Abstract
We previously generated an adenoassociated viral gene therapy vector, rAAV-Δ264 cystic fibrosis transmembrane conductance regulator (CFTR), missing the first four transmembrane domains of CFTR. When infected into monkey lungs, Δ264 CFTR increased the levels of endogenous wild type CFTR protein. To understand this process, we transfected Δ264 CFTR plasmid cDNA into COS7 cells, and we noted that protein expression from the truncation mutant is barely detectable when compared with wild type or ΔF508 CFTR. Δ264 CFTR protein expression increases dramatically when cells are treated with proteasome inhibitors. Cycloheximide experiments show that Δ264 CFTR is degraded faster than ΔF508 CFTR. VCP and HDAC6, two proteins involved in retrograde translocation from endoplasmic reticulum to cytosol for proteasomal and aggresomal degradation, coimmunoprecipitate with Δ264 CFTR. In cotransfection studies in COS7 cells and in transfection of Δ264 CFTR into cells stably expressing wild type and ΔF508 CFTR, Δ264 CFTR increases wild type CFTR protein and increases levels of maturation of immature band B to mature band C of ΔF508 CFTR. Thus the adenoassociated viral vector, rAAV-Δ264 CFTR, is a highly promising cystic fibrosis gene therapy vector because it increases the amount of mature band C protein both from wild type and ΔF508 CFTR and associates with key elements in quality control mechanism of CFTR.
The cystic fibrosis transmembrane conductance regulator (CFTR)2 is the Cl– channel defective in cystic fibrosis (CF) (1). The most common disease-causing mutation in CFTR is a missing phenylalanine at position 508 (ΔF508 CFTR) (2). Wild type (WT) CFTR is well known to function at the plasma membrane (1), whereas ΔF508 CFTR is recognized as a mutant protein by the cell, retained in the ER, incompletely glycosylated, and subsequently degraded (3). Because of this, ΔF508 CFTR cannot function at the plasma membrane. How WT and mutant CFTR are processed and trafficked to the plasma membrane has been studied extensively (4). Very early studies (3) identified three forms of CFTR, a fully glycosylated band C, an incompletely glycosylated band B, and a core-glycosylated band A. Very little core-glycosylated CFTR can be detected in the cells, but significant amounts of both B and C bands of WT CFTR can be detected especially in transfected cells (3). The presence of significant quantities of the immature B band of WT CFTR has been attributed to inefficient processing of WT CFTR to completely glycosylated forms (4). ΔF508 CFTR is not processed past the ER, so it is mostly detected as the immature band B. Processing of ΔF508 CFTR to mature band C can occur if cells containing this mutant are cultured at a lower temperature (5). Many avenues are being pursued as potential therapies for CF to correct mutant trafficking of ΔF508 CFTR (6).
Our group has been interested in another approach for a CF therapy, replacing the defective gene by gene therapy using adenoassociated viral vectors (7). A major limitation to the success of AAV gene therapy for CF is that the large size of the CFTR cDNA insert fills the packaging capacity of AAV viral particles precluding inclusion of a highly efficient promoter. To overcome this limitation, AAV2/5 pseudotyped vectors were designed to express a truncated version of CFTR (Δ264) missing the first 264 amino acids of CFTR, driven by a chicken β-actin promoter rAAV-CB-Δ264 CFTR (pTR2-CB-Δ264CFTR) (8). Our group showed previously that this truncated form was capable of generating CFTR channels when expressed in Xenopus oocytes with characteristics associated with WT CFTR (9). We also showed that Δ264 CFTR suppressed the inflammatory pathology induced by Pseudomonas aeruginosa beads or Aspergillis fumigatus in CFTR knock-out mouse models (8, 10, 11). We delivered this vector to the airways of Rhesus macaques, and to our surprise, rather than detecting robust expression of Δ264 CFTR, we observed an increase in the expression of band C protein originating from the endogenous wild type CFTR of the monkey (7). Fragments of WT CFTR are known to cause increased processing of ΔF508 CFTR band B to C (12) through a process called transcomplementation. However, the mechanism of how this occurs is poorly understood.
This study aims to understand how the quality control mechanism of the cell processes Δ264 CFTR and to show that Δ264 CFTR does indeed function to efficiently increase the amounts of both WT and ΔF508 CFTR protein detected in Western blot experiments. The dual purpose of this study is to gain insights into mechanisms that result in ΔF508 CFTR processing from immature to mature forms and to provide new strategies for gene therapy vectors for CF.
EXPERIMENTAL PROCEDURES
Cell Culture—African green monkey kidney cells (COS7) obtained from American Type Tissue Culture (ATCC) were maintained in 1× Dulbecco's modified Eagle's medium high glucose, penicillin (100 units/ml), streptomycin (100 μg/ml), and 10% fetal bovine serum. Media and other components were purchased from Invitrogen. The CFBE 41o– cell line was derived by Dieter Gruenert from a patient with CF and transfected with additional quantities of WT CFTR and selected for stable expression of WT CFTR (13). These cells were provided to us by Dieter Gruenert. The CFBE41o-ΔF508 CFTR cells originated from the parental CFBE 41o– cell line derived by Dieter Gruenert but were subsequently stably transduced with a lentivirus containing ΔF508 CFTR (14). These cells were provided to us by J. P. Clancy.
Plasmids and Constructs—The construct pEGFP WT CFTR mammalian expression vector was from Bruce A. Stanton (15). The ΔF508 CFTR mutation was generated by site-directed mutagenesis in pEGFP-WT CFTR by PCR using ΔF508 primers. WT CFTR, ΔF508 CFTR, and Δ264 CFTR were subcloned into pCDNA3.1 with CBA and cytomegalovirus promoters (Invitrogen). The plasmids were transfected into the cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After 48 h of transfection, the cells were harvested and used for immunoprecipitation and immunoblotting.
