Abstract
Airway epithelial Na-K-2Cl (NKCC1) cotransport is activated through hormonal stimulation and hyperosmotic stress via a protein kinase C (PKC) δ-mediated intracellular signaling pathway. Down-regulation of PKCδ prevents activation of NKCC1 expressed in Calu-3 cells. Previous studies of this signaling pathway identified coimmunoprecipitation of PKCδ with SPAK (Ste20-related proline alanine-rich kinase). We hypothesize that endogenous PKCδ activates SPAK, which subsequently activates NKCC1 through phosphorylation. Double-stranded silencing RNA directed against SPAK reduced SPAK protein expression by 65.8% and prevented increased phosphorylation of NKCC1 and functional activation of NKCC1 during hyperosmotic stress, measured as bumetanide-sensitive basolateral to apical 86Rb flux. Using recombinant proteins, we demonstrate direct binding of PKCδ to SPAK, PKCδ-mediated activation of SPAK, binding of SPAK to the amino terminus of NKCC1 (NT-NKCC1, amino acids 1–286), and competitive inhibition of SPAK-NKCC1 binding by a peptide encoding a SPAK binding site on NT-NKCC1. The carboxyl terminus of SPAK (amino acids 316–548) pulls down endogenous NKCC1 from Calu-3 total cell lysates and glutathione S-transferase-tagged NT-NKCC1 pulls down endogenous SPAK. In intact cells, hyperosmotic stress increased phosphorylated PKCδ, indicating activation of PKCδ, and activity of endogenous SPAK kinase. Inhibition of PKCδ activity with rottlerin blocked the increase in SPAK kinase activity. The results indicate that PKCδ acts upstream of SPAK to increase activity of NKCC1 during hyperosmotic stress.
Na-K-2Cl cotransporter (NKCC1)2 is expressed widely in plasma membranes of mammalian cells where it couples the transport of chloride to that of sodium and/or potassium, in a stoichiometry of 2:1:1 (1). In epithelial cells, NKCC1 is unique for its localization to the basolateral membrane of salivary glandular cells and epithelial cells lining the airways, intestinal tract, inner medullary collecting duct cells, and non-mammalian rectal gland cells, thus allowing efficient salt and water secretion and reabsorption and volume regulation (1, 2). In epithelia lining the large airways, efficient mucociliary clearance depends on functional NKCC1 for supplying Cl and osmotically active water to a periciliary fluid layer encompassing cilia in the apical or luminal side of the tissue. In the nervous system, NKCC1 is expressed in the apical membrane of choroid plexus, in neurons, oligodendrocyte, and dorsal root ganglion neurons (3–8). The most profound functional impact from disruption of the Nkcc1 gene in mice is a shaker/waltz phenotype due to the collapse of endolymphatic cavity. This defect is linked to loss of basolateral NKCC1 in stria vascularis cells, resulting in loss of K+ uptake for secretion with fluid into the cochlear chamber (9–12). In other neuronal cell types, NKCC1 maintains an optimal Cl- gradient necessary for optimal GABA responses.
There is compelling evidence implicating phosphorylation of NKCC1 as a key step in activation of NKCC1 ion transport function. Shark rectal gland and rat brain NKCC1 are each phosphorylated during activation of the cotransporter (13–16). Phosphorylation of serine and threonine on the N terminus of shark rectal gland NKCC1 has been correlated to cAMP-dependent activation (17). Stimulation of rat salivary gland by β-adrenergic agents leads to an increase in NKCC1 transport activity and to phosphorylation of NKCC1 (18). Rat parotid NKCC1 is, in addition, phosphorylated by endogenous kinases associated with the basolateral plasma membrane and possibly coprecipitated with NKCC1 (19). Extracellular signal-regulated kinase (ERK) 1/2 kinases are implicated in the activation of cardiomyocyte NKCC by α1-adrenergic agents (20) and human tracheal epithelial NKCC1 by hyperosmotic stress (21).
More recently, SPAK (Ste20-related proline alanine-rich kinase, or PASK)-mediated phosphorylation of heterologous NKCC1 has emerged as important during osmotic stress (14–16). Expression of catalytically inactive SPAK in Xenopus oocytes or human embryonic kidney HEK-293 cells results in a decrease of stimulated NKCC1 activity (14, 22). Localization of SPAK in neurons and secreting and absorbing epithelia is consistent with its role in modulation of ion transport (16, 23, 24). The initial observation by Piechotta et al. (15, 16) of an interaction between SPAK and the amino terminus of NKCC1 (NT-NKCC1) led to in vitro phosphorylation assays showing phosphorylation of NT-NKCC1 (aa 1–260) by GST-tagged SPAK (25). These observations and reports correlating phosphorylation of Thr residues Thr206 and Thr211 of NKCC1 indicate that NT-NKCC1 constitutes a phosphoregulatory domain of NKCC1 (13). Details of the molecular mechanism are still not known.
In a recent study, we established the interaction of human airway NT-NKCC (aa 1–286) with unique and diverse intracellular proteins, which we termed a regulatory NKCC1 proteome (26). This proteome is distinguished by protein interactions that include two serine/threonine protein kinases, PKCδ and SPAK, the scaffold protein actin, and the serine/threonine protein phosphatase PP2A. We have shown a critical role for activity of PKCδ during regulation of airway NKCC1 by intracellular Cl and hyperosmotic stress and during α1-adrenergic stimulation in airway epithelial cells (21, 27) and for stable binding of PKCδ to actin for activation of NKCC1 (28, 29). The identification of a regulatory proteome that includes PKCδ suggests PKCδ may regulate SPAK activity by phosphorylation and thus indirectly phosphorylate and regulate activation of NKCC1. Two studies provide evidence for PKC regulation of SPAK. One study demonstrates direct binding of SPAK to PKCθ in T-cells followed by SPAK phosphorylation on two specific Ser sites (30). A second study using recombinant enzymes reports marked inhibition of SPAK activity by a general PKC inhibitor (25).
In the current study, we examine the role for SPAK in a NKCC1 regulatory proteome as an effector kinase linking PKCδ to activation of NKCC1 by hyperosmotic stress. In contrast to model systems with heterologously expressed proteins, we use Calu-3 airway epithelial cells, which serve as a model for airway epithelial cells and that express the proteins of interest, as determined from previous studies (26, 28). In the present study, we characterize the interaction of endogenous proteins and consequences of hyperosmotic-mediated activation of PKCδ on SPAK kinase activity and SPAK protein expression on NKCC1 phosphorylation state and activation.
EXPERIMENTAL PROCEDURES
Materials
86Rb (specific activity 154 Bq/g Rb, 4200 Ci/g Rb), [γ-32P]ATP (specific activity 111 TBq/mmol, 3000 Ci/mmol), and glutathione-Sepharose beads were purchased from GE Healthcare (Piscataway, NJ). Baculovirus-expressed recombinant PKCδ was purchased from Pan Vera (Madison, WI). Goat polyclonal antibody to NKCC1 (N-16), rabbit polyclonal antibody to PKCδ, polyclonal anti-GST antibody, anti-GST conjugated to agarose beads, and horseradish peroxidase-coupled secondary antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal antibody to SPAK was obtained from Abgent (San Diego, CA) and mouse monoclonal antibody to actin from Cytoskeleton (Denver, CO). Bumetanide was purchased from Sigma, protease inhibitor mixture set III from Calbiochem-Novabiochem Corp. (San Diego, CA), Talon beads from Clontech (Mountain View, CA), and an enhanced chemiluminescence reagent from Denville Scientific, Inc. (South Plainfield, NJ). Tissue culture supplies were purchased from Invitrogen and pre-cast 4–15% gradient slab gels from Bio-Rad. All other chemicals were reagent grade.
Cell Isolation and Culture
Calu-3 cells were grown in cell culture on tissue culture plastic or on 24-mm diameter, 0.4-μm pore size, Transwell Clear polyester filter inserts. All cell cultures were incubated in a humidified CO2 incubator at 37 °C. Culture medium was changed at 48–72-h intervals until confluence was reached.
