Abstract
Excessive neuroinflammation contributes to many neurological disorders and is poorly controlled therapeutically. The signal transducer and activator of transcription (STAT) family of transcription factors has a central role in inflammatory reactions, being stimulated by multiple cytokines and interferons and regulating the expression of many proteins involved in inflammation. We found that STAT3 activation is highly dependent on glycogen synthase kinase-3 (GSK3). Inhibitors of GSK3 greatly reduced (>75%) the activating STAT3 tyrosine phosphorylation in mouse primary astrocytes, microglia, and macrophage-derived RAW264.7 cells induced by interferon-γ (IFNγ), IFNα, interleukin-6, or insulin. GSK3 inhibitors blocked STAT3 DNA binding activity and the expression of STAT3-induced GFAP and Bcl-3. GSK3 dependence was selective for activation of STAT3 and STAT5, whereas STAT1 and STAT6 activation were GSK3-independent. Knockdown of the two GSK3 isoforms showed STAT3 and STAT5 activation were dependent on GSK3β, but not GSK3α. The regulatory mechanism involved GSK3β binding STAT3 and promoting its association with the IFNγ receptor-associated intracellular signaling complex responsible for activating STAT3. Furthermore, GSK3β associated with the IFNγ receptor and was activated by stimulation with IFNγ. Thus, inhibitors of GSK3 reduce the activation of STAT3 and STAT5, providing a mechanism to differentially regulate STATs to modulate the inflammatory response.
The brain mounts a crucial inflammatory response to control the detrimental effects of injury, infection, and other insults. This neuroinflammatory response is mediated by astrocytes, the most numerous cells in the brain, and macrophage-derived microglia, which assume the immune surveillance role in the brain. If neuroinflammation is excessive or chronic, neuronal function and survival can be impaired, which contributes to many widespread neurodegenerative diseases, such as Alzheimer disease and multiple sclerosis (1–3). Therefore, clarifying inflammatory signaling pathways in the brain is critical for developing new methods to control the detrimental consequences of neuroinflammation.
A central component of inflammatory signaling is the Janus kinase (JAK)2/signal transducer and activator of transcription (STAT) cascade (4). Activated by cytokines and interferons, receptor-associated tyrosine kinase JAKs phosphorylate STATs on an activating tyrosine residue (e.g. Tyr701-STAT1 and Tyr705-STAT3). STATs are nucleocytoplasmic shuttling transcription factors that accumulate in the nucleus as a result of tyrosine phosphorylation increasing the STAT binding affinity to DNA, which slows dephosphorylation of STATs that is necessary for nuclear export, leading to regulation of gene expression (reviewed in Ref. 5). Besides regulation by tyrosine phosphorylation, the duration and degree of gene activation by STATs can be regulated by serine phosphorylation, by binding to transcriptional coactivators, and by modulation of the rate of nuclear export, which is required for renewing the non-phosphorylated pool of STATs available for reactivation (6, 7). This reflects the short half-life of activated STATs (∼15 min) even at optimal DNA binding sites (8). The rapid activation of STATs in response to inflammatory stimuli has heightened interest in developing strategies targeting STATs to control inflammatory responses in the periphery and the brain. In astrocytes, STAT3 is crucial for their differentiation (9, 10), and STAT3 is activated in numerous neuropathological conditions such as autoimmune encephalomyelitis (11) and ischemia (12) and has been implicated in reactive astrogliosis (13). The participation of STAT3 in neuroinflammation suggests that regulating STAT3 activation in astrocytes is a promising strategy for intervention.
Recently, glycogen synthase kinase-3 (GSK3) was identified as a crucial regulator of innate inflammatory processes (14, 15). GSK3 is a constitutively active Ser/Thr kinase consisting of two isoforms, GSK3α and GSK3β (16). GSK3 activity is tightly regulated, primarily by the phosphorylation of regulatory serines, Ser21 in GSK3α and Ser9 in GSK3β, that inhibit its activity, and also by its association in protein complexes and its subcellular localization (17). GSK3 was found to be a strong promoter of Toll-like receptor (TLR)-induced production of pro-inflammatory cytokines, including interleukin-6 (IL-6), tumor necrosis factor-α, IL-12p40, and interferon-γ (IFNγ), in part by promoting NF-κB activity (14), and inhibition of GSK3 protects rodents from a variety of peripheral inflammatory conditions (reviewed in Ref. 18).
As reviewed by Yoshimura (19), “three major transcription factors, including NF-κB, STAT3, and STAT1 have been shown to play major roles in transmitting inflammatory cytokine signals to the nucleus.” The recent revelations that GSK3 promotes inflammation and the activation of NF-κB (14, 20, 21) raised the question of whether GSK3 also promotes the activation of STAT3 or STAT1. Examination of this revealed that GSK3, particularly GSK3β, is required for the activating tyrosine phosphorylation of STAT3, but not STAT1, in astrocytes, microglia, and macrophages induced by IFNγ and other stimuli. Surprisingly, GSK3 was found to be associated with the IFNγ receptor and activated following stimulation with IFNγ. Thus, inhibition of GSK3 reduces activation of two critical pathways of the inflammatory response, NF-κB and STAT3.
