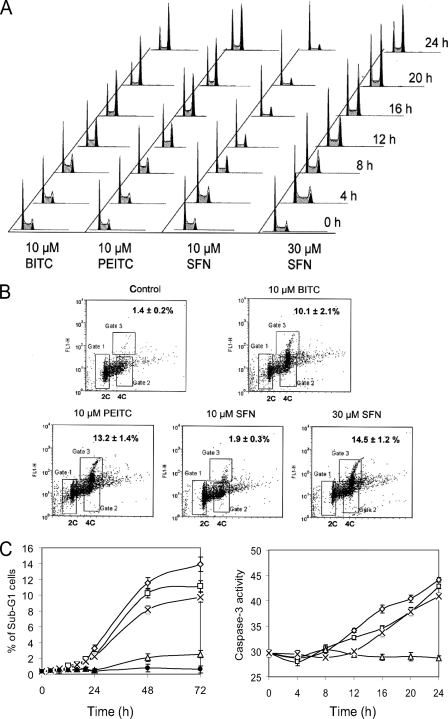
BITC, PEITC, and SFN Induced Cell Cycle Arrest and Apoptosis—We next examined the effects of ITCs on cell cycle regulation using flow cytometry. When cells were treated with 10 μm BITC and 10 μm PEITC for 24 h, the proportions of cells residing in the G1 phase decreased from 41 to 13 and 25%, respectively (Fig. 2A); the proportions of cells residing in the S phase decreased from 42 to 23 and 30%, and the cell populations in the G2/M phase markedly increased from 10 to 63 and 48%, respectively. After 12-h treatments, BITC caused a higher percentage of cells to be arrested in G2/M than PEITC. Unlike BITC and PEITC, the percentage of cells in the G2/M phase was essentially unaffected with 10 μm SFN treatment, although the percentage of cells in S phase was decreased, and the percentage of cells arrested in G1 phase increased to 64% in the G1 phase after 24 h of treatment compared with 41% at the start. Interestingly, 30 μm SFN increased the G2/M arrest population to 57%.
FIGURE 2.
BITC, PEITC, and SFN induce cell cycle arrest and apoptosis in A549 cells. A, histograms of ITC-treated cells indicating that 10 μm BITC, 10 μm PEITC, and 30 μm SFN induce substantial G2/M arrest (increase of the second peak), whereas 10 μm SFN induce G1 arrest. B, substantial amounts of mitotic cells were detected after treatments with 10 μm BITC, 10 μm PEITC, and 30 μm SFN for 24 h but not in cells treated with 10 μm SFN. The mitotic cells are identified as having a tetraploid DNA content and a high level of phosphorylated histone 3 as indicated in Gate 3. Gate 1 contains cells with diploid DNA, whereas Gate 2 contains cells with tetraploid DNA and low level of phospho-H3. Numbers in the upper right corners indicate percentages of mitotic cells in each sample. C, induction of apoptosis was indicated by increases in the relative size of sub-G1 fractions of cells (left) and increased caspases-3 activities (right) in cells treated with 10 μm BITC (⋄), 10 μm PEITC (□), and 30 μm SFN (×) but not in 10 μm SFN (▵) and DMSO (•).
To determine whether the G2/M arrest occurs in the G2 or mitosis phase, we used immunofluorescence to detect phosphorylated histone 3, a mitotic biomarker. Fig. 2B shows the sorting results of cells treated with ITCs. Gate 3 is the assay positive zone, containing cells with duplicated DNA and high levels of phosphor-H3. Compared with 1.4% of mitotic cells in the control sample (dimethyl sulfoxide, DMSO), BITC and PEITC at 10 μm induced increases to 10.1 and 13.2% in Gate 3, respectively, after 24 h of treatment. In contrast, 10 μm SFN did not significantly increase the Gate 3 population (only 1.9%) and had no effect on the condensed cell population in the diploid DNA region (Gate 1). However, 30 μm SFN caused 14.5% of the cells to be arrested in the mitotic phase. These results are consistent with the cell cycle findings shown in Fig. 2A and clearly show that ITC treatments caused cells to arrest in the mitotic phase.
To examine whether apoptosis could be involved in the cell morphology changes observed (Fig. 1C) and the reduced numbers of viable cells measured after ITC treatments, two different apoptosis assays were used. In the first assay, Fig. 2C (left), we followed the accumulation of cells in the sub-G1 fraction for 72 h of ITC treatment, 48 h longer than needed to detect the mitotic arrest with 10 μm BITC or PEITC, or 30 μm SFN. Each of these treatments caused a sharp increase in the sub-G1 population. These results indicate that apoptosis induced by ITCs is likely a consequence of the mitotic arrest. In contrast, treatment with 10 μm SFN did not show these effects. Apoptosis, as measured by caspases-3 activity, also increased dramatically only when cells were arrested in mitosis (Fig. 2C, right). The relative potency of the ITC compounds in both of these apoptosis assays, BITC > PEITC > SFN, are the same as found for the cell proliferation and cell cycle arrest assays.