Down-regulation of SPAK
SPAK (accession number NM_013233) was down-regulated using double-stranded siGENOME SMARTpool oligonucleotides siRNA (Dharmacon RNA Technologies, M-004875). Cells were grown in culture to 60–75% confluence, collected after trypsinization, pelleted, and washed twice with PBS at room temperature. Cells (9 × 106) were resuspended in 100 μl of transfection solution (Amaxa Biosystems, kit “V”), mixed with 0–7 pmol of siRNA/106 cells specific for SPAK (siSPAK) or, as a control, siCONTROL Non-Targeting siRNA pool (Dharmacon RNA Technologies, D-001206). The suspension was transferred to a cuvette and electroporated using program T-024 (Amaxa Biosystems). Cells were immediately diluted with 500 μl of culture medium, prewarmed to 37 °C, and plated onto cell culture plates or, for experiments with polarized monolayers, 24-mm diameter filter inserts at a seeding density of 3 × 106 cells/filter or plate. Preliminary experiments were performed to determine a 48-h incubation period as optimal for maximal loss of SPAK.
Measurement of in Vivo Kinase Activity
Calu-3 cells were grown to confluence on 100-mm tissue culture plastic dishes, serum-deprived overnight, then stimulated with or without (vehicle), methoxamine, or sufficient sucrose to increase medium osmolarity to 500 mOsm for 4 min. Final concentration of methoxamine was 10 μm. Cells were harvested in 1 ml of lysis buffer consisting of 100 mm NaCl, 50 mm sodium fluoride, 50 mm Tris-HCl, pH 7.5, 1% Nonidet P-40, 0.25% sodium deoxycholate, 1 mm EDTA, 1 mm EGTA, 1 mm sodium orthovanadate and protease and phosphatase inhibitor mixtures. Lysates were clarified by microcentrifugation at 4 °C for 20 min at 12,000 × g and incubated with antiserum against SPAK, as previously described (26). Immune complexes were recovered using Protein G-agarose beads that were prewashed and resuspended in lysis buffer. Kinase activity of SPAK was measured in immune complexes in an 30-μl assay buffer consisting of 25 mm HEPES, pH 7.5, 100 mm NaCl, 10 mm sodium fluoride, 10 mm MgCl2, 5 mm MnCl2, 1 mm β-mercaptoethanol, 1 mm sodium vanadate, protease inhibitor mixture, and phosphatase inhibitor mixture (24, 32). An aliquot of reaction mixture consisting of 1 μg of myelin basic protein (MBP) or 2 μg of CATCHtide peptide (amino acids RRHYYYDTHTNTYYLRTFGHNTRR, Ref. 33), 20 μm ATP, and 1.2 μCi of [γ-32P]ATP was added to SPAK and the incubation continued for 20 min at 30 °C. The reaction was terminated by addition of 25 μl of glacial acetic acid. An aliquot of 70 μl was transferred to spin columns (Pierce), centrifuged to recover radiolabeled substrate, washed three times with 5% phosphoric acid, and counted for radioactivity by Cerenkov counting. Alternatively, 70 μl was transferred to phosphocellulose discs, washed twice with 5% phosphoric acid, then twice with deionized water. The discs were transferred to scintillation vials containing 1 ml of deionized water and counted for radioactivity by Cerenkov counting.
Measurement of in Vitro SPAK Kinase Activity
In vitro SPAK kinase activity was measured using recombinant GST-tagged full-length SPAK suspended in 24 μl of 2× kinase buffer (50 mm HEPES, pH 7.5, 100 mm NaCl, 200 mm MgSO4, 20 μm β-mercaptoethanol, 1 μm KN93, 2 mm sodium vanadate, and protease inhibitor mixture). SPAK was preincubated without or with PKCδ in the absence or presence of 30 μg/ml phosphatidylserine, 2 μg/ml dioctanylglycerol (DiC8), and 25 ng of recombinant PKCδ for 15 min at 30 °C. An aliquot of reaction mixture consisting of 1 μg of MBP or 2 μg of CATCHtide peptide, 20 μm ATP, and 1.2 μCi of [γ-32P]ATP was added to SPAK, incubated for 20 min at 37 °C, and the reaction terminated by addition of 9 μl of glacial acetic acid. In some experiments, cleaved NT-NKCC1 (aa 1–286), prepared as previously described (26), was used as substrate. As a control, rottlerin was added to a 10 μm final concentration to block PKCδ activity prior to addition of substrate. Aliquots of 53 μl were transferred to spin columns (Pierce), centrifuged to recover radiolabeled substrate, washed three times with 5% phosphoric acid, and counted for radioactivity by Cerenkov counting. Alternatively, aliquots of acidified sample were applied to phosphocellulose discs, washed, and counted as described above.
Measurement of NKCC1 Activity
NKCC1 activity was measured as bumetanide-sensitive uptake of 86Rb, a congener of potassium, from a basolateral bathing medium in serum-deprived cells (27, 34). Serum-deprived cells were preincubated for 15 min at 32 °C following addition of vehicle or 10 μm bumetanide. To initiate radiotracer uptake, filters were transferred to a well of a 6-well tissue culture dish containing 1 μCi of 86Rb in HEPES-buffered Hanks' balanced salt solution (HPSS), consisting of 10 mm HEPES, 137 mm NaCl, 4.2 mm NaHCO3, 5.8 mm KCl, 0.3 mm Na2HPO4, 1.2 mm CaCl2, 0.4 mm MgSO4, and 10 mm glucose, pH 7.5, or HPSS supplemented with 10 μm bumetanide in a basolateral bathing solution. Influx was measured for a 4-min time interval then terminated by rapidly immersing filters four times in an ice-cold isotonic buffer consisting of 100 mm MgSO4 and 137 mm sucrose. Intracellular radioactivity was extracted by incubating cell monolayers in 0.1 n NaOH. Aliquots of cell extracts were assayed for protein content with a Pierce protein assay kit using bovine serum albumin as the standard and for radioactive counts by Cerenkov counting. Intracellular radioisotopic content was calculated as nanomole of potassium/mg of protein (86Rb).
Preparation and Expression of Recombinant Protein
Cytoplasmic RNA was isolated from the human airway epithelial cell line Calu3 using the RNAeasy Midi Kit (Invitrogen). Rapid amplification of cDNA ends-ready cDNA was prepared using the GeneRacer Kit (Invitrogen) with Superscript II Reverse Transcriptase according to the manufacturer's protocol.
CT-SPAK—A GST-tagged peptide consisting of the carboxyl terminus (CT) of human SPAK (GenBank™ accession number AF099989) from aa 315 to 548 (nucleotides 1119–1883) was constructed in a pGEX-4T vector (Amersham Biosciences). The pGEX-4T plasmid vector placed a GST tag at the NH2 terminus of the construct. First strand cDNA was synthesized using a SPAK-specific primer matching a sequence in the 3′-untranslated region of the SPAK mRNA (SPAK1883R, 5′-ATCGATGAGGGTTGAAGGGAG-3′) and Superscript III Reverse Transcriptase (Invitrogen). Amplification of the 3′ end of the SPAK open reading frame was carried out using gene-specific primers SPAK1119F (5′-GGATCCCTTTCACTGTGTCTTC-3′) and SPAK1883R with Platinum Taq DNA Polymerase (Invitrogen) and 10% dimethyl sulfoxide. Resulting PCR products were purified, cloned into the pCR4-TOPO vector (Invitrogen), and verified for sequence analysis with ABI Prism Big Dye Terminator (Applied Biosystems, Foster City, CA). The cDNA encoding the CT-SPAK was digested with BamHI and EcoRI for 1 h at 37 °C and ligated into a pGEX-4T expression vector using a T4 DNA ligase for 1 h at room temperature. The ligations were transformed into DH5α-competent cells and plated on 100 mg/ml ampicillin agar plates. cDNA sequenced confirmed the SPAK-CT cDNA in-frame with the plasmid vector.
NT-NKCC1—The full-length amino terminus (NT, aa 1–286, nucleotides -12–870) of human NKCC1 (GenBank accession number AY280459) was constructed as a GST-tagged protein in pGEX-6p-1, as described previously (26).