EXPERIMENTAL PROCEDURES
Reagents—Protein-free Escherichia coli (K235) lipopolysaccharide (LPS) was prepared as described before (22). IFNγ was obtained from R&D Systems, IFNα from PBL Biomedical Laboratories, IL-4 and GM-CSF from RayBiotech Inc., SB216763 and SB415286 from Tocris, kenpaullone, indirubin-3′-monoxime, 6-bromoindirubin-3′-oxime (BIO), TDZD-8 and GSK3 inhibitor II from Calbiochem, IL-6 from eBioscience, insulin and LiCl from Sigma, and JSI-124 (cucurbitacin) from the NCI Developmental Therapeutic Program, National Institutes of Health.
Cell Culture—Primary astrocytes were prepared from cerebral cortex of 1-day-old C57Bl/6 mice as described previously with minor modifications (23), cultured in Dulbecco's modified Eagle's medium/F-12 medium supplemented with 10% fetal bovine serum, 0.3% glucose, 2 mm l-glutamine, 5 units/ml penicillin, and 5 μg/ml streptomycin, and after 10 days of culture shaken 30 h at 250 rpm, resulting in >95% pure astrocytes as determined by immunostaining with the astrocyte marker glial fibrillary acidic protein. After the first hour of shaking, the medium containing microglia cells was collected. RAW264.7 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 50 units/ml penicillin, and 50 μg/ml streptomycin. IFNγ (1 ng/ml), LPS (100 ng/ml), or both, IFNα (103 units/ml), IL-4 (25 ng/ml), GM-CSF (25 ng/ml), IL-6 (50 ng/ml), or insulin (50 ng/ml) were added to cell culture medium without supplements for 5 min to 6 h before cells were harvested.
Western Blotting, Immunoprecipitation, and GSK3 Kinase Assay—Cells were lysed in 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm EDTA, 1% Triton X-100, 10% glycerol, 1 μg/ml of leupeptin, aprotinin, and pepstatin A, 1 mm orthovanadate, 10 mm NaF, 0.1 μm okadaic acid, 1 mm phenylmethylsulfonyl fluoride. Membrane and soluble fractions were separated using a membrane extraction kit (Calbiochem), and nuclear and cytosolic fractions were fractionated using a nuclear extraction kit (Active Motif). Proteins were resolved with SDS-PAGE, and nitrocellulose membranes were immunoblotted with antibodies to phospho-Tyr705-STAT3, phospho-Ser727-STAT3, phospho-Tyr701-STAT1, phospho-Tyr694-STAT5, phospho-Tyr641-STAT6, STAT3, STAT1, STAT5, STAT6, phospho-Tyr1022/1023-JAK1, phospho-Tyr1007/1008-JAK2, phospho-Tyr1054/1055-TYK2 (Cell Signaling Technology, Danvers, MA), total phosphotyrosine, glial fibrillary acidic protein (GFAP), GSK3α/β (Upstate/Millipore, Lake Placid, NY), TLR4 (eBioscience, San Diego, CA), CD119, JAK1 (BD Pharmingen, San Diego, CA), JAK2 (BIOSOURCE), Bcl-3 (Santa Cruz Biotechnology, Santa Cruz, CA), and β-actin (Sigma). For immunoprecipitation, samples were incubated with the indicated primary antibody overnight at 4 °C with gentle agitation, followed by incubation with protein G-Sepharose beads (Amersham Biosciences) for 2 h at 4 °C, and the immune complexes were washed three times. GSK3 activity was assayed in CD119 and TLR4 immunoprecipitants from primary astrocytes as described previously (24). As a negative control, GSK3 activity was inhibited with lithium (20 mm) in the kinase assay to ensure specificity.
STAT3 DNA Binding—Nuclei from mouse astrocytes were extracted with a nuclear extraction kit (Active Motif). Astrocyte nuclear extracts (2 μg) were assayed for STAT3 DNA binding activity using the TransAM kit (Active Motif) according to the manufacturer's instructions.
Adenoviral Transduction—Mouse astrocytes were rinsed twice with Dulbecco's modified Eagle's medium/F-12 medium without supplements, infected with 50 multiplicity of infection of the designated adenovirus construct for 30 min, and supplemental medium was added for incubation for 36–48 h. All infected cultures were examined for adequate infection efficiency (∼80%) as assessed by green fluorescent protein fluorescence.
siRNA Transfection—The prevalidated siRNA oligonucleotide sequence for STAT3 (25) and Silencer negative control were obtained from Ambion, and those of GSK3α and GSK3β (Smart pool) were purchased from Dharmacon Research, Inc. Cells were transfected using liposome-mediated transfection reagent Lipofectamine RNAiMAX (Invitrogen) with 50 nm siRNA according to the manufacturer's instructions.
Statistical Analysis—Statistical significance between groups was evaluated by analysis of variance or Student's t test.
RESULTS
GSK3 Inhibitors Block STAT3 Activation—Stimulation of TLR4 with LPS activates inflammatory signaling in responsive cells, and this inflammatory response can be amplified by costimulating the IFNγ receptor (26), which predominantly activates the JAK/STAT signaling pathway (27). Therefore, the kinetics of STAT3 activation in mouse primary astrocytes was determined in response to treatment with LPS, IFNγ, or both, at the minimal concentration of IFNγ sufficient to induce synergistic physiological responses. Stimulation with LPS plus IFNγ caused a rapid (15 min) and long lasting increase in phospho-Tyr705-STAT3 in primary astrocytes (Fig. 1A). Most of this STAT3 activation was due to IFNγ (28), because IFNγ alone increased phospho-Tyr705-STAT3 to a similar degree as attained following treatment with LPS plus IFNγ. The total level of STAT3 remained unaltered following each treatment. Thus, treatment with IFNγ induced a robust and long lasting activation of STAT3 in astrocytes.