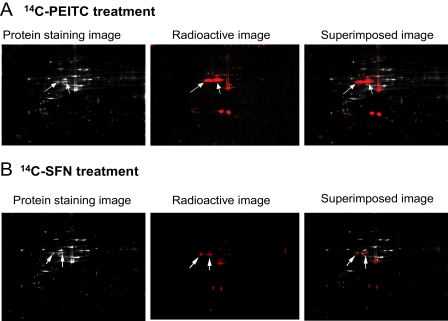
Tubulin Is a Major ITC Binding Target in Vivo—Next, we identified the protein targets of ITCs. For this we used proteins extracted from A549 cells treated with 14C-PEITC and 14C-SFN and then analyzed by two-dimensional gel electrophoresis and mass spectrometry. After superimposing the colloidal Coomassie Blue protein staining pattern with the pattern of radioactivity, obtained from x-ray films, it was clear that only a few proteins contained radioactivity, presumably resulting from selective binding with PEITC or SFN, via thiocarbamation (Fig. 3). The MALDI-TOF/TOF analysis of proteins extracted from the major radioactive-containing gel spots identified several proteins, including multiple human tubulin isoforms (Table 1). These results suggest that both α- and β-tubulins are potential ITC targets and thus carriers of radioactivity, i.e. they may have been modified covalently by the radioactive ITCs. Similar, but not identical, results were obtained with both ITCs.
FIGURE 3.
Two-dimensional gel electrophoresis images of A549 whole cell lysates after treatments with 20 μm14C-PEITC (A) and 14C-SFN (B) for 1 h. The colloidal Coomassie Blue staining images show the total protein profiles; the radioactive images obtained by exposing dried two-dimensional gel to x-ray film show the profiles of the 14C-ITC bound proteins; the superimposed images were used for identifying individual protein of ITCs in two-dimensional gel. The two arrows point to the spots containing α- and β-tubulin.
TABLE 1.
Peptides of tubulin were identified by mass spectrometry through radiolabeled-two-dimensional gel electrophoresis
| Protein name | NCBI access no. | Identified peptide sequence | No. of hits on the peptide |
|---|---|---|---|
| Tubulin α1 chain | P68366 | K.DVNAAIAAIK.T | 3 |
| R.AVFVDLEPTVIDEIR.N | 2 | ||
| K.VGINYQPPTVVPGGDLAK.V | 2 | ||
| R.LISQIVSSITASLR.F | 1 | ||
| R.PTYTNLNR.L | 1 | ||
| K.EDAANNYAR.G | 1 | ||
| R.LSVDYGKK.S | 1 | ||
| Tubulin α2 chain | Q13748 | K.DVNAAIATIK.T | 4 |
| K.TIGGGDDSFNTFFSETGAGK.H | 1 | ||
| Tubulin α3 chain | Q71U36 | R.AVFVDLEPTVIDEVR.T | 7 |
| K.EIIDLVLDR.I | 2 | ||
| Tubulin β1 chain | Q9H4B7 | R.FPGQLNADLR.K | 3 |
| R.FPGQLNADLRK.L | 4 | ||
| Tubulin β2 chain | P07437 | R.ISVYYNEATGGK.Y | 2 |
| R.ALTVPELTQQVFDAK.N | 2 | ||
| K.NSSYFVEWIPNNVK.T | 1 | ||
| R.YLTVAAVFR.G | 1 | ||
| R.ISVYYNEATGGK.Y | 1 | ||
| Tubulin β2C chain | P68371 | R.INVYYNEATGGK.Y | 2 |
| Tubulin β3 chain | Q13509 | R.YLTVATVFR.G | 2 |
| K.MSSTFIGNSTAIQELFK.R | 1 | ||
| K.VREEYPDR.I | 1 | ||
| R.ISVYYNEASSHK.Y | 1 | ||
| β-Tubulin 4Q | Q8WZ78 | R.INVYYNEASGGR.Y | 2 |
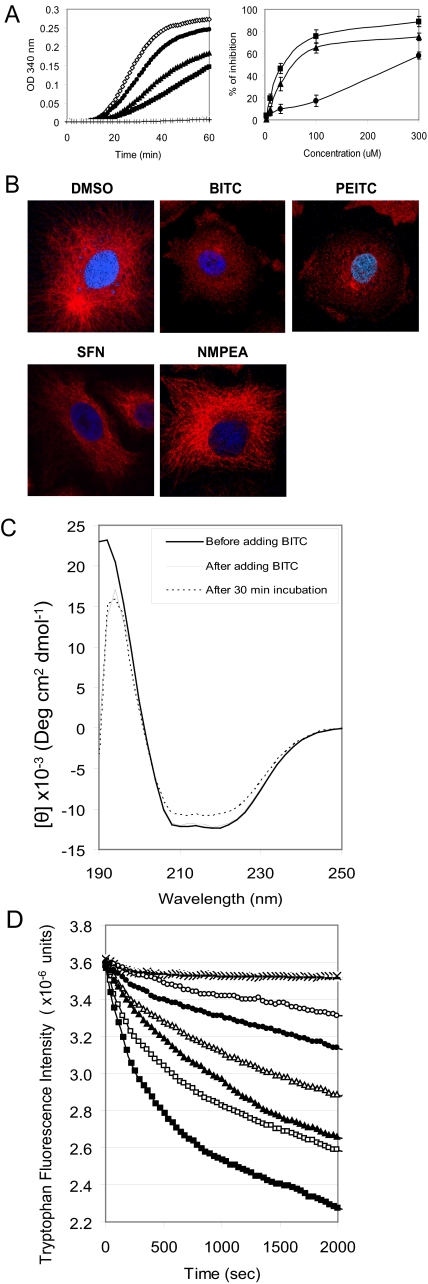
BITC, PEITC, and SFN Hindered Tubulin Polymerization in Vitro and Disrupted Microtubules in Vivo—To investigate if binding to tubulin by ITCs could be an early event leading to mitotic arrest, we first studied the effects of ITCs on tubulin polymerization in vitro. The turbidity (absorption at 340 nm) increase caused by tubulin polymerization was monitored continuously at 37 °C (Fig. 4A, left). All three ITCs inhibited tubulin polymerization in a dose-dependant manner; in a 1-h reaction, 30 μm BITC, PEITC, and SFN inhibited polymerization by 47, 33, and 10%, respectively, compared with the controls (curves obtained from DMSO and 30 μm NMPEA, which completely overlapped). As an assay-positive control, we used 1 μm vinblastine, which completely suppressed tubulin polymerization. Again, Fig. 4A (right), the relative inhibitory potencies of the ITCs at every concentration tested was BITC > PEITC > SFN.