Recombinant proteins were expressed in DH5α or BL21(DE3)-competent cells upon induction with 0.4 mm isopropyl β-d-thiogalactopyranoside for 3 h at 37°C. Cells were harvested and recombinant protein extracted from the bacterial cell pellet using B-PER Bacterial Protein Extraction Reagent (Pierce) and eluted in 2 ml of Elution Buffer. Peptides were purified using Centricon YM-30 extraction columns (Millipore), dialyzed against PBS, and stored at -80 °C. Protein content was determined using a BCA protein assay kit (Pierce) with bovine serum albumin as the standard. To verify the molecular mass of the recombinant proteins, samples were subjected to SDS-gel electrophoresis on 4–15% gradient slab gels under reducing conditions and probed with a polyclonal antibody specific to the GST tag. For some experiments, the GST tag was cleaved from SPAK using thrombin and from NT-NKCC1 using PreScission enzyme according to manufacturer's instructions.
Immunoprecipitation and Immunoblot Analysis
Cells were grown to confluence, serum deprived overnight, and washed with ice-cold PBS. For immunoblot analysis, cells were lysed and harvested in 1 ml of lysis buffer consisting of 100 mm NaCl, 50 mm sodium fluoride, 50 mm Tris-HCl, pH 7.5, 1% Nonidet P-40, 0.25% sodium deoxycholate, 1 mm EDTA, 1 mm EGTA, and protease inhibitor mixture. Aliquots of 20–30 μg of protein were added to Laemmli buffer, heated for 5 min in a boiling water bath, and cooled to room temperature. Samples were subjected to SDS-PAGE on 4–15% gradient slab gels. Protein bands were transferred to polyvinylidine fluoride membrane paper for immunoblot analysis and probed with specific antibodies for proteins of interest.
NKCC1 was immunoprecipitated from Calu-3 cells, as previously described (26). Briefly, cells were lysed in 1 ml of ice-cold 10 mm HEPES, pH 7.4, buffer supplemented with 3.5 mm MgCl2, 150 mm NaCl, 0.3% Triton X-100, 1 μm benzamide, and protease inhibitor mixture. NKCC1 was immunoprecipitated by overnight incubation with constant rotation at 4 °C with polyclonal antibody to NKCC1 (Santa Cruz, SC21545, N-16). Immune complexes were recovered with Protein G-agarose beads, centrifuged, and washed five times, incubated at 37 °C for 30 min with Laemmli buffer, and subjected to gel electrophoresis on 4–15% gradient slab gels. NKCC1 and co-immunoprecipitated proteins were detected by Western blot analysis using specific antibodies and enhanced chemiluminescence.
Pulldown of endogenous proteins was performed using GST- or His6-tagged recombinant proteins. Total cell lysates were prepared and incubated for 40 min at 30 °C with the indicated amounts of recombinant protein. GST-tagged proteins were pulled down using glutathione-Sepharose beads or anti-GST-agarose beads and His6-tagged proteins using Talon beads. Beads were recovered by centrifugation, washed extensively with PBS, and resuspended in Laemmli buffer. Samples were heated as described above, subjected to 4–15% SDS-PAGE and immunoblot analysis for proteins of interest, as specified in the figure legends.
NKCC1 Phosphorylation Analysis
We used a phospho-specific R5 antibody that was raised to detect phosphorylation of residues Thr212 and Thr217 in human NKKC1 (35). Calu-3 cells were grown to confluence on 100-mm tissue culture dishes, serum deprived overnight, and treated with vehicle of HPSS or with sufficient sucrose to increase medium osmolarity to 500 mOsm. To stop the incubation, the experimental medium was rapidly aspirated and the cell monolayer was washed rapidly with three rinses in ice-cold PBS. NKCC1 was immunoprecipitated from total cells lysates, as described above. Immune complexes were subjected to SDS-PAGE and probed with the R5 antibody for phosphorylated NKCC1, then reprobed for NKCC1 using a polyclonal antibody directed against NT-NKCC1. In some experiments, siSPAK was electroporated into Calu-3 cells using an AMAXA nucleofector, as described above. Exposed protein bands were quantitated by densitometer.
In Vitro Binding Assays
For solution binding assays, aliquots of recombinant His6-CT-SPAK (30 μg) were mixed without or with 25 ng of recombinant PKCδ in the absence or presence of the PKC activators in mixed micelles consisting of 30 μg/ml phosphatidylserine and 2 μg/ml DiC8. The mixture was incubated at 30 °C for 40 min. Prewashed and precooled Talon beads were added to the protein mixture and incubation continued for 1 h at 4°C. Beads were recovered by centrifugation, washed extensively using PBS, subjected to gel electrophoresis on 4–15% gel gradient slab gels, and immunoblotted for PKCδ. For experiments on binding of SPAK and NT-NKCC1, aliquots of GST-SPAK and NT-NKCC1 cleaved from a GST tag using Precision enzyme were incubated in the absence or presence of an inhibitory peptide, denoted spNT (aa 76–91, PSQSRFQVDPVSENAG), which is designed to encode a binding domain for SPAK on NKCC1 (16). Bound proteins were pulled down using glutathione-Sepharose beads and immunoblotted for NT-NKCC1 and the GST tag. Exposed bands were quantitated by laser densitometry.
Solid phase slot blot binding assays were performed by immobilizing 2 μg of GST-SPAK or GST-CT-SPAK (aa 316–548) onto polyvinylidine fluoride membrane paper in a slot blot apparatus then flooding the paper with varying amounts of protein of interest. For binding of PKCδ to SPAK, PKCδ was added in 50 μl of kinase buffer consisting of 50 mm Tris-HCl, pH 7.5, 10 mm β-mercaptoethanol, 10 mm MgSO4, 1 mm sodium orthovanadate plus protease inhibitor mixtures with or without 30 μg/ml phosphatidylserine and 2 μg/ml DiC8 in mixed micelles. For binding of GST-NT-NKCC1, varying amounts of His6-SPAK was immobilized on membrane paper and overlaid with 1 μg of GST-NT-NKCC1. Bound proteins were detected by immunoblot analysis and exposed bands were quantitated by laser densitometry. Effective binding constants (EC50) were determined using a GraphPad Prism software program.
Data Analysis
Immunoreactive protein bands detected by chemiluminescence were quantitated by laser densitometry. Data are reported as mean ± S.E. and represent at least three or more experiments, unless otherwise stated. Treatment effects were evaluated using a multiple regression analysis with Tukey post comparison test and two-sided Student's t test for unpaired samples using a GraphPad InStat, version 3.00, software program.
RESULTS
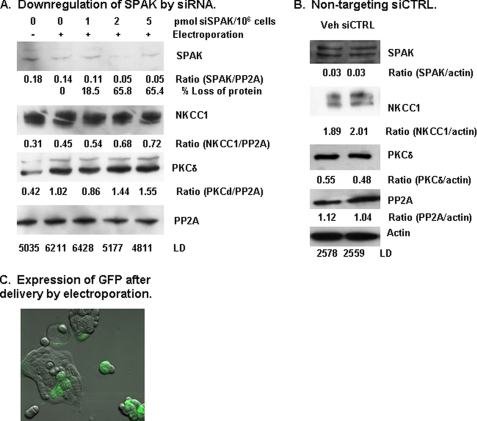
Down-regulation of SPAK—To ascertain the functional role of SPAK, expression of SPAK was manipulated using double-stranded siRNA directed against SPAK (siSPAK). We first performed experiments to optimize the amount of siSPAK to deliver into cells and the time after electroporation to achieve maximal loss of SPAK protein. Maximal loss of 65.8% SPAK was detected at 48 h after electroporation with 2 pmol of siSPAK per 1 × 106 cells (Fig. 1A). Expression of NKCC1, PP2A, PKCδ, and actin were not affected by electroporation alone or by treatment with siSPAK. As a control for the specificity of the siSPAK, cells were electroporated with a like amount of Non-Targeting siCONTROL RNA. Treatment with non-targeting siCONTROL RNA did not affect the amount of SPAK, NKCC1, PP2A, PKCδ, or actin detected in total cell lysates (Fig. 1B). As a control for delivery of nucleic acid into the cell nucleus, we simultaneously delivered a reporter vector pmaxGFP, which encoded the full-length cDNA for GFP. GFP expressing cells were detected by fluorescence microscopy after 48 (Fig. 1C) and 72 h incubation following electroporation.
FIGURE 1.