FIGURE 1.
GSK3 promotes STAT3 activation. A, mouse primary astrocytes were treated with 1 ng/ml IFNγ, 100 ng/ml LPS, or both for 15 min to 4 h, and cell lysates were immunoblotted for phospho-Tyr705-STAT3 and total STAT3. Mouse primary astrocytes were treated with 100 ng/mlLPS, 1 ng/ml IFNγ, or both, in the absence or presence of 20 mm lithium (LiCl) (B) for 15 min to 6 h or (C) for 30 min, followed by analysis of cell lysates by immunoblotting with phosphorylation-specific antibodies to assess the phosphorylation of STAT3 (Tyr705 and Ser727) (n = 4). D, mouse primary astrocytes were treated with 100 ng/ml LPS and/or 1 ng/ml IFNγ for 30 min without or with a pretreatment of 30 min with 20 mm lithium (LiCl) or 10 μm other GSK3 inhibitors (SB415286, SB216763, indirubin-3′-monoxime, and TDZD-8), and cell lysates were analyzed by immunoblotting. E, mouse primary astrocytes were treated with 1 ng/ml IFNγ for 30 min without or with a pretreatment of 30 min with 2, 5, 10, or 20 mm lithium, or 1, 2, 4, or 10 μm SB216763, and cell lysates were immunoblotted for phospho-Tyr705-STAT3 and total STAT3. RAW264.7 cells (F) or primary microglia (G) were treated with 100 ng/ml LPS and/or 1 ng/ml IFNγ for 30 min without or with a pretreatment of 30 min with 20 mm lithium (LiCl) or 10μm of other GSK3 inhibitors (SB415286, SB216763, indirubin-3′-monoxime, kenpaullone, and TDZD-8), and cell lysates were analyzed by immunoblotting. The ratio of phospho-Tyr705-STAT3 (PYSTAT3) to total STAT3 was calculated, and values represent the mean ± S.E.; n = 3–4. Immunoblots were reblotted for β-actin to ensure equal protein loading.
GSK3 was found to be crucial for the activation of STAT3 in primary astrocytes. Treatment with the selective GSK3 inhibitor lithium (29) reduced the initial tyrosine phosphorylation of STAT3 attained after 15 min of stimulation with LPS plus IFNγ, and the inhibition by lithium was maintained throughout 6 h of treatment (Fig. 1B). Lithium blocked by 77 ± 9% (mean ± S.E., n = 4) the increase in phospho-Tyr705-STAT3 induced by a 30-min treatment with LPS plus IFNγ (Fig. 1C) but did not markedly affect the phosphorylation of STAT3 on Ser727 or the total level of STAT3. To confirm that reduced STAT3 tyrosine phosphorylation caused by lithium treatment was due to GSK3 inhibition, four other structurally diverse, selective GSK3 inhibitors were tested, including SB415286, SB216763 (30), indirubin-3′-monoxime (31), and TDZD-8 (32). Along with lithium, all five GSK3 inhibitors greatly reduced the tyrosine phosphorylation of STAT3 induced by treatment with LPS plus IFNγ in primary astrocytes without modifying the total level of STAT3 (Fig. 1D). Furthermore, the concentration dependencies of lithium and SB216763 in reducing IFNγ-stimulated STAT3 tyrosine phosphorylation (Fig. 1E) were consistent with the original reports describing their inhibitions of GSK3 (30, 33).
To determine if GSK3 also is required for STAT3 activation in other types of cells besides primary astrocytes, macrophage-derived RAW264.7 cells and primary mouse microglia were examined. GSK3 inhibitors greatly reduced IFNγ-induced STAT3 tyrosine phosphorylation in both RAW264.7 cells (Fig. 1F) and mouse primary microglia (Fig. 1G), indicating that GSK3 is required for STAT3 activation induced by IFNγ in multiple cell types.
Tyr705 phosphorylation of STAT3 promotes its accumulation in the nucleus, and thus its activity as a transcription factor (34). Treatment of astrocytes with IFNγ caused a time-dependent increase in nuclear phospho-Tyr705-STAT3, which reached a maximum at 30 min (Fig. 2A). The nuclear accumulation of activated phospho-Tyr705-STAT3 was greatly reduced by inhibition of GSK3 with lithium throughout a 5- to 120-min stimulation with IFNγ (Fig. 2A). Stimulated nuclear accumulation of phospho-Tyr705-STAT3 also was largely eliminated by treatment with TDZD-8 (Fig. 2B). Consistent with the findings that GSK3 inhibitors reduced the activation of STAT3, treatment with lithium or TDZD-8 blocked the transcriptional activation of STAT3 as determined by using nuclear extracts from primary astrocytes to measure STAT3 binding to consensus site oligonucleotides, although they did not alter the basal level of STAT3 binding (Fig. 2C). Thus, inhibition of GSK3 depressed the tyrosine phosphorylation of STAT3, resulting in inhibition of STAT3 DNA binding activity.