FIGURE 4.
BITC, PEITC, and SFN inhibit tubulin polymerization in vitro and in vivo. A, left, in vitro tubulin polymerization inhibition by 30 μm BITC (▪), 30 μm PEITC (▴), and 30 μm SFN (•) with negative controls, DMSO and 30 μm NMPEA (⋄), and a positive control, 1 μm vinblastine (+); A, right, the dose-dependent inhibition by BITC (▪), PEITC (▴), and SFN (•). B, microtubule images of A549 cells after ITC treatments. The microtubule network was disrupted and degraded in cells treated with 5 μm BITC for 30 min and 5 μm PEITC for 1 h. The normal tubulin network was reduced to isolated spots in these cells. However, most of the microtubule network was still visible in cells treated with 10 μm SFN for 4 h. There was also no detectable change in microtubule network in cells treated with 10 μm NMPEA for 4 h. The α-tubulin was stained in red by indirect immunofluorescent staining and the nucleus in blue by 4′,6-diamidino-2-phenylindole. C, structural changes of tubulin upon binding with ITCs. The secondary structure changes of tubulin (2 μm) by 40 μm BITC are shown by circular dichroism (left). D, changes of tubulin intrinsic tryptophan fluorescent intensity during the treatments indicate the tertiary structural changes induced by 40 μm BITC (▪), 20 μm BITC (□), 40 μm PEITC (▴), 20 μm PEITC (Δ), 40 μm SFN (•), and 20 μm SFN (○). The curves of negative controls (treated with DMSO, NMPEA, and benzyl cyanide) overlap and are indicated by the ×.
We then examined the effects of ITCs on the microtubule network in vivo by indirect immunofluorescent staining of α-tubulin in cells (Fig. 4B). Compared with the normal microtubule distribution and its network-like structure in vehicle (DMSO)-treated cells, 5 μm BITC disrupted the network within 30 min, with similar results seen after 1 h with 5 μm PEITC. In contrast, some of the microtubule network remained intact after 4 h of treatment with 10 μm SFN. For all ITC treatments lasting for 24 h, the average cell spreading area was reduced, and the normal polarized cell shapes became almost round. Because the actin skeleton was not visibly affected by these concentrations of ITC for treatment as long as 24 h (data not shown), the observed cell shrinkage and depolarization induced by the ITCs were probably because of a complete collapse of the microtubule cytoskeleton. Treatment with 10 μm NMPEA for 24 h also did not show any effect on microtubule structure, indicating that the isothiocyanate functional group is essential for the inhibition of tubulin polymerization in vivo.
BITC, PEITC, and SFN Altered Tubulin Secondary and Tertiary Structures—Our next question was whether ITCs can cause any conformational changes in tubulin. To answer this, we employed circular dichroism and fluorometry to detect secondary and tertiary structural changes in tubulin after binding with ITCs. The CD spectra of native tubulin before and after treatment with BITC are shown in Fig. 4C. Native tubulin has two apparent minima, at 208 and 222 nm, and an apparent maximum at about 192 nm, indicating that tubulin has a high helical content. Comparing this spectrum with the spectrum obtained after a 30-min incubation at room temperature with 40 μm BITC (BITC:tubulin cysteine = 2:1), one obtains a decrease in ellipticity at both 208 and 222 nm, suggesting either an α-helical content decrease from 31 to 26% when calculated by an empirical method (16) or formation of tubulin aggregates. In either case, these results show that BITC binding induced tubulin conformational changes. The spectrum of the untreated tubulin as a control did not change under these incubation conditions.
To identify tertiary structural changes caused by ITC binding, the intrinsic tryptophan fluorescence was monitored at 37 °C for 2000 s right after the compounds were added. Fig. 4D shows that the tryptophan fluorescence intensity of untreated tubulin remained constant except for a slight drop at the beginning, possibly because of thermal effects. In contrast, the samples treated with ITCs showed significant fluorescence intensity decay in a dose-dependent manner. Consistent with the functional changes of tubulin, the greatest and fastest decay was observed following the BITC treatment, and the smallest and slowest decay occurred following the addition of SFN. Because all eight tryptophan residues (four in α-tubulin and four in β-tubulin) are buried and widely spaced in the three-dimensional structure (see Protein Data Bank code 1SA0) and because the majority of them are not in the vicinity of cysteine residues, the abnormal decay observed indicates that the global, rather than local, tubulin structure was changed dramatically as a result of ITC binding. As negative controls, tubulin treated with the same concentration of NMPEA (Fig. 4D) or benzyl cyanide (an analog to BITC) did not induce fluorescence decay. Interestingly, the decay caused by 20 μm cyclohexylmethyl-ITC, a nonaromatic ITC analog of BITC, was close to that of 20 μm BITC (data not shown), indicating that tryptophan fluorescence decay was not because of energy transfer of the benzene ring in BITC or PEITC, but was solely a result of tertiary structural changes caused by binding with the ITC group.