Down-regulation of SPAK using silencing RNA. A, down-regulation of endogenous SPAK. Double-stranded silencing RNA directed against SPAK (siSPAK) was delivered into Calu-3 cells by electroporation, as described under “Experimental Procedures.” Cells were incubated for 48 h after electroporation with 0, 1, 2, or 5 pmol of siSPAK per 1 × 106 cells and harvested as total cell lysate (TCL). As controls, equal aliquots of cells were not electroporated (0-) or electroporated without siSPAK (0+). Immunoblot analysis for SPAK was performed on a 30-μg aliquot of TCL and exposed bands were quantitated by laser densitometry (LD). Immunoblots were sequentially reprobed for NKCC1, PKCδ, and PP2A, which served as a loading control. Ratios are calculated as LD values divided by PP2A LD value for the specific experimental condition. Maximal loss of SPAK protein was 65.8% after a 48-h incubation immediately following electroporation. siSPAK did not affect the expression of NKCC1, PKCδ, and PP2A. B, non-targeting siCTRL. To test for nonspecific effects of silencing RNA, cells were electroporated with 2 pmol of non-targeting siCONTROL RNA (siCTRL)/106 cells. TCL were collected after 48 h incubation and analyzed for proteins of interest by immunoblot analysis. Exposed bands were quantitated by laser densitometry and normalized to values for actin that served as a loading control. Treatment with siCTRL did not alter expression of SPAK, NKCC1, PKCδ, PP2A, and actin. Typical results from three experiments are illustrated. C, expression of GFP 48 h after delivery of pmaxGFP into Calu-3 cells by electroporation. GFP is detected in multiple cells in this micrograph as a green fluorescent signal.
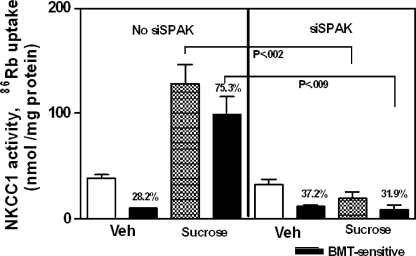
Effect of SPAK Down-regulation on Activation of NKCC1 by Hyperosmotic Stress—One predicted consequence of the loss of endogenous SPAK is the loss of activation of NKCC1 by hyperosmotic stress (14–16, 31). Fig. 2 shows that down-regulation of SPAK in Calu-3 cells using silencing RNA results in loss of NKCC1 function, measured as bumetanide-sensitive 86Rb uptake. In cells electroporated in the absence of siSPAK, elevating osmolarity to 500 mOsm using sucrose increased NKCC1 activity 89.9% from 10.0 ± 0.5 (n = 3) to 99.0 ± 16.6 (n = 4) nmol/mg of protein (p < 0.009). The bumetanide-sensitive portion of total 86Rb uptake also increased 63.9% from 28.2 ± 0.5% (n = 3) to 78.3 ± 3.2% (n = 7) (p < 0.0001). In cells electroporated with 2 pmol of siSPAK/106 cells, baseline NKCC1 activity, indicated in Fig. 2 as incubation with vehicle of HPSS, was not significantly different compared with cells electroporated without siSPAK. However, stimulation with sucrose to increase medium osmolarity to 500 mOsm did not significantly alter NKCC1 activity. In cells electroporated with non-targeting siCONTROL RNA, bumetanide-sensitive 86Rb uptake and % bumetanide-sensitive radiolabel uptake each increased 2.1-fold after elevating medium osmolarity to 500 mOsm, indicating the absence of off-target effects of siRNA. The loss of NKCC1 activation by hyperosmotic stress after down-regulation of SPAK indicates a role for SPAK in the regulation of endogenous NKCC1 during hyperosmotic stress in Calu-3 human airway epithelial cells.
FIGURE 2.
Down-regulation of SPAK prevents activation of NKCC1 in Calu-3 cells. Calu-3 cells were electroporated without or with 2 pmol of siSPAK/106 cells, seeded on filter inserts, and incubated in a humidified, 5% CO2 atmosphere for 48 h. Uptake of 86Rb was measured for a 4-min time period, as described under “Experimental Procedures.” In cells incubated with HPSS (Veh), baseline 86Rb uptake (white bars, Veh; strippled bars, sucrose) and bumetanide-sensitive 86Rb uptake (black bars) in cells electroporated without (left panel) or with siSPAK (right panel) were not significantly different. Stimulation of cells by imposing hyperosmotic stress significantly increased total and bumetanide-sensitive 86Rb uptake in cells electroporated without siSPAK (left panel) but had no effect on cells electroporated with siSPAK (right panel). Values are mean ± S.E. for four separate experiments.
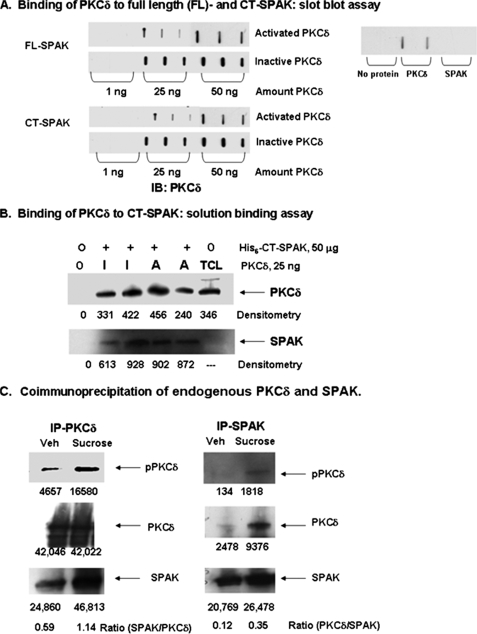
Interaction of SPAK with PKCδ—Previous studies demonstrated coimmunoprecipitation of SPAK and PKCδ in Calu-3 cells (26). This raised the possibility of a direct interaction between PKCδ and SPAK in the signaling pathway leading to activation of NKCC1. One question we addressed was whether activated or inactive PKCδ interacts with SPAK. To do this, we examined direct binding of the proteins in slot blot and solution binding assays. For a slot blot assay, His6-SPAK or GST-CT-SPAK (aa 316–548) was immobilized on paper then overlaid with solutions containing 1, 25, or 50 ng of inactive or preactivated recombinant PKCδ. We observed direct binding of inactive and preactivated PKCδ to full-length SPAK and to CT-SPAK (Fig. 3A). For a solution binding assay, His6-tagged CT-SPAK was mixed with inactive or activated PKCδ. Bound proteins were recovered on Talon beads and probed by immunoblot analysis for PKCδ, then reprobed for CT-SPAK. Active and inactivated PKCδ was pulled down with CT-SPAK, as illustrated in Fig. 3B. These results corroborated those of Fig. 3A. CT-SPAK was not detected in TCL, as expected.
FIGURE 3.
Interaction between SPAK and PKCδ. A, solid phase slot blot binding assay. Recombinant His6-full-length (FL)-SPAK or GST-CT-SPAK (aa 316–548), immobilized on paper, was overlaid with solutions containing 1, 25, or 50 ng of inactive or preactivated recombinant PKCδ (Pan Vera). To activate the enzyme, PKCδ was preincubated with 30 μg/ml phosphatidylserine and 2 μg/ml DiC8. We observed direct binding of PKCδ to FL-SPAK and CT-SPAK. The figure is representative of four experiments. B, solution binding assay. Fifty μg of recombinant His6-tagged CT-SPAK was mixed without (0, lane 1) or with 25 ng of inactive (I, lanes 2 and 3) or activated (A, lanes 4 and 5) PKCδ. The mixture was incubated for 40 min at 30 °C and bound proteins recovered by pulldown using Talon beads followed by immunoblot analysis for PKCδ. A 20-μg aliquot of Calu-3 total cell lysate (TCL) served as a positive control for the PKCδ immunoblot. Blots were reprobed with antibody to SPAK; His6-tagged CT-SPAK was detected as a 30-kDa band in lanes 1–5 and endogenous SPAK as a 70-kDa protein band in TCL (data not shown). Densitometry revealed binding of inactive and activated PKCδ to CT-SPAK. The figure is representative of four experiments. C, coimmunoprecipitation of endogenous PKCδ and SPAK. Calu-3 cells were serum-deprived overnight then treated with sufficient sucrose to increase medium osmolarity to 500 mOsm or with vehicle. SPAK or PKCδ was immunoprecipitated from TCL and immunoprecipitates were subjected to immunoblot analysis for pPKCδ, using phospho-PKCδ antibody (pPKCδ, Santa Cruz, sc-1177), then sequentially reprobed for PKCδ, which served as a loading control, and SPAK. Typical results for three experiments are shown. The ratio of SPAK to PKCδ and PKCδ to SPAK was calculated from densitometry values. Hyperosmotic stress increased the amount of pPKCδ detected in immunoprecipitates of PKCδ and SPAK and increased the coimmunoprecipitation of SPAK and PKCδ.