FIGURE 2.
GSK3 inhibitors block STAT3 transcription factor activity. Mouse primary astrocytes were treated (A) for 5, 30, 60 or 120 min with 1 ng/ml IFNγ without or with 20 mm lithium (LiCl), or (B) for 30 min with 1 ng/ml IFNγ, 100 ng/ml LPS, or both, without or with 10 μm TDZD-8, and nuclear fractions were immunoblotted for phospho-Tyr705-STAT3 (PYSTAT3) (n = 3). C, STAT3 DNA binding activity was measured in astrocytes after 30-min stimulation with 1 ng/ml IFNγ modulated by treatments with 20 mm lithium or 10 μm TDZD-8. Values represent the mean ± S.E.; n = 5, *, p < 0.05 compared with samples not treated with a GSK3 inhibitor. Constitutively active STAT3C was expressed after infection with the FLAG-tagged STAT3 adenovirus (Ad5STAT3C) in mouse primary astrocytes for 24 h without or with 20 mm lithium (LiCl), and cell lysates were analyzed by immunoblotting for phospho-Tyr705-STAT3 (D) or GFAP or Bcl-3 (E). The ratio of phospho-Tyr705-STAT3 (PYSTAT3) to total STAT3 or the total levels of GFAP or Bcl-3 were calculated, and values represent the mean ± S.E.; n = 3–4. F, mouse primary astrocytes were treated for 24 h, 48 h, or 72 h without or with 20 mm lithium (LiCl), and cell lysates were analyzed by immunoblotting. The level of GFAP was calculated and values represent the mean ± S.E.; n = 3.
The GSK3 dependence of STAT3 tyrosine phosphorylation and DNA binding activity indicated that GSK3 inhibitors would block STAT3-induced gene expression. To test this, we examined the effect of inhibiting GSK3 on STAT3-induced increases in levels of proteins known to be STAT3 targets. STAT3-mediated expression was attained by expressing an active form of STAT3, STAT3C, in which covalent bonds between cysteines establishes the active dimer conformation (35). Although originally thought to be constitutively active, recent evidence shows that tyrosine phosphorylation of STAT3C is necessary for it to be active (36). Expressed STAT3C was tyrosine phosphorylated in the absence of a stimulus, and this was effectively blocked by inhibiting GSK3 with lithium (Fig. 2D). STAT3C expression in primary astrocytes was sufficient to increase the levels of GFAP and Bcl-3, proteins widely reported to be induced by active STAT3 (9, 10, 13, 37–40), and these increases were blocked by lithium treatment (Fig. 2E). We noted that lithium reduced GFAP to below the control level and found that lithium treatment time-dependently reduced GFAP levels in primary astrocytes independent of any additional treatment (Fig. 2F). Thus, the inhibition of STAT3C activation by GSK3 inhibitors was sufficient to block its induction of several proteins.
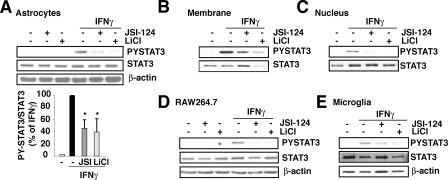
The regulation of STAT3 activation by GSK3 was evaluated further by comparing the effect of inhibiting GSK3 with lithium to the effect of a direct inhibitor of STAT3 activation, JSI-124 (also called cucurbitacin) (41). Treatment of primary astrocytes with 10 μm JSI-124 effectively blocked IFNγ-induced tyrosine phosphorylation of STAT3 measured in cell extracts (Fig. 3A), membrane fractions (Fig. 3B), or nuclear fractions (Fig. 3C), and inhibition of GSK3 with lithium produced comparable, or greater, inhibition of STAT3 activation in each preparation. Treatment with JSI-124 or lithium also caused similar reductions of STAT3 activation in RAW264.7 cells (Fig. 3D) and mouse primary microglia (Fig. 3E). These findings demonstrate that GSK3 inhibitors have a strong ability to inhibit STAT3 activation induced by IFNγ.
FIGURE 3.
GSK3 inhibitors block phospho-Tyr705-STAT3. Mouse primary astrocytes were treated with 1 ng/ml IFNγ for 30 min, with or without a 30-min pretreatment with 10 μm JSI-124 or 20 mm lithium, and cell lysates (A), membrane (B), or nuclear fractions (C) were analyzed by immunoblotting. The ratios of phospho-Tyr705-STAT3 (PYSTAT3) to total STAT3 were calculated and values represent the mean ± S.E.; n = 4, *, p < 0.05 compared with IFNγ alone. RAW264.7 cells (D) or microglia (E) were treated with 1 ng/ml IFNγ for 30 min, with or without a 30-min pretreatment with 10 μm JSI-124 or 20 mm lithium, and cell lysates were analyzed by immunoblotting. Immunoblots were reblotted for β-actin for cell lysates or CREB for nuclear lysates to ensure equal protein loading.