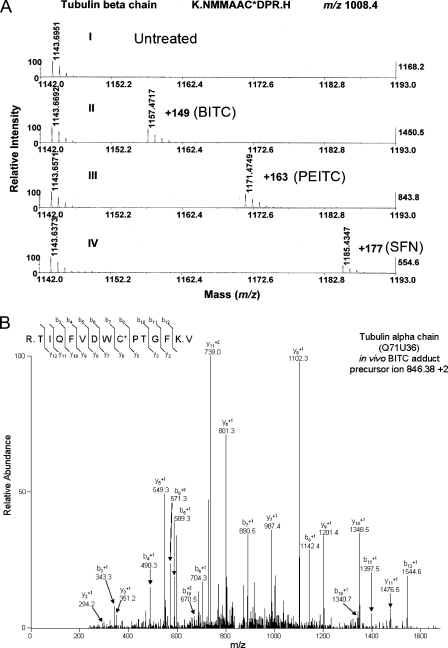
Covalent Binding and Differential Modification of Thiols in Tubulin by ITCs—To demonstrate that ITCs can covalently modify tubulin and to identify the binding sites, we trypsin-digested purified protein that had been treated in vitro with and without ITCs and then analyzed the digests by both MALDI-TOF/TOF and nanoRPLC-MS/MS. Fig. 5A shows a typical cysteine-containing tryptic peptide, 298NMMAACDPR306 ([M + H]+ of m/z 1008.4), that was obtained from β-tubulin treated with any of the three ITCs. The tandem MS spectra of this peptide, shown in supplemental Figs. A–C, indicate that the modification was on Cys303 with mass increments of 149, 163, and 177 for BITC, PEITC, and SFN, respectively, as indicated by an increase in m/z of the b- and y-type fragment ions. These results showed that different ITCs can covalently modify tubulin on the same cysteine residues.
FIGURE 5.
Covalent binding of ITCs to cysteine residues in tubulin is indicated by both MALDI-TOF/TOF and nanoRPLC-MS/MS. A, covalent binding is indicated by a mass increment of 149, 163, and 177 (molecular weight of BITC, PEITC, and SFN, respectively) in a typical cysteine-containing peptide, 298NMMAACDPR306 ([M + H]+ of m/z 1008.4). The peptides were obtained by trypsin digestion of purified porcine tubulin treated with each ITC individually and identified by MALDI-TOF/TOF. The individual peptides were further analyzed by tandem mass spectrometry. The spectra are shown in the supplemental material. The data base access number for porcine β-tubulin is P02554. B, tandem mass spectrometry (MS/MS) spectrum of a tubulin α chain peptide isolated from the insoluble fraction of A549 cells treated with BITC (20 μm) showing that a cysteinyl residue (Cys347) is covalently modified by BITC. A549 cells were treated and lysed as described under “Materials and Methods.” Tubulin-containing cellular fractions were identified by Western blot and subjected to tryptic digestion prior to analysis using MS/MS. The BITC-modified cysteinyl residue was identified by the presence of a mass of 149.03 Da added to this residue. The data base access number for human α-tubulin is Q71U36.
To determine the levels of ITC binding to tubulin, we performed Ellman assays, which can quantify the number of free thiols on tubulin, both before and after reaction with ITCs. The number of free thiols in proteins was determined by measuring the absorbance at 410 nm after reaction with 5,5′-dithiobis(2-nitrobenzoic acid). The results (Table 2) indicated that the thiols of the purified tubulin were modified differentially by the ITCs. Using 80 and 160 μm of ITC (ITC: tubulin cysteine = 1:1 and 2:1), the numbers of thiols modified by BITC were 9.5 and 11.7, PEITC 6.2 and 9.1, and SFN 2.6 and 3.8, respectively. We used diallyl trisulfide as an internal control, and the results were in excellent agreement with that reported previously (17).
TABLE 2.
Number of thiols in tubulin modified by ITC compounds (Ellman assay)
|
Total no. of free thiols/pair of tubulin
|
No. of free thiols modified/pair of tubulin
|
|||||
|---|---|---|---|---|---|---|
|
BITC
|
PEITC
|
SFN
|
||||
| 80 μm (1:1)a | 160 μm (2:1) | 80 μm | 160 μm | 80 μm | 160 μm | |
| 14.7 ± 0.2 | 9.5 ± 0.2 | 11.7 ± 0.4 | 6.2 ± 0.3 | 9.1 ± 0.3 | 2.6 ± 0.1 | 3.8 ± 0.1 |
The stoichiometric ratio between compound to cysteines in tubulin. The tubulin cysteine concentration was 40 μm
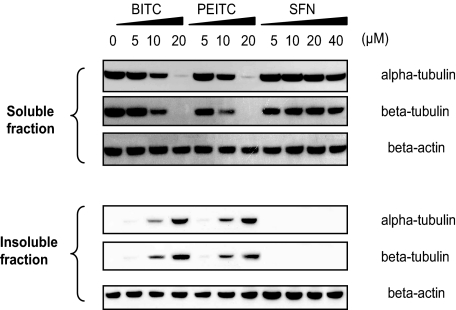
BITC and PEITC Caused Tubulin Precipitation in Cells—Surprisingly, we found (Fig. 6) that substantial amounts of tubulin were rapidly precipitated in A549 cells treated with BITC and PEITC, but not in SFN-treated cells. The concentration used (10 μm) giving this effect was the same as was used to induce cell cycle arrest and substantial apoptosis. Western blot analysis after 4-h treatments showed that >50% of the total cellular α- and β-tubulin could not be recovered from the soluble fraction (defined as the supernatant recovered when using Nonidet P-40 or Triton X-100 containing lysis buffers). Instead, the majority of the tubulin appeared in the insoluble fraction, defined as insoluble in nonionic detergent lysis buffers but soluble in ionic detergent such as SDS. We found that this tubulin precipitation occurred in a dose- and time-dependent manner and in the same order of potency BITC > PEITC > SFN (data not shown). To our knowledge, tubulin precipitation in whole cells has not been previously described and thus may be a unique response to treatment with ITCs, suggesting that tubulin precipitation is a result of the structural misfolding caused by binding with ITCs.