Treatment—After transfection, cells were treated either with proteasome inhibitors, MG132 (Calbiochem), PS-341/Bortezomib (Millennium Pharmaceuticals), and lactacystin (Calbiochem) or the lysosomal inhibitor E64 (Calbiochem) for 16 h unless indicated otherwise. Cycloheximide (Sigma) was used at 25 μg/ml for various times as indicated.
Immunoprecipitation—Cells were lysed directly on plates using M-PER protein lysis buffer (Pierce) containing protease inhibitor mixture (Pierce) after washing with ice-cold phosphate-buffered saline. Total protein extracts (500 μg/ml) were incubated with 50 μl of protein A/G-agarose beads (Santa Cruz Biotechnology, Inc.) for 3 h at 4 °C. After preclearing, 2.5 μg of both CFTR 169 and M3A7 (R & D Systems) antibodies were added to each tube. After 1 h, protein A/G-agarose beads (50 μl) were added to each tube, and tubes were incubated overnight at 4 °C. Beads were washed once with lysis buffer, followed with two washes with phosphate-buffered saline. The beads were suspended in Laemmli sample buffer (30 μl) containing β-mercaptoethanol, vortexed for 1 min, resolved by 4–10% SDS-PAGE, and detected using the respective primary antibodies as described below.
Immunoblotting—Cells were harvested and processed as described previously (16). Briefly, cells were solubilized in lysis buffer (50 mm NaCl, 150 mm Tris-HCl, pH 7.4, 1% Nonidet P-40) and in complete protease inhibitors (Roche Applied Science). The cell lysates were spun at 14,000 × g for 15 min at 4 °C to pellet-insoluble material. The supernatants were subjected to SDS-PAGE and Western blotting followed by enhanced chemiluminescence (Amersham Biosciences). The chemiluminescence signal on the polyvinylidene difluoride membrane was directly captured by FujiFilm LAS-1000 plus system with a cooled CCD camera. Quantification was carried out within the linear range using the ImageGauge version 3.2 software (FujiFilm). CFTR was detected with monoclonal anti-human CFTR (C terminus) antibody (1:1500; R & D Systems, Inc.). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), used as a loading control, was detected with monoclonal GAPDH antibody (1:10,000; US Biological). Rabbit polyclonal VCP and HDAC6 antibodies were purchased from Santa Cruz Biotechnology, Inc., and used at final concentration of 2 μg/ml.
RESULTS
Processing of Δ264 CFTR—When Δ264 CFTR cDNA was transfected into COS7 cells, Δ264 CFTR protein expression is barely detectable (Fig. 1). This could be the consequence of an inefficient promoter driving only small amounts of gene transcription or of enhanced protein degradation resulting in reduced steady state amounts of detectable protein. The former is unlikely because we utilized a powerful CBA promoter with a cytomegalovirus enhancer known to express mRNA in high levels in these cells (8). To evaluate this, we transfected into COS7 cells the same amount of CBA-WT CFTR, CBA-ΔF508 CFTR, or CBA-Δ264 CFTR cDNA. In Western blot experiments, WT and ΔF508 CFTR proteins are detectable at very high levels, whereas Δ264 CFTR is barely detectable. The results verified that CBA is a powerful promoter and is not the cause of low amounts of detectable Δ264 CFTR protein in transfection experiments.
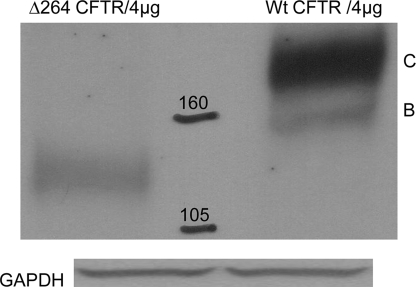
FIGURE 1.
Δ264 CFTR (left) runs at a molecular weight lower than that of wild type CFTR (right). COS7 cells were transfected with 4 μg of pTR2-CB-Δ264CFTR and 4 μg of pCDNA3.1-WT CFTR. After 48 h, cells were lysed, and the total lysate was analyzed by Western blot using anti-human CFTR antibodies (C terminus-specific) fromR&D Systems. Loading was evaluated with GAPDH antibodies (US Biological) in all experiments even if data are not shown. (n = 8).
To evaluate the degradation of Δ264 CFTR, we treated cells with the inhibitor, MG132, a nonspecific inhibitor of proteasomal degradation. Fig. 2 shows that in the absence of MG132 (lane 5), Δ264 CFTR is barely detectable under these exposure conditions. In sharp contrast, in the presence of MG132, large amounts of Δ264 CFTR can be detected. We have obtained the same results with the proteasomal inhibitors, lactacystin (data not shown) and PS341 (discussed later), confirming that the data were indeed the result of proteasome inhibition.
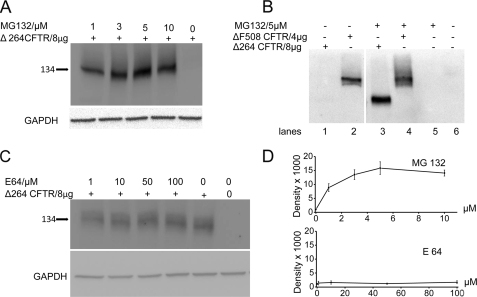
FIGURE 2.