The GSK3 dependence of IFNγ-induced STAT3 activation raised the question of whether GSK3 also is required for other stimuli to activate STAT3. IL-6 (50 ng/ml) (42) induced a large increase in phospho-Tyr705-STAT3 in primary astrocytes, and this was blocked by inhibition of GSK3 with lithium or by JSI-124 (Fig. 4A). Similarly, GSK3 inhibitors also greatly reduced the stimulation of phospho-Tyr705-STAT3 induced by IFNα (103 units/ml) (43) or by insulin (50 ng/ml) (44) in both primary astrocytes and RAW264.7 cells (Fig. 4B). These findings indicate that GSK3 is required for STAT3 tyrosine phosphorylation induced by a variety of stimuli in multiple types of cells.
FIGURE 4.
GSK3 promotes the activation of STAT3 induced by other stimulants. A, mouse primary astrocytes were treated with 50 ng/ml IL-6 for 30 min, in the absence or presence of a 30-min pretreatment with 10 μm JSI-124 or 20 mm lithium (LiCl), and lysates were immunoblotted for phospho-Tyr705-STAT3 and STAT3. B, mouse primary astrocytes and RAW264.7 cells were treated with 103 units/ml IFNα or 50 ng/ml insulin for 30 min in the absence or presence of a 30-min pretreatment with 20 mm lithium (LiCl) or 10 μm SB216763, and lysates were immunoblotted for phospho-Tyr705-STAT3 and STAT3 (n = 3). Immunoblots were reblotted for β-actin to ensure equal protein loading.
GSK3 Selectively Regulates STAT3 Activation—Because GSK3 strongly promoted STAT3 activation, we examined if this effect is selective or if GSK3 is also required for activation of other members of the STAT family. Treatment of primary astrocytes or RAW264.7 cells with LPS plus IFNγ increased the tyrosine phosphorylation of STAT1 (27, 28, 43), and this was unaffected by GSK3 inhibitors (Fig. 5, A and B). Moreover, the absence of effect of GSK3 inhibitors on STAT1 activation was not dependent on the stimulus, because the phosphorylation of Tyr701-STAT1 induced by IFNα (43) was not affected by GSK3 inhibitors (Fig. 5B). Thus, GSK3 inhibitors did not completely block IFNγ-induced signaling but selectively inhibited IFNγ-induced activation of STAT3 but not of STAT1. Activation of STAT6 also was independent of GSK3. IL-4 was used to activate STAT6 (45) in primary astrocytes, microglia, and RAW264.7 cells, and GSK3 inhibitors did not reduce the stimulated production of phospho-Tyr641-STAT6 (Fig. 5C). However, STAT5 (STAT5A and STAT5B) activation demonstrated GSK3 dependence in primary astrocytes, because stimulation of STAT5 tyrosine phosphorylation by either IFNα or IFNγ (43) was blocked by inhibitors of GSK3 (Fig. 5D). GSK3 inhibitors also inhibited the production of phospho-Tyr694-STAT5 induced by treatment with GM-CSF (46) or IFNγ in primary microglia (Fig. 5E). Thus, the activation of STAT3 and STAT5 displayed a dependence on GSK3, whereas STAT1 and STAT6 were GSK3-independent.
FIGURE 5.
GSK3 is not a strong modulator of STAT1 and STAT6 but activates STAT5. A, mouse primary astrocytes (left panel) or RAW264.7 cells (right panel) were treated with 100 ng/ml LPS and 1 ng/ml IFNγ for 30 min without or with a pretreatment of 30 min with 20 mm lithium (LiCl) or 10 μm other GSK3 inhibitors (SB415286, SB216763, indirubin-3′-monoxime, kenpaullone, TDZD-8, and GSK3 inhibitor II), and cell lysates were analyzed by immunoblotting for phospho-Tyr701-STAT1 and STAT1 (n = 3). B, RAW264.7 cells (top panel) and primary microglia (bottom panel) were treated with 103 units/ml IFNα or 1 ng/ml IFNγ for 30 min without or with a pretreatment of 30 min with 20 mm lithium (LiCl) or 10 μm SB216763, and cell lysates were analyzed by immunoblotting for phospho-Tyr701-STAT1 and STAT1 (n = 2). C, mouse primary astrocytes (left panel), RAW264.7 cells (middle panel), and mouse primary microglia (right panel) were treated with 25 ng/ml IL-4 for 30 min without or with a pretreatment of 30 min with 20 mm lithium (LiCl) or 10 μm other GSK3 inhibitors (SB415286, SB216763, kenpaullone, TDZD-8, and GSK3 inhibitor II), and cell lysates were analyzed by immunoblotting for phospho-Tyr641-STAT6 and STAT6 (n = 4). D, mouse primary astrocytes were treated with 103 units/ml IFNα or 1 ng/ml IFNγ for 30 min without or with a pretreatment of 30 min with 20 mm lithium (LiCl) or 10 μm other GSK3 inhibitors (SB216763, BIO, and TDZD-8), and cell lysates were analyzed by immunoblotting for phospho-Tyr694-STAT5 and STAT5. The ratio of phospho-Tyr694-STAT5 (PYSTAT5) to total STAT5 was calculated, and values represent the mean ± S.E. n = 3. E, mouse primary microglia were treated with 1 ng/ml IFNγ or 25 ng/ml GM-CSF for 30 min without or with a pretreatment of 30 min with 20 mm lithium (LiCl) or 10 μm other GSK3 inhibitors (BIO and TDZD-8), and cell lysates were analyzed by immunoblotting for phospho-Tyr694-STAT5 and STAT5 (n = 3). Immunoblots were reblotted for β-actin to ensure equal protein loading.