In vitro assays indicate that inactive and active PKCδ can bind to recombinant SPAK. The next question was whether this applied to endogenous proteins. In the next series of experiments, we examined endogenous PKCδ and its interaction with endogenous SPAK in Calu-3 cells. Activated PKCδ was quantitated using an antibody that recognizes PKCδ phosphorylated at Thr507, a site critical for activation of the enzyme. The portion of PKCδ that is activated is calculated as the ratio of phospho-PKCδ (pPKCδ) to total PKCδ from densitometry. Calu-3 cells were stimulated by increasing medium osmolarity to 500 mOsm using sucrose, PKCδ or SPAK were immunoprecipitated, and immune complexes were probed for pPKCδ and total PKCδ. Hyperosmotic stress increased the amount of pPKCδ detected in immunoprecipitates of PKCδ 2.7-fold and in immunoprecipitates of SPAK 13.1-fold, indicating activation of PKCδ (Fig. 3C). The amount of SPAK associated with PKCδ also increased 1.9-fold from 0.59 to 1.14 (ratio = SPAK/PKCδ) (Fig. 3D, left panel). Fig. 3C, right panel, illustrates results with immunoprecipitates of SPAK. Hyperosmotic stress increased the association of PKCδ with SPAK as seen in a 2.9-fold increase in the ratio of PKCδ/SPAK from 0.12 to 0.35. Overall, these results demonstrate that hyperosmotic stress increases activity of PKCδ and increases the association of SPAK with PKCδ and activated (phosphorylated) PKCδ.
To ascertain whether expression of SPAK is necessary for activation of PKCδ, SPAK was down-regulated using siSPAK in cells that were subsequently exposed to hyperosmotic stress. Treatment with sucrose increased the ratio of pPKCδ/PKCδ by 1.28 ± 0.13 (n = 3)-fold in cells electroporated with siSPAK and by 1.31 ± 0.03 (n = 3)-fold in cells treated with siCONTROL. For comparison, in non-electroporated cells, sucrose increased the ratio of pPKCδ/PKCδ by 1.33 ± 0.07 (n = 5)-fold. There is no significant difference in the activation of pPKCδ among these experimental treatments. These results indicate that down-regulation of SPAK did not alter activation of PKCδ by hyperosmotic stress and, therefore, activation of PKCδ is upstream of SPAK in the signaling complex.
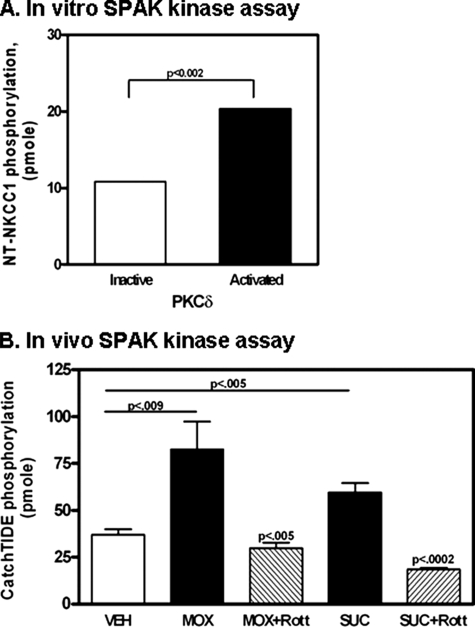
In Vitro and in Vivo Kinase Activity of SPAK—SPAK regulation of NKCC1 function is thought to depend on phosphorylation of SPAK, which apparently depends at least in part on autophosphorylation and on another kinase that coimmunoprecipitates with SPAK. One candidate enzyme is PKC (25, 30). To ascertain whether PKCδ activates airway epithelial SPAK, we first performed in vitro SPAK kinase assays with MBP, with cleaved NT-NKCC1 (aa 1–286), or with the SPAK substrate peptide CATCHtide (33), as substrate to examine SPAK kinase activity with or without preincubation with inactive or activated PKCδ. In these experiments, after pretreatment of SPAK with PKCδ, PKCδ was blocked by addition of rottlerin, just prior to addition of substrate and radiolabeled ATP. CATCHtide is a peptide encoding aa 198–218 of human NKCC1 (GenBank Protein ID AAP33908), which comprises two threonine residues that are phosphorylated by SPAK. Baseline phosphorylation was defined as phosphorylation detected in the presence of inactive PKCδ. A baseline phosphorylation of 10.3 ± 1.0 (n = 3) pmol of MBP increased 2.8-fold from 28.6 ± 1.7 (n = 3) pmol of MBP after preincubation with activated PKCδ. Similarly, using CATCHtide peptide as substrate, SPAK kinase activity increased from 50.7 ± 3.4 (n = 3) to 98.4 ± 23.2 (n = 3). Using cleaved NT-NKCC1 (aa 1–286) as substrate, a baseline phosphorylation of 10.8 ± 1.3 (n = 3) pmol increased 2.0-fold to 20.4 ± 0.1 (n = 3) pmol after pretreatment of recombinant SPAK with activated PKCδ (Fig. 4A). The consistent pattern of increased substrate phosphorylation after preincubation of recombinant SPAK with activated PKCδ indicates that PKCδ increases activity of SPAK.
FIGURE 4.
In vitro and in vivo SPAK kinase activity. A, in vitro SPAK kinase activity was measured using 3.45 μg of NT-NKCC1 as substrate, as described under “Experimental Procedures.” GST-SPAK was pretreated with activated PKCδ for 15 min at 30 °C. Rottlerin was then added to the mixture to block PKCδ activity. An aliquot of kinase reaction mixture, containing NT-NKCC1, ATP, and [γ-32P]ATP, was added to SPAK, incubated for 20 min at 37 °C, and the reaction terminated by addition of 9 μl of glacial acetic acid. Phosphorylated NT-NKCC1 was recovered on phosphocellulose discs, washed with phosphoric acid and deionized water, and subjected to Cerenkov counting. Pretreatment of SPAK with activated PKCδ significantly increased phosphorylation of NT-NKCC1. The results also indicate that NKCC1 is a substrate for SPAK. B, in vivo SPAK kinase activity. Calu-3 cells were grown to confluence, serum-deprived overnight, then stimulated with or without vehicle (Veh), 10 μm methoxamine (MOX), or sufficient sucrose (SUC) to increase medium osmolarity to 500 mOsm. Cells were lysed in 1 ml of lysis buffer and SPAK immunoprecipitated, as described under “Experimental Procedures.” SPAK kinase activity was measured in immune complexes using 2 μg of CATCHtide peptide as substrate. Stimulation of Calu-3 cells with MOX or SUC significantly increased SPAK kinase activity. Preincubation of cells with the PKCδ inhibitor rottlerin (Rott) blocked the stimulatory effects of MOX and SUC, indicating that PKCδ activity is necessary for SPAK kinase activity.
To determine whether endogenous PKCδ regulates SPAK activity, SPAK kinase activity was measured in immune complexes recovered from Calu-3 cells after treatment with vehicle, methoxamine, or sufficient sucrose to increase medium osmolarity to 500 mOsm. Methoxamine, a α1-adrenergic agonist, has been shown to increase PKCδ activity and NKCC1 transport function in Calu-3 cells and in primary human tracheal epithelial cells (21, 27). As seen in Fig. 4B, baseline (vehicle) SPAK kinase activity of 36.9 ± 3.9 (n = 9) pmol increased to 82.4 ± 15.5 (n = 8, p < 0.009) pmol after stimulation for 4 min with 10 μm methoxamine and to 59.5 ± 5.2 (n = 5, p < 0.005) pmol after induction of hyperosmotic stress. Preincubation with rottlerin, a PKCδ inhibitor, decreased the MOX-mediated SPAK kinase activity by 63.8% to 29.8 ± 3.6 pmol (n = 8, p < 0.005, compared with MOX alone) and hyperosmotic-induced SPAK kinase activity by 69.0% to 18.4 ± 1.0 pmol (n = 4, p < 0.0002, compared with hyperosmotic stress alone). These results indicate that treatment of cells with NKCC1 stimulants increases endogenous SPAK activity and that activity of PKCδ is necessary for activation of endogenous SPAK.