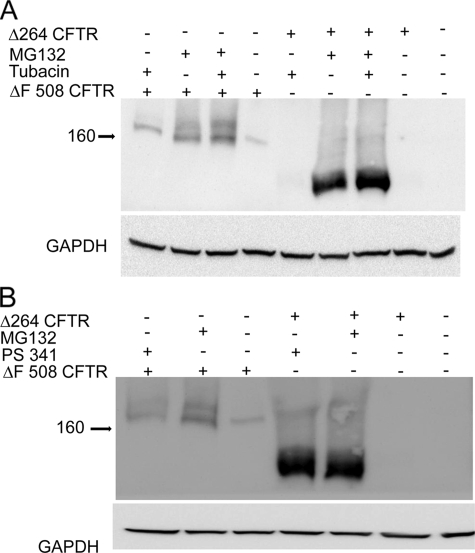
A, proteasome inhibition is shown. Cells were transfected with Δ264 cDNA and treated with MG132. Δ264 CFTR protein expression is not detectable in the absence but is increased dramatically in the presence of proteasome inhibitors (upper panel)(n = 4). B, cells were transfected with Δ264 or ΔF508 CFTR and treated with MG132. Δ264 CFTR protein expression is not detectable in the absence of MG132 (lane 1) but is increased dramatically by proteasome inhibition (lane 3). Note that ΔF508-CFTR is detectable in the absence of MG132 (lane 2) and increases modestly in the presence of MG132 (lane 4); lanes 5 and 6 are negative controls (n = 7). The two panels are from the same gel. C, COS7 cells were transfected with Δ264 CFTR and treated with lysosome inhibitor (E64) for 16 h. There is very little change in band density among all the treatment groups and Δ264 CFTR in the absence of the inhibitor. D, summary data for proteasomal and lysosomal inhibition experiments is shown. Only MG132 significantly increases Δ264 CFTR protein expression.
To study this further, we compared Δ264 to ΔF508 CFTR protein levels in the presence of proteasome inhibitors. As shown in Fig. 2A, in the absence of MG132, again Δ264 CFTR protein is barely detectable. In contrast, even though only half the amount of ΔF508 cDNA was transfected compared with that of Δ264 CFTR, ΔF508 CFTR detectable protein is much higher than the protein detected from Δ264 CFTR. Comparing the sensitivities of Δ264 and ΔF508 CFTR to MG132, Fig. 2B shows clearly that ΔF508 CFTR is not as sensitive to MG132 (compare lanes 4 with 2) as Δ264 CFTR (compare lanes 3 with 1). To evaluate how much is degraded by the lysosome, we utilized the inhibitor E64. As shown in Fig. 2C, the amounts of Δ264 CFTR protein detected is not significantly affected by lysosomal inhibition. Summary data are shown in Fig. 2D.
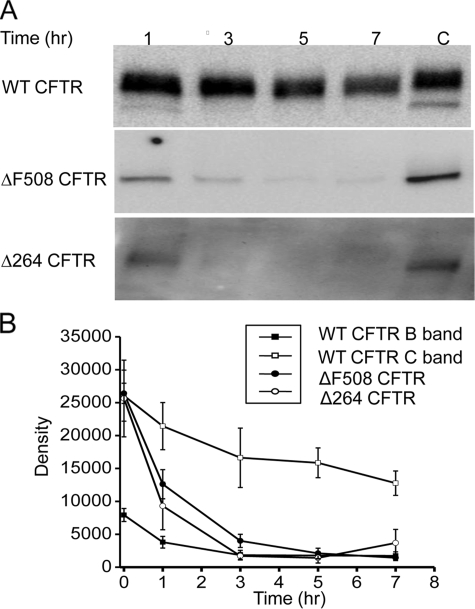
To evaluate how fast Δ264 protein is degraded compared with ΔF508 and WT CFTR, we treated transfected cells with cycloheximide. As shown in Fig. 3A, WT CFTR protein is relatively stable following cycloheximide treatment. This is in sharp contrast to ΔF508 and Δ264 CFTR whose proteins are degraded much faster. Among the three forms shown in Fig. 3, A and B, Δ264 CFTR protein is degraded the fastest.
FIGURE 3.
A, degradation assayed by inhibition of protein synthesis. COS7 cells were transfected with either WT, ΔF508, or Δ264 CFTR and treated with cycloheximide (25 μg/ml) for the indicated times (n = 4 for WT CFTR and ΔF508 CFTR, n = 3 for Δ264 CFTR). In this experiment, exposure times for the Fuji density measurements were 3 s for WT CFTR, 40 s for ΔF508 CFTR, and 5 min for Δ264 CFTR. This gave approximately equal density values at time 0 for the graph in B. Please note that the exposure times were varied in this experiment so that we could accurately measure the change in density following exposure to cycloheximide treatment for the various times especially for Δ264 CFTR. C indicates control. B, summary data of cycloheximide experiments. The differences in the rate of decay between Δ264 CFTR and ΔF508 CFTR are significantly different using the mean at time point 0–5, p < 0.05.
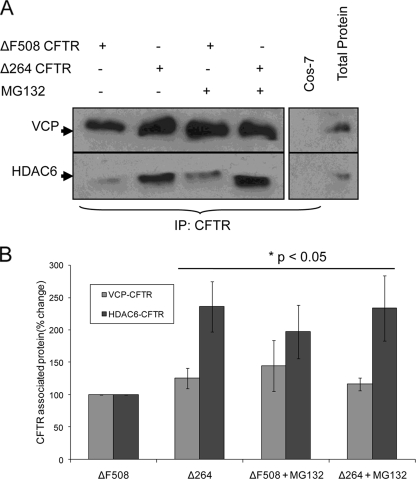
Because Δ264 is more efficiently degraded compared with ΔF508 CFTR, we hypothesized that it would interact with key components involved in CFTR degradation. CFTR can be degraded either in the proteasome (17) or sequestered and perhaps degraded in aggresomes (18). To evaluate these two routes, we tested for possible interactions of Δ264 CFTR with VCP, a member of the protein complex involved in extraction of CFTR from the ER membrane and retrograde translocation of CFTR to the proteasome (19), and with HDAC6, the ubiquitin interacting deacetylase that promotes the movement of CFTR to aggresomes (20). As expected, both Δ264 and ΔF508 CFTR do indeed associate both with VCP and HDAC6 (Fig. 4). We also observed that even though steady state levels of Δ264 CFTR protein are hardly detectable by immunoblotting and immunoprecipitation with anti-CFTR antibody (supplemental Fig. 1), we were still able to pull down both VCP and HDAC6 by immunoprecipitation. Pull down of both VCP and HDAC6 by Δ264 CFTR indicates that Δ264 CFTR uses a similar degradation pathway as reported earlier for ΔF508 and WT CFTR (20). To evaluate this further, we utilized the HDAC6 inhibitor, tubacin (21). Tubacin is a recently identified and highly specific small molecule inhibitor of HDAC6-mediated aggresome formation (21). We compared the effects of tubacin on both Δ264 and ΔF508 CFTR protein levels to those obtained with the more specific proteasome inhibitor, PS-341/Bortezomib (Fig. 5). As expected, the steady state amounts of Δ264 CFTR protein detected by Western blotting are again dramatically increased (∼60-fold) either by MG132 or PS-341, showing again that the amount of Δ264 CFTR protein is exquisitely sensitive to proteasome inhibitors. Tubacin, on the other hand, has a very small effect on the amount of Δ264 CFTR protein (∼40-fold less than PS-341). The small effect of tubacin on the amounts of Δ264 CFTR protein demonstrates that the majority of this truncated CFTR protein is processed through proteasomal degradation.