STAT3 Tyr705 Phosphorylation Is Dependent on the GSK3β Isoform—Both isoforms of GSK3, GSK3α and GSK3β, are expressed and constitutively active in most cells and are blocked by all GSK3 inhibitors. To test if either isoform was predominant in facilitating STAT3 or STAT5 activation in astrocytes, the level of each isoform of GSK3 was selectively reduced by ∼70% using siRNA (Fig. 6A). IFNγ-induced tyrosine phosphorylations of STAT3 and of STAT5 were reduced by knocking down GSK3β comparably with the knockdown efficiency, consistent with the effects of GSK3 inhibitors on the activation of STAT3 and STAT5 (Fig. 6, B and C). Surprisingly, the IFNγ-induced activation of STAT3 and STAT5 were increased by knockdown of GSK3α. In contrast, knocking down either isoform of GSK3 was without effect on the activation of STAT1 or on the total levels of the STATs (data not shown). Thus, GSK3β is essential for the activation of STAT3 or STAT5 by tyrosine phosphorylation, whereas GSK3α has a different regulatory role. Reinforcing these conclusions, expression of constitutively active S9A-GSK3β in astrocytes was sufficient to induce tyrosine phosphorylation of STAT3 in the absence of additional stimuli (Fig. 6D). In contrast, STAT3 tyrosine phosphorylation was not affected by expression of constitutively active S21A-GSK3α.
FIGURE 6.
GSK3β isoform is responsible for the phospho-Tyr705-STAT3. Mouse primary astrocytes were treated with control siRNA (Ctl) or siRNA for GSK3α and GSK3β for 48 h followed by stimulation with 1 ng/ml IFNγ for 30 min, and cell lysates were analyzed by immunoblot. The level of GSK3α (left) and GSK3β (right) are shown in A. Cell lysates were analyzed by immunoblot. The ratio of phospho-Tyr705-STAT3 (PYSTAT3) to total STAT3 (B) and phospho-Tyr694-STAT5 to STAT5 (C) was calculated, and values represent the mean ± S.E.; n = 5; *, p < 0.05 compared with control. D, control green fluorescent protein or constitutively active S21A-GSK3α and S9A-GSK3β were expressed in mouse primary astrocytes for 48 h. Cell lysates were analyzed by immunoblot (n = 3). Immunoblots were reblotted for β-actin to ensure equal protein loading.
STAT3 Inhibition by Blocking GSK3 Is Not Mediated by Phosphatases or JAKs—Because full activation of STAT3 is dependent on GSK3, we examined if GSK3 regulates phosphatase or kinase actions associated with STAT3 tyrosine phosphorylation (42). The inhibitory effect of lithium on IFNγ-induced phospho-Tyr705-STAT3 was still evident after inhibition of phospho-tyrosine phosphatases with pervanadate (Fig. 7A). Both nuclear and cytosolic phospho-Tyr705-STAT3 were increased by pervanadate and reduced by lithium, but lithium did not cause a general reduction of protein tyrosine phosphorylation induced by pervanadate (bottom panel), indicating that GSK3 selectively modulates phosphorylation, rather than dephosphorylation, of phospho-Tyr705-STAT3. We next examined tyrosine kinases that may be linked to STAT3 activation (27, 43, 47, 48). Src did not contribute to IFNγ-induced phospho-Tyr705-STAT3 in primary astrocytes, because the Src family inhibitor PP-2 did not affect phospho-Tyr705-STAT3 induced by IFNγ (data not shown), suggesting a JAK family kinase likely mediates GSK3-dependent STAT3 tyrosine phosphorylation. However, inhibitors of GSK3 did not affect the phosphorylation of JAK2 induced by treatment with LPS plus IFNγ in primary astrocytes (Fig. 7B), RAW264.7 cells (Fig. 7C), or primary microglia (Fig. 7D). GSK3 inhibitors also did not alter IFNγ-induced activation of TYK2 or JAK1 in astrocytes, RAW264.7 cells, or microglia (Fig. 7E). These results, taken together with the lack of effect on STAT1 activation, demonstrate that GSK3 does not function upstream of STAT3-activating tyrosine kinases, but instead regulates their ability to phosphorylate STAT3, raising the possibility that GSK3 may be associated with STAT3 to modulate its activation at the receptor.
FIGURE 7.
STAT3 inhibition by GSK3 is not mediated by phosphatases or JAKs. A, mouse primary astrocytes were treated for 30 min with 0.1 mm pervanadate, 1 ng/ml IFNγ, and 20 mm lithium (LiCl) as indicated, followed by immunoblotting nuclear or cytosolic extracts (PY indicates total phosphotyrosine immunoreactivity) (n = 2). B, mouse primary astrocytes; C, RAW264.7 cells; or D, mouse primary microglia were treated for 30 min with 1 ng/ml IFNγ and 100 ng/ml LPS, without or with 20 mm lithium (LiCl) or 10 μm other GSK3 inhibitors (SB415286, SB216763, kenpaullone, TDZD-8, and GSK3 inhibitor II). Cell lysates were analyzed by immunoblot for phospho-Tyr1007/1008-JAK2 and JAK2 (n = 4). E, mouse primary astrocytes, microglia, and RAW264.7 cells were treated for 30 min with 1 ng/ml IFNγ, without or with 20 mm lithium (LiCl) or 10 μm SB216763. Cell lysates were analyzed by immunoblot for phospho-Tyr1022/1023-JAK1 and JAK1 or phospho-Tyr1054/1055-TYK2. Immunoblots were reblotted with β-actin to ensure equal protein loading.