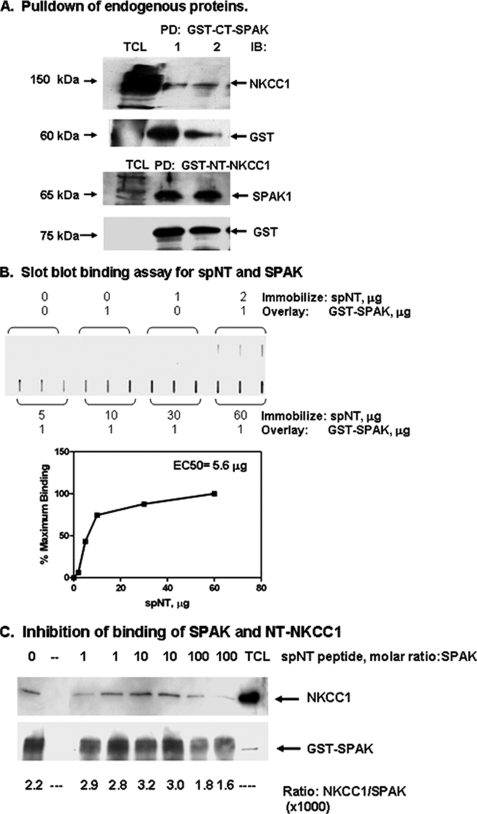
Binding of Endogenous SPAK to NKCC1—To determine whether endogenous SPAK and NKCC1 interact, we performed pull down assays with Calu-3 total cell lysates by adding either GST-tagged CT-SPAK (aa 316–548) or GST-tagged NT-NKCC1 (aa 1–286). GST-tagged proteins were pulled down using glutathione-Sepharose beads and associated proteins identified from immunoblot analysis. Fig. 5A illustrates typical results for three separate experiments. NKCC1 was detected in pulldowns of GST-CT-SPAK and SPAK was detected in pulldowns of GST-NT-NKCC1, indicating binding of NKCC1 and SPAK in Calu-3 cells. Solid phase binding assays with the recombinant proteins also indicated direct binding of the proteins (data not shown). One test of specificity of binding is inhibition of binding by a peptide encoding the binding domain on one of the two proteins. The binding domain for SPAK on NT-NKCC1 was identified from a yeast two-hybrid analysis (16). A peptide, denoted spNT, which encodes a SPAK binding domain (aa 76–91, PSQSRFQVDPVSENAG), was synthesized and used first in solid phase binding assays to test its binding to recombinant SPAK. As shown in Fig. 5B, spNT binds to SPAK in a dose-dependent manner with an EC50 of 5.6 μg. spNT was next used as a competitive inhibitor in a SPAK-NKCC1 solution binding assay. GST-SPAK and NT-NKCC1, cleaved from a GST tag, were incubated without or with varying molar ratios of spNT for 20 min at 30 °C. The molar ratio of spNT peptide to NT-NKCC1 was varied from 1:1 to 100:1. GST-SPAK was pulled down using glutathione-Sepharose beads and proteins bound to SPAK were immunoblotted for NT-NKCC1. In the absence of spNT, the binding ratio of NT-NKCC1 to SPAK was 2.69; incubation with 100:1 molar excess spNT reduced the binding ratio by 37.0%, indicating inhibition of binding. These results are consistent with binding of SPAK to human NT-NKCC1.
FIGURE 5.
Interaction between SPAK and NKCC1. A, Calu-3 cells were grown to confluence and serum-deprived overnight. TCL was prepared and used for pull-down assays. Pulldowns were performed by adding 50 μg of GST-CT-SPAK (aa 316–548) or 30 μg of GST-NT-NKCC1 (aa 1–286) to 1 ml of TCL followed by a 40-min incubation at 30 °C. GST-tagged protein was recovered using anti-GST-agarose beads to the reaction mixture. Samples were incubated for 60 minat 4 °C. Beads were recovered by centrifugation and washed five times with PBS. Interacting proteins were detected by immunoblot analysis. Typical results for three experiments each using six 1-ml aliquots of TCL are shown. NKCC1 was detected in pulldowns using GST-CT-SPAK and SPAK was detected in pulldowns using GST-NT-NKCC1. B, solid phase slot blot binding assay. Varying amounts (0–60 μg) of the peptide spNT was immobilized on membrane paper and overlaid with 1 μg of GST-SPAK. Samples were incubated for 25 min at room temperature. Unbound protein was removed by repeated washes with PBS. Bound GST-SPAK was detected by immunoblot analysis using anti-GST antibody. Protein bands were detected by chemiluminescence and quantitated by densitometry. Densitometry values were used to calculate an EC50 using the GraphPad Prism software program. C, competitive inhibition of binding of SPAK and NKCC1. GST-SPAK (10 μg, 0.12 nmol) and NT-NKCC1 cleaved from a GST tag (7 μg, 0.32 nmol) were incubated without or with peptide spNT at various molar ratios to GST-SPAK for 20 min at 30 °C. GST-SPAK was pulled down using glutathione-Sepharose beads and proteins bound to SPAK were immunoblotted with antibody to the amino terminus of NKCC1. Immunoblots were reprobed for the GST tag using an anti-GST antibody. Protein bands were detected by chemiluminescence and quantitated by densitometry. In the absence of spNT, the mean binding ratio of NT-NKCC1 to SPAK was 2.7 (lane 1, no spNT); incubation with 100:1 molar excess spNT reduced the binding ratio to 1.7, a 37.0% decrease in binding. Controls consisted of loading spNT alone (lane 2), which was not detected with the antibodies used in these immunoblots, and TCL. A protein band immunoreactive with antibody to NKCC1 was detected at the expected molecular mass of 150 kDa (TCL, lane 9).
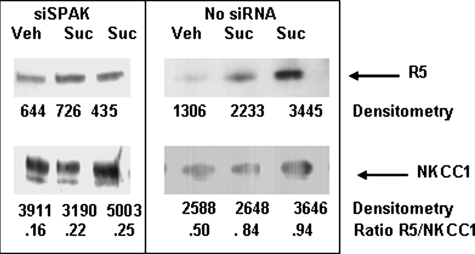
In Vivo Phosphorylation of NKCC1—Phosphorylation of NKCC1 is associated with its activation. To determine whether SPAK is necessary for phosphorylation of NKCC1 in Calu-3 cells, we used a R5 antibody as a probe for phosphorylated NKCC1 (35). Calu-3 cells were stimulated with sucrose to induce hyperosmotic stress and endogenous NKCC1 was immunoprecipitated and probed with a R5 antibody. Fig. 6 illustrates typical experiments. Sucrose treatment increased endogenous NKCC1 phosphorylation by 68% in cells not treated with siSPAK (right panel). In three experiments, sucrose stimulated NKCC1 phosphorylation by 1.81 ± 0.07 (n = 3)-fold. Down-regulation of endogenous SPAK using siSPAK led to sucrose-induced NKCC1 phosphorylation of 37.5%, a 1.8-fold reduction compared with cells not electroporated with siRACK1 (left panel). In three experiments with siSPAK-treated cells, sucrose induced a significantly lower 1.38 ± 0.09 (n = 3)-fold (p < 0.02) phosphorylation of NKCC1. These results are consistent with SPAK-mediated phosphorylation of NKCC1 and suggest that endogenous SPAK phosphorylates residues Thr212 and Thr217 on endogenous NT-NKCC1 expressed in Calu-3 cells.
FIGURE 6.