FIGURE 4.
Both ΔF508 CFTR and Δ264 CFTR are associated with VCP and HDAC6 protein complex. Cos-7 cells were transfected with ΔF508 or Δ264 CFTR and treated with 10 μm MG-132 for 8 h. A, CFTR was immunoprecipitated (IP) from 500 μg of total protein and immunoblotted for VCP or HDAC6 (left panel). Protein from non-CFTR transfected Cos-7 cells was used as a negative control for CFTR immunoprecipitation, whereas total protein extracts (25 μg) were loaded as positive control (right panel). Higher amounts of both VCP and HDAC6 are coimmunoprecipitated with Δ264, as compared with ΔF508 CFTR. B, VCP or HDAC6 immunoprecipitated with ΔF508 or Δ264 CFTR for each group is shown as percentage change (mean ± S.D.) from ΔF508 protein complex. Statistics were also compared with the ΔF508 protein complex.
FIGURE 5.
Δ264 CFTR is degraded primarily by the proteasome. Cells were transfected either with Δ264 CFTR or ΔF508 CFTR and were treated either with MG132, tubacin alone, or MG132 plus tubacin at 10 μm for 16 h (A). B, cells transfected either with Δ264 CFTR or ΔF508 CFTR were treated either with MG132 or PS341. Δ264 CFTR or ΔF508 CFTR control bands (absence of inhibitors) were quantified and normalized to control = 1. The following fold increases were obtained for Δ264 CFTR with inhibitors: MG132 = 54 ± 22 (p < 0.001); PS341 = 62 ± 21 (p < 0.001); tubacin alone = 1.5 ± 0.08 (p < 0.001); tubacin plus MG132 = 59 ± 8 (p < 0.001). The following fold increases were obtained for ΔF508 CFTR. Band B: MG132 = 4 ± 0.04 (p < 0.001); PS341 = 2.7 ± 0.05 (p < 0.001); tubacin alone = 1.8 ± 0.3 (p < 0.005); tubacin plus MG132 = 5.4 ± 0.2 (p < 0.001). The following fold increases were obtained for ΔF508 CFTR. Band C: MG132 = 7 ± 2 (p < 0.001); PS341 = 2 ± 0.5 (p < 0.001); tubacin alone = 2 ± 0.5 (p < 0.01); tubacin plus MG132 = 11 ± 3 (p < 0.001). All fold increases were log-transformed and tested for significance and found to be significantly different from control. Note that the fold changes following inhibition of the proteasome either with MG132 or PS341 are much greater for Δ264 CFTR than for ΔF508 CFTR. Also note that there is a highly significant increase in band C of ΔF508 CFTR compared with control when cells are treated with MG132, PS341, or tubacin plus MG132. n = 3 for all experiments.
Neither proteasome nor HDAC6 inhibition has large effects on the B band of ΔF508 CFTR suggesting that at the steady state a significant pool of the immature band of ΔF508 CFTR protein resides in the ER. In contrast, the most dramatic effect is on band C. When MG132 is applied alone, there is an ∼7-fold increase, and when both MG132 and tubacin are applied together, there is an 11-fold increase in the steady state levels of band C of ΔF508 CFTR detected by Western blot. This indicates that if both proteasomal degradation and aggresome formation pathways are inhibited, we rescue the ΔF508 CFTR targeted for degradation and at the same time favor the processing of ΔF508 to mature band C.
Taken together, our data suggest that Δ264 CFTR is efficiently degraded by the proteasome as compared with ΔF508 CFTR and lacks the biosynthetic arrest in the ER. The mechanism of the efficient degradation of Δ264 CFTR involves interactions with key elements of the quality control mechanism of the cell such as VCP and HDAC6.
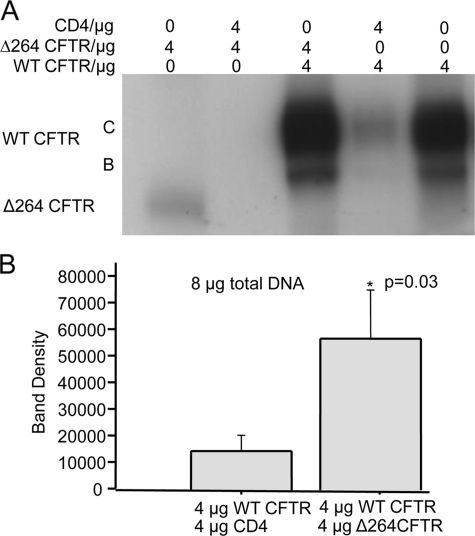
Δ264 CFTR Increases Expression of WT CFTR—In our previous experiments, when we infected the lungs of Rhesus monkeys with AAV5 Δ264 CFTR virus, we expected to detect robust expression of Δ264 CFTR protein, instead, to our surprise we saw increased amounts of the monkey's endogenous WT CFTR protein (7). We tested this further in transfection and cotransfection experiments of WT and Δ264 CFTR cDNA into COS7 cells. When Δ264 CFTR cDNA is transfected alone, Δ264 CFTR protein is again barely detectable (Fig. 6A, 1st lane) compared with the transfection of WT CFTR cDNA alone (2nd and 4th lanes). Note the increase in mature band C protein when WT CFTR is cotransfected with Δ264 CFTR cDNA (Fig. 6A, 3rd and 5th lanes). One possibility is that the increase in detectable WT CFTR protein is caused simply by transfecting two genes into the cells. To eliminate this possibility, we cotransfected WT CFTR and Δ264 CFTR with CD4 cDNA, a non-CFTR membrane protein.