FIGURE 6.
ITC treatment induced tubulin precipitation, indicated by altered levels of α- and β-tubulin in both the soluble and insoluble fractions of A549 cells, as determined on Western blots. Cells were treated and lysed as described under “Materials and Methods.” Membranes were probed for α- and β-tubulin, whereas β-actin was used as the loading control.
BITC-modified Cys347 of α-Tubulin in Vivo—Tubulin in both the soluble and insoluble fractions of the BITC treated-cell lysate were purified and analyzed by mass spectrometry. The results (Fig. 5B) show that Cys347, a conserved cysteine in all α-tubulin isoforms, was covalently modified by BITC as indicated by the m/z ratio increments of 149. The modification was found only in tubulin from the insoluble fraction suggesting modification by BITC triggers the precipitation. Mass spectrometry analysis also showed that none of the 20 cysteines in both α- and β-tubulin was modified in any form of oxidation in either the soluble or insoluble fractions. Because no reducing agents were used throughout the sample preparation, the results suggest that the changes related to tubulin are irrelevant to oxidative stress induced by BITC.
DISCUSSION
Previously we showed that proteins are the major target of PEITC and SFN in cells and concluded that direct binding of ITCs to intracellular proteins may trigger apoptosis (6). In this study, we identified tubulin as one of these in vivo targets for ITC binding, demonstrated covalent binding of BITC, PEITC, and SFN to tubulin, and showed that this binding correlated well with their ability to induce mitosis arrest and apoptosis. We found that the relative potency of ITCs to inhibit growth of A549 cells was BITC > PEITC > SFN, the same relative potencies for cell growth inhibition reported by others for a variety of cell lines (2, 18–21). A recent study showed that SFN treatments can disrupt tubulin polymerization and spindle assembly, which in turn caused mitotic arrest (22). We demonstrated here that ITCs inhibit tubulin polymerization and disrupt microtubule networks in the same relative order of potency, providing evidence for models in which ITC binding to tubulin disrupts microtubule formation, which may then provoke cell cycle arrest and apoptosis. Importantly, we have for the first time demonstrated in vivo adduct formation between an ITC and any intracellular human protein. Finally, this study also shows that, although the isothiocyanate functionality seems essential for adduct formation and the observed biological effects, all ITCs are not created equal; their affinities for covalent binding to tubulin, specifically cysteines, and the functional consequences of these adducts depend largely on the individual ITC structures. This information is essential for increasing our understanding of the mechanisms underlying ITC-induced apoptosis and will help in designing more efficacious ITC-related compounds for the prevention and therapy of cancer.
As electrophiles, ITCs are capable of binding to protein thiols and amino groups, and these modifications may alter protein functions. The binding observed under our conditions appears to be selective as our two-dimensional gel electrophoresis using cells treated with 14C-labeled ITCs yielded only a relatively small number of spots with Coomassie staining that were radioactive. It is conceivable that the structure of individual ITC compounds may influence their binding preferences as it dictates hydrophobicity, size, shape, and electrophilicity. The selective interactions obtained with intracellular proteins were suggested by previous studies in which ITCs were shown to inhibit preferentially, presumably via binding to enzymes, specific cytochrome P450 isozymes, depending on their structures (22, 23–26). A recent study showed that two SFN derivatives, SFN-N-acetylcysteine and SFN-cysteine, were effective histone deacetylase (HDAC) inhibitors, whereas the SFN parent compound had little effect (27). Our in vivo and in vitro studies showed little or no effects of BITC, PEITC, and SFN on the activities of class I and class II HDACs; however, BITC and PEITC, but not SFN, had moderate inhibitory effects on the activities of some class III HDACs, such as Sirt1 and Sirt2.5 The possibility that these inhibitory effects on class III HDACs are a result of direct modifications on cysteines at the catalytic sites of these two class III HDACs (class I and II HDACs have no cysteines at their active sites) requires further investigation.
More than 30 proteins from A549 cells have been identified thus far, by fingerprinting peptides by mass spectrometry, as potential targets of PEITC and SFN.6 Tubulin was chosen for this initial study for several reasons. It is an abundant protein; it plays a pivotal role in cell division; it offers a useful model for studying the biological consequences of protein modifications by ITCs; and it might offer insight into the mechanistic basis for ITC-induced apoptosis. The radioactivity in the protein spots detected in the two-dimensional gel electrophoresis study was insufficient, by itself, for proving that tubulin-specific ITC adducts had been formed because other proteins in the tubulin-containing spots could also contain cysteines that can react with the ITC. However, tubulin is far more abundant than the proteins sharing the tubulin-containing spots. We also made an interesting, unexpected, and novel observation that substantial amounts of tubulin precipitated under our conditions (Fig. 6) and were degraded in a variety of cell lines treated with ITCs.7 The details of these results, to be reported separately, provide additional evidence supporting our conclusion that tubulin conformation changes as a result of ITC binding. Finally, we identified tubulin-BITC adducts by using mass spectrometry analysis of the precipitated, purified tubulin. Our biophysical evidence combined with the evidence of rapid microtubule disruption (Fig. 4B) suggests that this binding is likely to be relevant to the observed tubulin-related biological effects.