GSK3 Is Associated with STAT3—Because GSK3 is required for STAT3 activation but not for the kinases that phosphorylate it, we speculated that GSK3 may directly associate with STAT3 to facilitate its delivery to the IFNγ receptor-associated signaling complex. Co-immunoprecipitation measurements showed that endogenous GSK3 was associated with endogenous STAT3, because STAT3 co-immunoprecipitated with both GSK3α and GSK3β in primary astrocytes and stimulation with IFNγ rapidly increased their association (Fig. 8A). In contrast, no STAT1 was found to be associated with GSK3 (data not shown). Conversely, each GSK3 isoform also co-immunoprecipitated with STAT3, and examination of the subcellular localization of these GSK3·STAT3 complexes showed that GSK3 co-immunoprecipitated with STAT3 from membrane (Fig. 8B) and nuclear (Fig. 8C) fractions prepared from primary astrocytes. Notably, GSK3β was primarily associated with STAT3 in membranes (Fig. 8B), consistent with its regulation of the initial activation of STAT3, whereas GSK3α was primarily associated with STAT3 in the nucleus (Fig. 8C), suggesting that GSK3α may regulate nuclear STAT3, such as its nucleocytoplasmic shuttling. Stimulation with LPS plus IFNγ increased the association of GSK3β with STAT3 (Fig. 8B), consistent with evidence that GSK3β promotes the activation of STAT3 at the membrane, and increased the association of GSK3α with STAT3 in the nucleus (Fig. 8C).
FIGURE 8.
GSK3 associates with STAT3. A, GSK3α or GSK3β were immunoprecipitated (IP) from cell lysates of primary astrocytes treated for 30 min with 1 ng/ml IFNγ, and immunoprecipitated lysates were immunoblotted for STAT3 and GSK3α/β. The level of STAT3 associated with each isoform of GSK3 was evaluated, and values represent the mean ± S.E. (n = 3). STAT3 was immunoprecipitated from membrane (B) or nuclear fractions (C) prepared from primary astrocytes treated for 30 min with 1 ng/ml IFNγ, 100 ng/ml LPS, or both, and immunoprecipitated lysates were immunoblotted for GSK3α/β and STAT3 (n = 4). To ensure the efficiency of the immunoprecipitation, the recovery in the supernatant after immunoprecipitation (Sup.) was performed. D, the IFNγ receptor α-chain (CD119) or STAT3 (E) was immunoprecipitated from membrane fractions prepared from mouse primary astrocytes after infection with adenovirus for the expression of green fluorescent protein (control), constitutively active S9A-GSK3β, or constitutively active S21A-GSK3α, without or with constitutively active FLAG-tagged STAT3C (Ad5STAT3C), for 48 h, and immunoprecipitated lysates were immunoblotted for GSK3α/β, CD119, or STAT3. (n = 3). To ensure the specificity and the efficiency of the immunoprecipitation, an immunoprecipitation with a nonspecific isotypic IgG and the recovery in the supernatant after immunoprecipitation (Sup) were performed. F, astrocytes were treated with control siRNA (Ctl) or siRNA for GSK3α and GSK3β for 48 h followed by a pretreatment for 2 h with 10 ng/ml leptomycin B (LMB) and then addition of IFNγ (1 ng/ml) for 2 h, and nuclear fractions were immunoblotted. The level of phospho-Tyr705-STAT3 (PY-STAT3) was calculated, and values represent the mean ± S.E. (n = 5). Immunoblots were reblotted with CREB to ensure equal protein loading.
The localization of GSK3β associated with STAT3 in the membrane fraction raised the possibility that GSK3 may associate with receptors for inflammatory molecules. In primary astrocytes, expressed active mutants of each GSK3 isoform, S9A-GSK3β and S21A-GSK3α, co-immunoprecipitated with the IFNγ receptor (Fig. 8D). The associations of expressed active GSK3α and GSK3β with the IFNγ receptor were increased by expression of constitutively active STAT3C. Additionally, expression of S9A-GSK3β induced a marked increase in the membrane fraction of the amount of STAT3 associated with GSK3β, and more expressed STAT3C was recruited to the membrane fraction upon expression of active GSK3 isoforms (Fig. 8E). These results indicate that GSK3 and STAT3 cooperatively recruit each other to the membrane fraction containing the IFNγ receptor.
Whereas GSK3β clearly associates with STAT3 at the membrane-localized receptor where it promotes STAT3 tyrosine phosphorylation, the effects of GSK3α differed from GSK3β. The increased IFNγ-induced phospho-Tyr705-STAT3 obtained with knockdown of GSK3α and the predominant association of GSK3α with STAT3 in the nucleus indicated that GSK3α may control the nucleocytoplasmic shuttling of STAT3. Therefore, the effects of GSK3 manipulations on nuclear STAT3 were examined using leptomycin B to inhibit CRM1-dependent export of STAT3 (6). Knocking down GSK3β reduced the nuclear level of IFNγ-induced phospho-Tyr705-STAT3 in the absence or presence of leptomycin B, as would be expected by the requirement for GSK3β at the membrane to promote IFNγ-induced STAT3 activation. In contrast, knocking down of GSK3α had the opposite effect of increasing nuclear phospho-Tyr705-STAT3 (Fig. 8F). These results suggest that, although GSK3β is required for the activation of STAT3 at the membrane, GSK3α may promote dephosphorylation and nuclear export of STAT3 to promote the recycling of STAT3.