FIGURE 6.
A, cells were transfected or cotransfected either with WT or Δ264 CFTR or with CD4. Note that cotransfection of Δ264 CFTR or WT CFTR with CD4 reduces detectable Δ264 CFTR (compare 1st lane (Δ264 CFTR alone) with 2nd lane (Δ264 CFTR + CD4)) and WT CFTR protein (compare 5th lane (WT CFTR alone) with 4th lane WT CFTR + CD4). This is in sharp contrast to the result with Δ264 CFTR; when cotransfected with WT CFTR, Δ264 CFTR increases detectable CFTR protein (compare 5th with 3rd lane). GAPDH experiments showed equal loading (data not shown), n = 5. B, summary data is shown; only C band density was measured. Data are significantly different.
Fig. 6A shows that CD4 cDNA cotransfection reduces both Δ264 CFTR (compare lane 1 to 2) and WT CFTR (lane 5) protein, and cotransfection with Δ264 CFTR cDNA increases the amount of WT CFTR protein detected. Fig. 6B shows the summary data where the total amount of cDNA transfected was kept constant at 8 μg. Note that when coexpressed with Δ264 CFTR, there is a significant increase in detectable C band of WT CFTR.
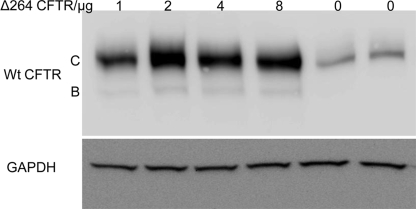
The result shows that cotransfection of Δ264 cDNA significantly increases WT CFTR protein. The observation that cotransfection with CD4 cDNA produces a reduction in detectable protein for both WT and Δ264 CFTR is strong evidence that the enhanced effect of Δ264 on WT CFTR is not caused simply by transfecting two plasmids. However, to be sure that this is not the case, we transfected CFBE41o– cells that were stably transduced previously with WT CFTR cDNA. Fig. 7 clearly shows that WT CFTR protein is increased when Δ264 CFTR cDNA is transiently transfected into the cells. Thus, the conclusion from this work is that Δ264 CFTR does indeed enhance the processing of WT CFTR.
FIGURE 7.
CFBE41o– WT CFTR cells were transfected with Δ264 CFTR. These human bronchial epithelial cells were derived from a CF patient but stably express WT CFTR (created by and a gift from Dieter Gruenert (13)). Note that transfection with Δ264 CFTR increases detectable WT CFTR protein. The increase peaks when 2 μg of Δ264 CFTR is transfected (p < 0.004 for 2 μg), n = 8.
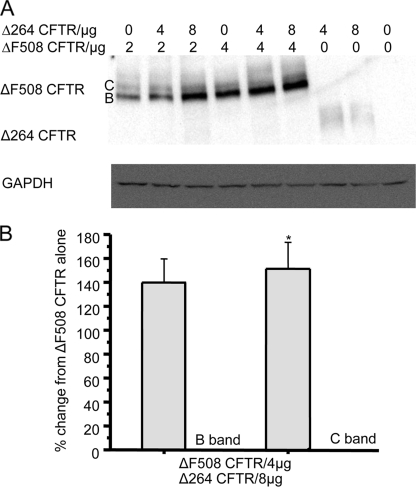
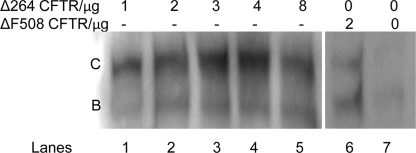
Δ264 CFTR Rescues ΔF508 CFTR—To study whether Δ264 CFTR affects the maturation of ΔF508 CFTR, additional transfection and cotransfection experiments in COS7 cells were performed (Fig. 8, A and B). Mature C band from ΔF508 CFTR was significantly increased by 55% in cells cotransfected with ΔF508 CFTR and Δ264 CFTR versus cells transfected with ΔF508 CFTR cDNA alone (Fig. 8, A and B). Note that when cotransfected with ΔF508 CFTR cDNA, Δ264 CFTR protein is not detectable (Fig. 8A).
FIGURE 8.
A, COS7 cells were transfected either with Δ264 CFTR or ΔF508 CFTR alone or with both plasmids in combination. Δ264 CFTR increases expression of mature band C and immature band B of ΔF508 CFTR. Legend at the top represents micrograms of cDNA of each construct that was transfected. B, summary data is shown. Both C and B band densities were measured. Data are significantly different only for an increase in C band. The control (no Δ264 CFTR transfected) is normalized to 100% compared with the % increase with Δ264 CFTR. *, data are significantly different for the increase in C band only.
Fig. 9 shows the results of experiments on CFBE41o-ΔF508. The CFBE41o-ΔF508 CFTR cells were previously stably transduced with a lentivirus containing ΔF508 CFTR cDNA (14). Fig. 9, lane 7, note that even though the cells were transduced prior to our experiments with a lentivirus containing ΔF508 CFTR, in the absence of Δ264 CFTR bands B and C of ΔF508 CFTRs are barely detectable in cells. When the cells were transiently transfected with an additional amount of ΔF508 CFTR cDNA (Fig. 9, lane 6), the primary effect is an increase in band B and a small increase in detectable band C protein. In contrast, when increasing amounts of Δ264 CFTR cDNA were transiently transfected into these cells (Fig. 9, lanes 1–5), the detectable band C protein originating from the stably expressed ΔF508 CFTR is increased to a much greater extent than band B.