Our analysis of the reaction products revealed the chemical nature of the tubulin-ITC interactions. There are 20 cysteine residues in tubulin (12 in α-tubulin and 8 in β-tubulin); almost all are highly conserved (28). Most of the cysteines are buried within the native molecule and have small cavities surrounding them, making them accessible only to small molecules (29). Modifications of tubulin cysteines often result in loss of the ability to polymerize (30). The results of the Ellman assay suggest that only six cysteines, possibly Cys316–Cys376 in α-tubulin and Cys129–Cys131 and Cys241–Cys256 in β-tubulin (29), are likely to exist as disulfides, and that 12 of the remaining 14 cysteines could be modified by BITC, 9 by PEITC, and 4 by SFN (Table 2) in vitro. These results indicate that most cysteines in tubulin are accessible to ITCs and that BITC and PEITC are better able to gain access to these cysteines than SFN. Therefore, the relative potencies in all the phenotypes observed, including collapse of the tubulin network, cell growth arrest, and apoptosis can be readily explained by a mechanism in which greater conformation changes occur in tubulin as a direct result of greater numbers of the free cysteines modified by ITCs, and that the largest conformation changes induce the most rapid and severe biological effects. The results from the Ellman assay also indicate that the number of modified thiols depends on the relative concentrations of ITCs and tubulin. Furthermore, mass spectrometric data confirmed that Cys303 and Cys347 are covalently modified in vitro and in vivo, respectively (Fig. 5). The differences between in vitro and in vivo mass spectrometry data may reflect different relative concentrations of ITCs and tubulin, or the different conformations of purified porcine tubulin and intracellular human tubulin. Conformational differences may affect the accessibility of individual cysteine residues to ITCs in porcine and human tubulins. Additionally, the thiocarbamates formed between ITCs and cysteines may not be stable when exposed to the analytical conditions such as the pH required during mass spectrometric sample preparation (5). The adduct formation may be reversed during this process, making it difficult to compare ITC-modified cysteines identified in vitro and in vivo. Nonetheless, efforts are currently being made to systematically investigate ITC-modified amino acids in vivo.
The differential affinities indicated by the Ellman assay may be explained by structure fit of ITCs and tubulin. First, the hydrophobicity of the side group of ITCs appears to be important in binding site accessibility and selectivity. Our structural analysis of all the cysteine binding pockets revealed that most cysteines in tubulin are buried in pockets that are surrounded by hydrophobic groups.8 This explains why the aromatic ring-containing BITC and PEITC would have better access to cysteines than SFN. Second, the size and shape of ITCs may influence binding affinity. BITC has a shorter “arm” than PEITC (methylene versus ethylene); therefore, it may be more accessible to smaller pockets than PEITC. It was also observed in our earlier studies that the alkyl chain length in aryl-ITCs can influence the inhibitory activity of ITCs toward specific human cytochrome P-450 isozymes for metabolic activation NNK (4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone) and lung carcinogenesis (31). Modification by ITCs at any single site could lead to slight local structural change, helix unwinding and entropy change. It is plausible that the global structure and energy changes observed in tubulin are the collective result of ITCs binding to multiple sites. The forced structural “distortions” caused by the stiffer side group of bound BITC may exert a greater steric effect than the PEITC side group.
Like other anti-tubulin compounds, ITCs arrest cells at mitosis by inhibiting tubulin polymerization. ITCs are less potent disrupters of microtubule structure than clinical therapeutic drugs such as vinblastine and colchicine; however, this less potent effect may be desirable for agents designed for use in prevention or treatment of the earlier stages of cancer. Because tubulin is an essential protein for cell division, it is reasonable to speculate that ITCs may induce more cell growth arrest and apoptosis in fast-growing, initiated, pre-neoplastic, or cancer cells than in normal cells, as has been reported (32, 33). However, additional studies are needed. In addition to the phospho-H3 marker used in this study, other surrogate markers for mitotic arrest have been used in cells treated with ITCs. For example, cyclin D1 levels were markedly elevated in several bladder cancer cell lines (34), and increased phosphorylated Bcl-2 was observed in cells treated with BITC and PEITC (BITC was a stronger inducer than PEITC) (2, 35). In human prostate cancer cells, both cdk1 and cdc25c were greatly reduced by PEITC treatment, leading to increased phospho-Cdk1 (Tyr15) (36). In addition to mitotic arrest, arrest at the G1 phase has been observed several times, in human colon carcinoma HT-29 under serum-stimulated conditions (37), in human prostate cancer cells LNCaP treated with low concentrations of SFN (38), and in human leukemia HL60/S treated with allyl-ITC (18). In this study, we found that SFN induces G1 arrest in A549 cells. Contrary to the increased cyclin D1 expression associated with mitotic arrest, the G1 phase arrest induced by SFN was associated with decreased cyclin D1 expression (37, 38), indicating that different cell cycle regulation pathways may operate at a lower dose of SFN.
Another signature event of anti-tubulin compounds is the induction of apoptosis following mitotic arrest (39, 40). Although the exact molecular basis of apoptosis induced by anti-tubulin agents is still unclear, mechanisms involving mitochondrial tubulin and Bcl-2 family members (as the direct ITC targets) have been suggested (28, 41). Similarly, effects on the mitochondria-related apoptosis pathway have also been demonstrated in cells treated with BITC (18, 19, 42), PEITC (18, 43, 44), and SFN (18, 19, 45, 46); however, in these studies the molecular targets were not identified. Although several mechanisms have been proposed (47), few studies have focused on the role of ITCs binding to target proteins on cell cycle arrest and apoptosis. This study is the first to provide evidence supporting a model in which the covalent binding of ITC to a specific protein (tubulin) could, by itself, be sufficient to cause cell growth inhibition.