NKCC1 phosphorylation detected using a R5 antibody. Calu-3 cells were electroporated with 2 pmol of siSPAK/106 cells, seeded onto 100-mm dishes (6 × 106 cells per dish), and incubated in a humidified, 5% CO2 atmosphere for 48 h. As a control, an aliquot of Calu-3 was directly seeded onto 100-mm dishes without electroporation. On the day of sucrose stimulation, cell culture medium was discarded and replaced with HPSS, preheated to 35 °C. Cells were treated with HPSS (Veh) or stimulated by imposing hyperosmotic stress for 4 min (Suc). Stimulation was stopped by transferring dishes to an ice bath and washing the cell monolayer rapidly three times with ice-cold PBS. NKCC1 was immunoprecipitated, as described under “Experimental Procedures,” and immune complexes probed for phosphorylated NKCC1 using the R5 antibody then reprobed for NKCC1. Exposed bands were quantified by densitometry and densitometry values used to calculate a ratio of R5 (phosphorylated NKCC1) to total NKCC1. The panels illustrate typical results from three experiments each in which each experimental condition was done in replicates of 2–4. Sucrose increased phosphorylation of NKCC1 by 1.81 ± 0.07 (n = 3)-fold in cells not treated with siSPAK (right panel) and by 1.38 ± 0.09 (n = 3)-fold in cells electroporated with siSPAK (left panel). Treatment with silencing RNA significantly decreased hyperosmotic induced phosphorylation of NKCC1 (p < 0.02).
DISCUSSION
Activation of a Na-K-2Cl cotransporter expressed in the basolateral membrane of epithelial cells, denoted as NKCC1, is linked to its phosphorylation at the amino terminus (13–16, 18, 19). Several protein kinases have been associated with activation of NKCC1 in diverse cells types, including PKCδ, an effector kinase in the activation of airway epithelial NKCC1 by hyperosmotic stress, low intracellular Cl concentration, and hormonal stimulation (21, 27). The intracellular signaling mechanism for PKCδ-mediated regulation of NKCC1 requires its binding to the actin cytoskeleton (28, 29); however, the molecular mechanism for PKCδ regulation of NKCC1 has not been completely solved. In this study, we ask whether SPAK, a Ste20-related proline alanine-rich kinase, acts as an effector kinase linking activated PKCδ to activated NKCC1.
Using a yeast two-hybrid approach, SPAK was initially identified as a protein kinase that interacts with NKCC1 and activates NKCC1 heterologously expressed in Xenopus oocytes and HEK293 cell lines (14, 16, 22). Perhaps the most compelling evidence for a critical role of SPAK is the loss of NKCC1 phosphorylation and activation after overexpression of dominant negative SPAK in a HEK293 stable clonal cell line (14, 22). Regulation of SPAK kinase activity has been explored intensely after the discovery in a hybrid screen, of an interaction between SPAK and WNK4 (with no lysine kinase) (31). WNK4 is a member of a family of four mammalian serine threonine kinases (WNK1-WNK4) (36). Mutations in WNK4 and WNK1 have been linked to familial hyperkalemic hypertension, a human autosomal dominant disorder (37). WNK1 is regulated by hyperosmotic stress (38) and phosphorylates and regulates SPAK and OSR1 (oxidative stress response kinase-1), a protein kinase closely related to SPAK (16, 39). WNK4 prevents or slows down trafficking of thiazide-sensitive NCC expressed in COS-7 cells (40) and, when coexpressed with NKCC1 in Xenopus oocytes, suppressed NKCC1 activity (41, 42). One model emerging from these studies is a linear array of proteins with WNK1/WNK4 interaction with SPAK leading to an increase in kinase activity followed by SPAK-mediated phosphorylation of NKCC1.
Our previous report of an association between SPAK and PKCδ in Calu-3 human airway epithelial cells is an indicator that regulation of SPAK kinase activity and subsequent activation of native epithelial NKCC1 by hyperosmotic stress is complicated (29). Indeed, in the study reported here, we present evidence that PKCδ binds to SPAK in the inactive and activated, i.e. phosphorylated, state (Fig. 3, A and B) and that stimulation of epithelial cells by hyperosmotic stress increases bound activated PKCδ and binding of PKCδ and SPAK (Fig. 3C). Activation of PKCδ by hyperosmotic stress is not altered after down-regulation of SPAK using double-stranded silencing RNA, indicating that PKCδ acts upstream of SPAK. Thus, SPAK acts as a link between PKCδ and NKCC1. This model for a physiological function for SPAK in epithelial cells is analogous to a recently reported model depicting SPAK as a link between PKCθ and activator protein-1 in T-cell receptor response (30). Our data argue for a functional interaction between PKCδ and SPAK, which involves direct binding, phosphorylation of SPAK by PKCδ, and SPAK-mediated activation of NKCC1.
Our data also suggest that binding of activated PKCδ to SPAK is necessary for increased SPAK kinase activity. In vitro SPAK kinase activity was specifically increased after preincubation with activated PKCδ. The substitution of NKCC1-derived peptides for myelin basic protein as substrate for in vitro SPAK kinase activity corroborated reports of SPAK-mediated phosphorylation of heterologously expressed NKCC1 in Xenopus oocytes and HEK293 cells (25, 33). Using these unique substrates, we show that immunoprecipitated native SPAK phosphorylates NT-NKCC1 (aa 1–286) and CATCHtide (aa 198–218, NKCC1) (Fig. 4), most likely at threonine residues predicted as phosphorylation sites for SPAK (33). Others have shown SPAK interaction with WNK4 in NKCC1-injected Xenopus oocytes expressing SPAK and WNK4 (31). Thus, WNK4 alone apparently interacts with SPAK to enhance NKCC1 and KCC2 cotransporter function in this heterologous expression system. Whether WNK4 and PKCδ act cooperatively to modulate native Calu-3 NKCC1 function has yet to be determined.
One model for SPAK activation emerging from the current study reported here and from reports of PKCθ-mediated phosphorylation of SPAK in T-cells (30) and inhibition of SPAK activity by a general PKC inhibitor (25) is a cascade of protein kinases involved in increasing SPAK kinase activity. Alternatively, SPAK and its regulatory kinases, such as PKCδ or WNK isoforms, may be localized to discrete intracellular pools with each pool targeting NKCC1. How each pool is recruited to activate NKCC1 is still unknown. For PKCδ-mediated NKCC1 activation, the proximity of potentially interacting proteins may be a determining factor in the increased association and phosphorylation of SPAK by activated PKCδ. Learning more about the site(s) of interaction between PKCδ and SPAK and the regulation of the interaction may provide clues for refining this model of NKCC1 activation.
In summary, the study reported here expands a model of a regulatory NKCC1 proteome to define the role of SPAK as an activator of NKCC1 via PKCδ-mediated phosphorylation. These results indicate a key role for PKC isoforms in the regulation of endogenous SPAK activity. In addition, our observation of increased binding of SPAK with phosphorylated (activated) PKCδ and vice versa during hyperosmotic stress indicates highly regulated binding of the two proteins. Our findings also imply that activation of epithelial NKCC1 is more complex than a linear array of protein kinases. The possibility of multiple protein complexes linked to activation of NKCC1 suggest a permissive binding of specific protein kinases and phosphatases, which may be stimulated by selective environmental or hormonal stimuli. This expansion in the diversity of regulatory mechanisms awaits further study, in particular, to link regulatory proteomes with human diseases.
Acknowledgments
We thank Dr. B. Forbush (Yale University) for the generous gift of the R5 antibody.
This work was supported, in whole or in part, by National Institutes of Health Grants CA-35299 (to A. A.) and HL-58598 (to C. M. L.). This work was also supported by a Cystic Fibrosis Foundation Research Development Grant. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: NKCC1, Na-K-2Cl cotransporter; HPSS, HEPES-buffered Hank's balanced salt solution; MBP, myelin basic protein; PKCδ, protein kinase C δ isoform; siSPAK, double-stranded silencing RNA directed against SPAK; SPAK, Ste20-related proline alanine-rich kinase; aa, amino acid(s); PBS, phosphate-buffered saline; GST, glutathione S-transferase; DiC8, dioctanylglycerol; NT, NH2-terminal; CT, COOH-terminal.