FIGURE 9.
CFBE41o– ΔF508 CFTR cells were transfected with Δ264 or with ΔF508 CFTR. These human bronchial epithelial cells were derived from a CF patient but stably expressing ΔF508 CFTR transduced with lentivirus (gift from J. P. Clancy (14)). Note that transfection with Δ264 CFTR increases detectable band C (p < 0.02 for 8 μg of Δ264 CFTR transfected). Lane 6 shows the same cells transfected with an additional amount of ΔF508 CFTR to indicate the size of the B and C bands. Lane 7 depicts the cells that express ΔF508 CFTR as they were received from J. P. Clancy. n = 5.
To check the difference in mass between C and B bands, we used two different glycosidases (data not shown). Peptide N-glycosidase F (N-glycanase), which completely removes core and complex glycosylation from CFTR, and endoglycosidase H, which removes only unprocessed core oligosaccharides, were used (22). Endoglycosidase H was very effective in removing the B band. Peptide N-glycosidase F in our experiments reduces C band (data not shown).
DISCUSSION
Δ264 CFTR Is Functional—A full structure of CFTR has yet to be solved, but homology modeling based upon the structural information for other ABC transporters such as SAV1866 (23) suggests that cytosolic loops 1 and 2 may interact with one or two of the nucleotide binding domains of CFTR (24). Because Δ264 CFTR is missing cytosolic loops 1 and 2 as well as the first four membrane-spanning domains, the question then can be raised about the protein conformation and functionality of Δ264 CFTR.
Functional data are available from electrophysiological experiments in Xenopus oocytes expressing CFTR truncation mutants where membrane-spanning domains 1–4 were progressively removed (9). Functional Cl– channels with ion selectivity identical to wild type CFTR were generated by all of these mutants, including Δ264 CFTR. On the other hand, removing more than four membrane-spanning segments did not produce functional channels. Thus, despite missing these predicted domains and inter-domain interactions with nucleotide binding domains, these mutants are still capable of forming selective ion channels (9).
We also observed in previous studies single channel activity similar to wild type CFTR when Δ264 CFTR cDNA was transfected into IB3-1 cells (8). IB3-1 are CF bronchial epithelial cells containing two mutant alleles of CFTR, ΔF508/W1282X. There are low levels of ΔF508 CFTR protein expression but no expression of W1282X protein (25). Based upon the experiments reported here, the channel activity noted in the IB3-1 cell experiments could have come either directly from Δ264 CFTR or from ΔF508 rescued through transcomplementation of the endogenous ΔF508 CFTR. It is difficult to ascribe the channel activity measured in the IB3-1 cells transfected with Δ264 CFTR cDNA to channels expressed from Δ264 CFTR protein. However, ΔF508 CFTR is well known to have a very low single channel activity when rescued to the cell surface (26). In contrast, the single channel activity of Δ264 CFTR observed in Xenopus oocytes injected with Δ264 CFTR mRNA is closer to that of WT CFTR than to ΔF508-CFTR (9). Thus, based upon these distinct differences in open probabilities of ΔF508 CFTR versus Δ264 CFTR, we surmise that the single channel recordings in the IB3-1 cells transfected with Δ264 CFTR cDNA are indeed from Δ264 CFTR channels at the cell surface or at least a combination of channel activity both from Δ264 and ΔF508 CFTR.
In another functional assay, a model of airway inflammation in a CFTR knock-out mouse was created utilizing A. fumigatus crude protein extract (Af-cpe) to mimic allergic bronchopulmonary aspergillosis. Intratrachial infection of these mice with AAV5 Δ264 CFTR partially corrected aberrant cytokine signaling and ameliorated the allergic bronchopulmonary aspergillosis in gut-corrected CFTR knock-out mice (11). Thus, despite our observations that Δ264 CFTR is rapidly degraded, it has been shown in CF knock-out mice that Δ264 CFTR is capable of at least of partial correction of the induced inflammatory pathology. How Δ264 CFTR is functioning was not studied. But because these knock-out mice did not have ΔF508 CFTR, it is likely that the functional correction is indeed coming from Δ264 CFTR protein expression at the surface of the mice airway cells.
Δ264 and ΔF508 CFTR Degradation—Our data show that when large amounts of Δ264 CFTR cDNA are transfected into cells, the mutant protein is barely detectable. This is in contrast to ΔF508 CFTR whose immature B band is readily detectable. It is known that newly synthesized core-glycosylated ΔF508 CFTR is produced at the same rate as core-glycosylated WT CFTR band B (27). This suggests that the quality control mechanism is not able to distinguish between WT and ΔF508 CFTR before the formation of band B. The impact of the ΔF508 CFTR mutation is to limit the conversion of immature band B into mature band C, the more complex-glycosylated form of CFTR.
In contrast to ΔF508 CFTR, which in the steady state we show resides in the ER, Δ264 CFTR is recognized by the quality control mechanism and rapidly degraded. As shown by others, portions of transmembrane segments 1–4 do play a role in the stability of CFTR (28). Thus, the maturation of ΔF508 CFTR toward biosynthetic arrest in the ER is likely to involve the formation of a stable intermediate early in biosynthesis (27), a situation that does not occur for Δ264 CFTR. The relative stability of the immature band of ΔF508 CFTR compared with other more efficiently degraded forms of CFTR, such as Δ264 CFTR, makes it possible to develop CF therapeutics targeted specifically at continuing the processing of ΔF508 CFTR to the plasma membrane (4).