Supplementary Material
Acknowledgments
We thank Dr. Karen Creswell for flow cytometry and Susette Mueller for microscopy imaging (both are heads of the respective core facilities of Lombardi Comprehensive Cancer Center at Georgetown University). We also thank Dr. Steven J. Metallo and Ariele G. Viacava Follis of the Department of Chemistry at Georgetown University for circular dichroism and fluorometry. We are grateful to Dr. Stephen Hecht of University of Minnesota and Dr. Shantu Amin of Pennsylvania State University for providing SFN and 14C-SFN, respectively. We thank Dr. Jan Wolff at NIDDK, National Institutes of Health, for fruitful discussions and suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grant CA100853 from NCI. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. A–C.
Footnotes
The abbreviations used are: ITC, isothiocyanate; BITC, benzyl-ITC; PEITC, phenethyl-ITC; SFN, sulforaphane; MALDI-TOF/TOF, matrix-assisted laser desorption/ionization-time-of-flight/time-of-flight; NMPEA, N-methyl phenethylamine; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt; BSA, bovine serum albumin; PBS, phosphate-buffered saline; PI, propidium iodide; RPLC, reversed-phase liquid chromatography; MS/MS, tandem mass spectrometry; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; MOPS, 4-morpholinepropanesulfonic acid; PIPES, 1,4-piperazinediethanesulfonic acid; HDAC, histone deacetylase.
L. Liebes, S. S. Hecht, and F.-L. Chung, unpublished results.
L. Mi and F.-L. Chung, unpublished data.
L. Mi, B. L. Hood, X. Wang, S. Govind, T. D. Veenstra, T. P. Conrads, and F.-L. Chung, unpublished data.
L. Mi, N. Gan, X. Wang, S. Govind, and F.-L. Chung, unpublished data.
S. Dakshanamurthy, L. Mi, and F.-L. Chung, unpublished data.
References
- 1.WHO (2004) IARC Handbook on Cancer Prevention, Vol. 9, pp. 43-171, Lyon, France [Google Scholar]
- 2.Tang, L., and Zhang, Y. (2005) Mol. Cancer Ther. 4 1250-1259 [DOI] [PubMed] [Google Scholar]
- 3.Singh, S. V., Herman-Antosiewicz, A., Singh, A. V., Lew, K. L., Srivastava, S. K., Kamath, R., Brown, K. D., Zhang, L., and Baskaran, R. (2004) J. Biol. Chem. 279 25813-25822 [DOI] [PubMed] [Google Scholar]
- 4.Zhang, Y., and Talalay, P. (1998) Cancer Res. 58 4632-4639 [PubMed] [Google Scholar]
- 5.Jiao, D., Conaway, C. C., Wang, M. H., Yang, C. S., Koehl, W., and Chung, F. L. (1996) Chem. Res. Toxicol. 9 932-938 [DOI] [PubMed] [Google Scholar]
- 6.Mi, L., Wang, X., Govind, G., Hood, B. L., Veenstra, T. D., Conrads, T. P., Saha, D. T., Goldman, R., and Chung, F. L. (2007) Cancer Res. 67 6409-6416 [DOI] [PubMed] [Google Scholar]
- 7.Nogales, E. (2000) Annu. Rev. Biochem. 69 277-302 [DOI] [PubMed] [Google Scholar]
- 8.Jordan, M. A., and Wilson, L. (2004) Nat. Rev. Cancer 4 253-265 [DOI] [PubMed] [Google Scholar]
- 9.Widrow, R. J., and Laird, C. D. (2000) Cytometry 39 126-130 [DOI] [PubMed] [Google Scholar]
- 10.Simpson, R. J. (2003) Proteins and Proteomics, pp. 143-218, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 11.Candiano, G., Bruschi, M., Musante, L., Santucci, L., Ghiggeri, G. M., Carnemolla, B., Orecchia, P., Zardi, L., and Righetti, P. G. (2004) Electrophoresis 25 1327-1333 [DOI] [PubMed] [Google Scholar]
- 12.Mi, L., Fischer, S., Chung, B., Sundelacruz, S., and Harden, J. L. (2006) Biomacromolecules 7 38-47 [DOI] [PubMed] [Google Scholar]
- 13.Conrads, T. P., Tocci, G. M., Hood, B. L., Zhang, C. O., Guo, L., Koch, K. R., Michejda, C. J., Veenstra, T. D., and Keay, S. K. (2006) J. Biol. Chem. 49 37836-37843 [DOI] [PubMed] [Google Scholar]
- 14.Suh, S. K., Hood, B. L., Kim, B. J., Conrads, T. P., Veenstra, T. D., and Song, B. J. (2004) Proteomic 4 3401-3412 [DOI] [PubMed] [Google Scholar]
- 15.Ellman, G. L. (1958) Arch. Biochem. Biophys. 74 443-450 [DOI] [PubMed] [Google Scholar]
- 16.Chen, Y. H., Yang, J. T., and Chau, K. H. (1974) Biochemistry 13 3350-3359 [DOI] [PubMed] [Google Scholar]
- 17.Hosono, T., Fukao, T., Ogihara, J., Ito, Y., Shiba, H., Seki, T., and Ariga, T. (2005) J. Biol. Chem. 280 41487-41493 [DOI] [PubMed] [Google Scholar]