References
- 1.Russell, J. M. (2000) Physiol. Rev. 80 211-276 [DOI] [PubMed] [Google Scholar]
- 2.Hebert, S. C., Mount, D. B., and Gamba, G. (2004) Pflugers Arch. Eur. J. Physiol. 447 580-593 [DOI] [PubMed] [Google Scholar]
- 3.Chen, H., Luo, J., Kinter, D. B., Shull, G. E., and Sun, D. (2005) J. Cereb. Blood Flow Metab. 25 54-66 [DOI] [PubMed] [Google Scholar]
- 4.Galan, A., and Cervero, F. (2005) Neuroscience 133 245-252 [DOI] [PubMed] [Google Scholar]
- 5.Lenart, B., Kintner, D. B., Shull, G. E., and Sun, D. (2004) J. Neurosci. 24 9585-9597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plotkin, M. D., Kaplan, M. R., Peterson, L. R., Gullans, S. R., Hebert, S. C., and Delpire, E. (1997) Am. J. Physiol. 272 C173-C183 [DOI] [PubMed] [Google Scholar]
- 7.Plotkin, M. D., Snyder, E. Y., Hebert, S. C., and Delpire, E. (1997) J. Neurobiol. 33 781-795 [DOI] [PubMed] [Google Scholar]
- 8.Sung, K. W., Kirby, M., McDonald, M. P., Lovinger, D. M., and Delpire, E. (2000) J. Neurosci. 20 7531-7538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crouch, J. J., Sakaguchi, N., Lytle, C., and Schulte, B. A. (1997) J. Histochem. 45 773-778 [DOI] [PubMed] [Google Scholar]
- 10.Delpire, E., Lu, J., England, R., Dull, C., and Thorne, T. (1999) Nat. Genet. 22 192-195 [DOI] [PubMed] [Google Scholar]
- 11.Goto, S., Oshima, T., Ikeda, K., Ueda, N., and Takasaka, T. (1997) Brain Res. 765 324-326 [DOI] [PubMed] [Google Scholar]
- 12.Mizuta, K., Adachi, F., and Isawa, K. H. (1997) Hearing Res. 106 154-162 [DOI] [PubMed] [Google Scholar]
- 13.Darmon, R. B., and Forbush, B. (2002) J. Biol. Chem. 277 37542-37550 [DOI] [PubMed] [Google Scholar]
- 14.Dowd, B. F. X., and Forbush, B. (2003) J. Biol. Chem. 278 27347-27353 [DOI] [PubMed] [Google Scholar]
- 15.Piechotta, K., Jianming, L., and Delpire, E. (2002) J. Biol. Chem. 277 50812-50819 [DOI] [PubMed] [Google Scholar]
- 16.Piechotta, K., Garbarini, N., England, R., and Delpire, E. (2003) J. Biol. Chem. 278 52848-52856 [DOI] [PubMed] [Google Scholar]
- 17.Lytle, C., and Forbush, B. (1992) J. Biol. Chem. 267 25438-25443 [PubMed] [Google Scholar]
- 18.Kurihara, K., Moore-Hoon, M. L., Saitoh, M., and Turner, R. J. (1999) Am. J. Physiol. 277 C1184-C1193 [DOI] [PubMed] [Google Scholar]
- 19.Kurihara, K., Nakanishi, N., Moore-Hoon, M. L., and Turner, R. J. (2002) Am. J. Physiol. 282 C817-C823 [DOI] [PubMed] [Google Scholar]
- 20.Andersen, G. Ø., Skomedal, T., Enger, M., Fidjeland, A., Brattelid, T., Levy, F. O., and Osmes, J.-B. (2004) Am. J. Physiol. 286 H1354-H1360 [DOI] [PubMed] [Google Scholar]
- 21.Liedtke, C. M., and Cole, T. S. (2002) Biochim. Biophys. Acta 1589 77-88 [DOI] [PubMed] [Google Scholar]
- 22.Gagnon, K. B. E., England, R., and Delpire, E. (2007) Cell Physiol. Biochem. 20 131-142 [DOI] [PubMed] [Google Scholar]
- 23.Marshall, W. S., Ossum, C. G., and Hoffmann, E. K. (2005) J. Exp. Biol. 208 1063-1077 [DOI] [PubMed] [Google Scholar]
- 24.Ushiro, H., Tsutsumi, T., Suzuki, K., Kayahara, T., and Nakano, K. (1998) Arch. Biochem. Biophys. 355 233-240 [DOI] [PubMed] [Google Scholar]
- 25.Gagnon, K. B. E., England, R., and Delpire, E. (2006) Mol. Cell Biol. 26 689-698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liedtke, C. M., Wang, X., and Smallwood, N. S. (2005) J. Biol. Chem. 280 25491-25498 [DOI] [PubMed] [Google Scholar]
- 27.Liedtke, C. M., and Cole, T. (1997) Am. J. Physiol. 273 C1632-C1640 [DOI] [PubMed] [Google Scholar]
- 28.Liedtke, C. M., Hubbard, M., and Wang, X. (2003) Am. J. Physiol. 284 C487-C496 [DOI] [PubMed] [Google Scholar]
- 29.Smallwood, N., Hausman, B. S., Wang, X., and Liedtke, C. M. (2005) Am. J. Physiol. 288 C903-C912 [DOI] [PubMed] [Google Scholar]
- 30.Li, Y., Hu, J., Vita, R., Sun, B., Tabata, H., and Altman, A. (2004) EMBO J. 23 1112-1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gagnon, K. B. E., England, R., and Delpire, E. (2006) Am. J. Physiol. 290 C134-C142 [DOI] [PubMed] [Google Scholar]
- 32.Johnston, A. M., Naselli, G., Gonez, L. J., Martin, P. M., Harrison, L. C., and DeAizpurua, H. J. (2000) Oncogene 19 4290-4297 [DOI] [PubMed] [Google Scholar]
- 33.Vitari, A. C., Thastrup, J., Rafiqi, F. H., Deak, M., Morrice, N. A., Karlsson, H. K. R., and Alessi, D. R. (2006) Biochem. J. 397 223-231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liedtke, C. M., Cody, D., and Cole, T. S. (2001) Am. J. Physiol. 280 L739-L747 [DOI] [PubMed] [Google Scholar]
- 35.Flemmer, A. W., Gimenez, I., Down, B. F. X., Darman, R. B., and Forbush, B. (2002) J. Biol. Chem. 277 37551-37558 [DOI] [PubMed] [Google Scholar]
- 36.Verissimo, F., and Jordan, P. (2001) Oncogene 20 5562-5569 [DOI] [PubMed] [Google Scholar]
- 37.Wilson, F. H., Dusse-Nicodeme, S., Choate, K. A., Ishikawa, K., Nelson-Williams, C., Desitter, I., Gunel, M., Milford, D. V., Lipkin, G. W., Achard, J. M., Feely, M. P., Dussol, B., Berland, Y., Unwin, R. J., Mayan, H., Simon, D. B., Farfel, Z., Jeunemaitre, X., and Lifton, R. P. (2001) Science 293 1107-1112 [DOI] [PubMed] [Google Scholar]
- 38.Zagorska, A., Pozo-Guisado, E., Boudeau, J., Vitari, A. C., Rafiqi, F. H., Thastrup, J., Dek, M., Campbell, D. G., Morrice, N. A., Prescott, A. R., and Alessi, D. R. (2007) J. Cell Biol. 176 89-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moriguchi, T., Urushiyama, S., Hisamoto, N., Iemura, S., Uchida, S., Natsume, T., Matsumoto, K., and Shibuya, H. (2005) J. Biol. Chem. 280 42685-42693 [DOI] [PubMed] [Google Scholar]
- 40.Cai, H., Cebotaru, V., Wang, Y.-H., Zhang, X.-M., Cebotaru, L., Guggino, S. E., and Guggino, W. E. (2006) Kidney Int. 69 2162-2170 [DOI] [PubMed] [Google Scholar]
- 41.Kahle, K. T., Gimenez, I., Hassan, H., Wilson, F. H., Wong, R. D., Forbush, B., Aronson, P. S., and Lifton, R. P. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2064-2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang, C.-L., Angell, J., Mitchell, R., and Ellison, D. H. (2003) J. Clin. Investig. 111 1039-1045 [DOI] [PMC free article] [PubMed] [Google Scholar]