Our data show that Δ264 CFTR is very sensitive to proteasome inhibitors. To understand this phenomenon, we showed that Δ264 is associated with VCP and HDAC6, both known to be involved in the ER-associated degradation of CFTR (20). p97/VCP and gp78 form complexes with polyubiquinated, misfolded proteins for translocation from the ER for proteasomal degradation. Interference in the VCP-CFTR complex leads to accumulation of immature ΔF508 CFTR in the ER and partial rescue of ΔF508 CFTR to the cell surface (29). HDAC6, on the other hand, is the microtubule-associated deacetylase, which by coupling with dynein motors translocates polyubiquitinated misfolded proteins to aggresomes (30). HDAC6 is known to bind to polyubiquitinated CFTR (20). p97/VCP and HDAC6 work together in controlling polyubiquinated CFTR with p97/VCP enhancing chain turnover favoring proteasome degradation and HDAC6 inhibiting turnover to promote aggresome accumulation (20). Both p97/VCP and HDAC6 associate with Δ264 CFTR and with ΔF508 CFTR suggesting that they share similar degradation pathways. Our data also indicate that the majority of the Δ264 CFTR is likely to process primarily through the proteasomal degradation pathway because proteasomal inhibition results in the appearance of much more detectable protein, whereas inhibiting HDAC6 with tubacin has little effect.
Transcomplementation—Previous studies have reported transcomplementation of ΔF508 CFTR in cells transfected with only parts of CFTR (12). These fragments are themselves extremely efficiently degraded and thereby improve the maturation of ΔF508 CFTR from B to C bands. We show here that Δ264 CFTR increases the amount of WT CFTR protein expression and causes maturation of immature band B to mature band Cof ΔF508 CFTR. How does Δ264 CFTR allow for the processing of mutant CFTR? Our data show that one way to increase processing of bands B to C of ΔF508 CFTR is to inhibit both proteasomal degradation and HDAC6 together. Perhaps by associating with both p97/VCP and HDAC6, Δ264 CFTR affects both proteasomal degradation and aggresomal accumulation of ΔF508 CFTR allowing for the processing of bands B to C. Consistent with this is that inhibition of 97/VCP is already known to rescue ΔF508 CFTR (29). We hypothesize that by engaging the processes involved in extraction of ΔF508 CFTR from the ER membrane, necessary for proteasome degradation, Δ264 CFTR redirects a pool of ΔF508 CFTR toward maturation of the C band.
It is possible that Δ264 CFTR also engages the quality control machinery prior to the proteasome and thus induces the maturation of the ER-localized form of ΔF508 CFTR. Alterations of components of early quality control, involved in recognition of misfolded protein, are known to rescue ΔF508 CFTR. One of the first CFTR binding partners to be identified and documented to affect CFTR processing was the molecular chaperone Hsp70 (31). It was later demonstrated that Hsp70 disassociates from WT CFTR during its movement to the Golgi. In contrast, Hsp70 remains associated with ΔF508 CFTR in the ER suggesting that Hsp70 plays a role in blocking ΔF508 CFTR transport out of the ER (32). Several other early quality control proteins have more recently been added to the list. For example, the co-chaperone HspBP1 inhibits the C terminus of Hsp70-interacting protein (CHIP) to stimulate CFTR maturation (33). Thus, it is clearly possible that Δ264 CFTR engages other components of quality control involved in the recognition of misfolded proteins to allow for the continued processing of ΔF508 CFTR out of the ER.
In summary, our data suggest that transcomplementation of ΔF508 CFTR by Δ264 CFTR most likely occurs because Δ264 CFTR interacts with proteins in the ER-associated degradation pathway. Our previous data show that Δ264 CFTR can function as an ion channel at the plasma membrane in Xenopus oocytes (9) and can correct the inflammatory lung disease phenotype induced by presensitizing CF of knockout animals to A. fumigatus and then instilling Pseudomonas-laden beads (8, 11). Our new data show that the Δ264 CFTR viral vector has a dual benefit. When rAAV-Δ264 CFTR is transfected into CF cells, Δ264 CFTR itself can function as a Cl– channel (9), and at the same time it can promote the expression of Δ508 CFTR band C. This dual effect makes the rAAV-Δ264 CFTR a highly promising CF gene therapy vector.
Supplementary Material
Acknowledgments
We thank Dr. Stuart L. Schreiber (Massachusetts Institute of Technology) for providing tubacin. We also thank Jie Cheng for his discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant PO1 HL51811-06. This work was also supported by Cystic Fibrosis Foundation Grants cebota05F0 and VIJ07I0. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: CFTR, cystic fibrosis transmembrane conductance regulator; WT, wild type; CF, cystic fibrosis; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; CBA, chicken β-actin; ER, endoplasmic reticulum; AAV, adenoassociated virus.