- 18.Zhang, Y., Tang, L., and Gonzalez, V. (2003) Mol. Cancer Ther. 2 1045-1052 [PubMed] [Google Scholar]
- 19.Jakubikova, J., Bao, Y., and Sedlak, J. (2005) Anticancer Res. 25 3375-3386 [PubMed] [Google Scholar]
- 20.Tseng, E., Scott-Ramsay, E. A., and Morris, M. E. (2004) Exp. Biol. Med. 229 835-842 [DOI] [PubMed] [Google Scholar]
- 21.Bonnesen, C., Eggleston, I. M., and Hayes, J. D. (2001) Cancer Res. 61 6120-6130 [PubMed] [Google Scholar]
- 22.Jackson, S. J., and Singletary, K. W. (2004) Carcinogenesis 25 219-227 [DOI] [PubMed] [Google Scholar]
- 23.Conaway, C. C., Jiao, D., and Chung, F. L. (1996) Carcinogenesis 17 2423-2427 [DOI] [PubMed] [Google Scholar]
- 24.Moreno, R. L., Goosen, T., Kent, U. M., Chung, F. L., and Hollenberg, P. F. (2001) Arch. Biochem. Biophys. 391 99-110 [DOI] [PubMed] [Google Scholar]
- 25.von Weymarn, L. B., Chun, J. A., and Hollenberg, P. F. (2006) Carcinogenesis 27 782-790 [DOI] [PubMed] [Google Scholar]
- 26.von Weymarn, L. B., Chun, J. A., Knudsen, G. A., and Hollenberg, P. F. (2007) Chem. Res. Toxicol. 20 1252-1259 [DOI] [PubMed] [Google Scholar]
- 27.Myzak, M. C., Karplus, P. A., Chung, F. L., and Dashwood, R. H. (2004) Cancer Res. 64 5767-5774 [DOI] [PubMed] [Google Scholar]
- 28.Cowan, N. (1984) in Oxford Surveys on Eukaryotic Genes (Maclean, N., ed) pp. 26-60, University of Oxford Press, Oxford, UK
- 29.Lowe, J., Li, H., Downing, K. H., and Nogales, E. (2001) J. Mol. Biol. 313 1045-1057 [DOI] [PubMed] [Google Scholar]
- 30.Britto, P. J., Knipling, L., Mcphie, P., and Wolff, J. (2005) Biochem. J. 389 549-558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morse, M. A., Eklind, K. I., Amin, S. G., Hecht, S. S., and Chung, F. L. (1989) Carcinogenesis 10 1757-1759 [DOI] [PubMed] [Google Scholar]
- 32.Yang, Y. M., Jhanwar-Uniyal, M., Schwartz, J., Conaway, C. C., Halicka, H. D., Traganos, F., and Chung, F. L. (2005) Cancer Res. 65 8538-8547 [DOI] [PubMed] [Google Scholar]
- 33.Trachootham, D., Zhou, Y., Zhang, H., Demizu, Y., Chen, Z., Pelicano, H., Chiao, P. J., Achanta, G., Arlinghaus, R. B., Liu, J., and Huang, P. (2006) Cancer Cell 10 241-252 [DOI] [PubMed] [Google Scholar]
- 34.Yao, S., Zhang, Y., and Li, J. (2006) Mol. Carcinog. 45 605-612 [DOI] [PubMed] [Google Scholar]
- 35.Ling, Y. H., Tornos, C., and Perez-Soler, R. (1998) J. Biol. Chem. 273 18984-18991 [DOI] [PubMed] [Google Scholar]
- 36.Xiao, D., Johnson, C. S., Trump, D. L., and Singh, S. V. (2004) Mol. Cancer Ther. 3 567-575 [PubMed] [Google Scholar]
- 37.Shen, G., Xu, C., Chen, C., Hebbar, V., and Kong, A. N. (2006) Cancer Chemother. Pharmacol. 57 317-327 [DOI] [PubMed] [Google Scholar]
- 38.Chiao, J. W., Chung, F. L., Kancherla, R., Ahmed, T., Mittelman, A., and Conaway, C. C. (2002) Int. J. Oncol. 20 631-636 [DOI] [PubMed] [Google Scholar]
- 39.Jordan, M. A., Wendell, K., Gardiner, S., Derry, W. B., Copp, H., and Wilson, L. (1996) Cancer Res. 56 816-825 [PubMed] [Google Scholar]
- 40.Ling, Y. H., Jiang, J. D., Holland, J. F., and Perez-Soler, R. (2002) Mol. Pharmacol. 62 529-538 [DOI] [PubMed] [Google Scholar]
- 41.Carre, M., Andre, N., Carles, G., Borghi, H., Brichese, L., Briand, C., and Braguer, D. (2002) J. Biol. Chem. 277 33664-33669 [DOI] [PubMed] [Google Scholar]
- 42.Nakamura, Y., Kawakami, M., Yoshihiro, A., Miyoshi, N., Ohigashi, H., Kawai, K., Osawa, T., and Uchida, K. (2002) J. Biol. Chem. 277 8492-8499 [DOI] [PubMed] [Google Scholar]
- 43.Rose, P., Armstrong, J. S., Chua, Y. L., Ong, C. N., and Whiteman, M. (2005) Int. J. Biochem. Cell Biol. 37 100-119 [DOI] [PubMed] [Google Scholar]
- 44.Xiao, D., Zeng, Y., Choi, S., Lew, K. L., Nelson, J. B., and Singh, S. V. (2005) Clin. Cancer Res. 11 2670-2679 [DOI] [PubMed] [Google Scholar]
- 45.Gamet-Payrastre, L., Li, P., Lumeau, S., Cassar, G., Dupont, M. A., Chevolleau, S., Gasc, N., Tulliez, J., and Terce, F. (2000) Cancer Res. 60 1426-1433 [PubMed] [Google Scholar]
- 46.Choi, S., and Singh, S. V. (2005) Cancer Res. 65 2035-2043 [DOI] [PubMed] [Google Scholar]
- 47.Zhang, Y., Yao, S., and Li, J. (2006) Proc. Nutr. Soc. 65 68-75 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.