References
- 1.Fuller, C. M., and Benos, D. J. (1992) Am. J. Physiol. 263 C267–C286 [DOI] [PubMed] [Google Scholar]
- 2.Rommens, J. M., Iannuzzi, M. C., Kerem, B.-S., Drumm, M. L., Melmer, G., Dean, M., Rozmahel, R., Cole, J. L., Kennedy, D., Hidaka, N., Zsiga, M., Buckwald, M., Riordan, J. R., Tsui, L.-C., and Collins, F. S. (1989) Science 245 1059–1065 [DOI] [PubMed] [Google Scholar]
- 3.Cheng, S. H., Gregory, R. J., Marshall, J., Paul, S., Souza, D. W., White, G. A., O'Riordan, C. R., and Smith, A. E. (1990) Cell 63 827–834 [DOI] [PubMed] [Google Scholar]
- 4.Amaral, M. D. (2004) J. Mol. Neurosci. 23 41–48 [DOI] [PubMed] [Google Scholar]
- 5.Denning, G. M., Anderson, M. P., Amara, J. F., Marshall, J., Smith, A. E., and Welsh, M. J. (1992) Nature 358 761–764 [DOI] [PubMed] [Google Scholar]
- 6.Zeitlin, P. L. (2003) Exp. Opin. Emerg. Drugs 8 523–535 [DOI] [PubMed] [Google Scholar]
- 7.Fischer, A. C., Smith, C. I., Cebotaru, L., Zhang, X., Askin, F. B., Wright, J., Guggino, S. E., Adams, R. J., Flotte, T., and Guggino, W. B. (2007) Mol. Ther. 15 756–763 [DOI] [PubMed] [Google Scholar]
- 8.Sirninger, J., Muller, C., Braag, S., Tang, Q., Yue, H., Detrisac, C., Ferkol, T., Guggino, W. B., and Flotte, T. R. (2004) Hum. Gene Ther. 15 832–841 [DOI] [PubMed] [Google Scholar]
- 9.Carroll, T. P., Morales, M. M., Fulmer, S. B., Allen, S. S., Flotte, T. R., Cutting, G. R., and Guggino, W. B. (1995) J. Biol. Chem. 270 11941–11946 [DOI] [PubMed] [Google Scholar]
- 10.Muller, C., Braag, S. A., Herlihy, J. D., Wasserfall, C. H., Chesrown, S. E., Nick, H. S., Atkinson, M. A., and Flotte, T. R. (2006) Lab. Investig. 86 130–140 [DOI] [PubMed] [Google Scholar]
- 11.Mueller, C., Torrez, D., Braag, S., Martino, A., Clarke, T., Campbell-Thompson, M., and Flotte, T. R. (2008) J. Gene Med. 10 51–60 [DOI] [PubMed] [Google Scholar]
- 12.Cormet-Boyaka, E., Jablonsky, M., Naren, A. P., Jackson, P. L., Muccio, D. D., and Kirk, K. L. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 8221–8226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gruenert, D. C., Willems, M., Cassiman, J. J., and Frizzell, R. A. (2004) J. Cyst. Fibros. 3 Suppl. 2, 191–196 [DOI] [PubMed] [Google Scholar]
- 14.Bebok, Z., Collawn, J. F., Wakefield, J., Parker, W., Li, Y., Varga, K., Sorscher, E. J., and Clancy, J. P. (2005) J. Physiol. (Lond.) 569 601–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moyer, B. D., Loffing, J., Schwiebert, E. M., Loffing-Cueni, D., Halpin, P. A., Karlson, K. H., Ismailov, I. I., Guggino, W. B., Langford, G. M., and Stanton, B. A. (1998) J. Biol. Chem. 273 21759–21768 [DOI] [PubMed] [Google Scholar]
- 16.Cheng, J., Moyer, B. D., Milewski, M., Loffing, J., Ikeda, M., Mickle, J. E., Cutting, G. R., Li, M., Stanton, B. A., and Guggino, W. B. (2002) J. Biol. Chem. 277 3520–3529 [DOI] [PubMed] [Google Scholar]
- 17.Gelman, M. S., Kannegaard, E. S., and Kopito, R. R. (2002) J. Biol. Chem. 277 11709–11714 [DOI] [PubMed] [Google Scholar]
- 18.Johnston, J. A., Ward, C. L., and Kopito, R. R. (1998) J. Cell Biol. 143 1883–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein, R. F., Niraj, A., Sanderson, T. P., Wilson, L. S., Rab, A., Kim, H., Bebok, Z., and Collawn, J. F. (2007) Am. J. Respir. Cell Mol. Biol. 36 706–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyault, C., Gilquin, B., Zhang, Y., Rybin, V., Garman, E., Meyer-Klaucke, W., Matthias, P., Muller, C. W., and Khochbin, S. (2006) EMBO J. 25 3357–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haggarty, S. J., Koeller, K. M., Wong, J. C., Grozinger, C. M., and Schreiber, S. L. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 4389–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farinha, C. M., Penque, D., Roxo-Rosa, M., Lukacs, G., Dormer, R., McPherson, M., Pereira, M., Bot, A. G., Jorna, H., Willemsen, R., Dejonge, H., Heda, G. D., Marino, C. R., Fanen, P., Hinzpeter, A., Lipecka, J., Fritsch, J., Gentzsch, M., Edelman, A., and Amaral, M. D. (2004) J. Cyst. Fibros. 3 Suppl. 2, 73–77 [DOI] [PubMed] [Google Scholar]
- 23.Dawson, R. J., and Locher, K. P. (2007) FEBS Lett. 581 935–938 [DOI] [PubMed] [Google Scholar]
- 24.Mendoza, J. L., and Thomas, P. J. (2007) J. Bioenerg. Biomembr. 39 499–505 [DOI] [PubMed] [Google Scholar]
- 25.Zeitlin, P. L., Lu, L., Rhim, J., Cutting, G., Stetten, G., Kieffer, K. A., Craig, R., and Guggino, W. B. (1991) Am. J. Respir. Cell Mol. Biol. 4 313–319 [DOI] [PubMed] [Google Scholar]
- 26.Dalemans, W., Barby, P., Champigny, G., Jallat, S., Dott, K., Dreyer, D., Crystal, R. G., Pavirani, A., Lecocq, J.-P., and Lazdunski, M. (1992) Nature 354 526–528 [DOI] [PubMed] [Google Scholar]
- 27.Du, K., Sharma, M., and Lukacs, G. L. (2005) Nat. Struct. Mol. Biol. 12 17–25 [DOI] [PubMed] [Google Scholar]
- 28.Lu, Y., Xiong, X., Helm, A., Kimani, K., Bragin, A., and Skach, W. R. (1998) J. Biol. Chem. 273 568–576 [DOI] [PubMed] [Google Scholar]
- 29.Vij, N., Fang, S., and Zeitlin, P. L. (2006) J. Biol. Chem. 281 17369–17378 [DOI] [PubMed] [Google Scholar]
- 30.Kawaguchi, Y., Kovacs, J. J., McLaurin, A., Vance, J. M., Ito, A., and Yao, T. P. (2003) Cell 115 727–738 [DOI] [PubMed] [Google Scholar]
- 31.Yang, Y., Janich, S., Cohn, J. A., and Wilson, J. M. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 9480–9484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brooks, D. A. (1999) Semin. Cell Dev. Biol. 10 441–442 [DOI] [PubMed] [Google Scholar]
- 33.Alberti, S., Bohse, K., Arndt, V., Schmitz, A., and Hohfeld, J. (2004) Mol. Biol. Cell 15 4003–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.