GSK3 Is Active at the IFNγ Receptor—We examined further the association of GSK3 with plasma membrane receptor complexes involved in inflammatory signaling in astrocytes, which express both IFNγ and TLR4 receptors (49–51), to test if receptor stimulation modulated receptor-associated GSK3 activity. The α-chain of the IFNγ receptor was immunoprecipitated from primary astrocytes and both GSK3α and GSK3β co-immunoprecipitated with the IFNγ receptor and were recruited to the receptor after stimulation with LPS plus IFNγ (Fig. 9). Membrane CD119 levels tended to decrease after IFNγ stimulation, suggesting a possible internalization of these receptors after IFNγ stimulation, as reported previously (52). In contrast, little change was observed in GSK3 isoforms associated with TLR4 after stimulation (Fig. 9).
FIGURE 9.
GSK3 is active at the IFNγ receptor. Co-immunoprecipitation (IP) of GSK3α/β with IFNγ receptor α-chain (CD119) (A), or TLR4 from membrane fractions (B) of mouse primary astrocytes, including untreated cells and following 30-min stimulation with 1 ng/ml IFNγ, 100 ng/ml LPS, or both. GSK3 activity was measured in CD119 or TLR4 immunoprecipitants from primary astrocytes. To ensure the specificity and the efficiency of the immunoprecipitation, an immunoprecipitation with a nonspecific isotypic IgG and the recovery in the supernatant after immunoprecipitation (Sup) were performed. Quantified data are presented as mean ± S.E.; n = 5, *, p < 0.05 compared with control. Primary mouse astrocytes were infected with the constitutively active, FLAG-tagged STAT3 adenovirus (Ad5STAT3C) for 24 h, and CD119 from cell lysates was immunoprecipitated (IP) after treatment with 20 mm lithium (LiCl) (C); 20 mm lithium, 10 μm TDZD-8, or 10 μm SB415286 for 4 h (D); 1 ng/ml IFNγ for 30 min (E); or 1 ng/ml IFNγ and 20 mm lithium (F), and immunoblotted for FLAG and CD119 (n = 4).
To test if the IFNγ receptor-recruited GSK3 was functionally active, GSK3 activity and STAT3 detection assays were carried out with receptor immunoprecipitates. The activity of GSK3 that co-immunoprecipitated with the IFNγ receptor approximately doubled after stimulation of astrocytes with IFNγ alone or in combination with LPS (Fig. 9A), a particularly notable increase considering that GSK3 is a constitutively active kinase, whereas stimulants did not significantly alter the GSK3 activity that co-immunoprecipitated with TLR4 (Fig. 9B). These results suggest that active GSK3 associated with the IFNγ receptor binds and promotes IFNγ-induced STAT3 activation. This conclusion was confirmed by the finding that GSK3 inhibitors blocked the association of STAT3 with the IFNγ receptor. To detect IFNγ receptor-bound STAT3, STAT3C was expressed, and its association was measured in IFNγ receptor immunoprecipitants. Treatment with lithium inhibited STAT3C binding to the IFNγ receptor by 77 ± 10% (n = 4) (Fig. 9C). Similar inhibitions of the STAT3C·IFNγ receptor complex formation were achieved by two other GSK3 inhibitors, TDZD-8 and SB415286, as well as lithium, confirming the requirement for GSK3 (Fig. 9D). Stimulation with IFNγ increased the association of STAT3C with the IFNγ receptor (Fig. 9E), and this also was blocked by inhibition of GSK3 (Fig. 9F). Thus, GSK3β is bound the IFNγ receptor and activated by IFNγ treatment, and GSK3 is necessary for the recruitment of STAT3 to the receptor to undergo tyrosine phosphorylation-mediated activation.
DISCUSSION
STAT3 is a key transcription factor involved in inflammatory responses, especially those mediated by cytokines, as well as in cellular proliferation, differentiation, and survival (5). The results reported here demonstrate a strong dependence on GSK3 for the activation of STAT3 induced by several stimuli in astrocytes, microglia, and macrophages, revealing a new mechanism that regulates this key inflammatory signaling pathway. STAT family members were differentially dependent on GSK3, because the activations of STAT3 and STAT5 were blocked by GSK3 inhibitors, but STAT1 and STAT6 activation were not. Inhibition of GSK3 largely impeded the activating tyrosine phosphorylation of STAT3 without altering activation of the upstream kinases JAK1, JAK2, or TYK2, or activation of STAT1 induced by IFNγ, indicating a STAT3-targeted action of GSK3 in regulating the response to IFNγ stimulation. This action of GSK3 was predominantly mediated by GSK3β, rather than GSK3α, and was accompanied by the association of GSK3β with STAT3 in the membrane fraction where IFNγ stimulation increased active GSK3 associated with the IFNγ receptor and GSK3 inhibitors blocked the recruitment of STAT3 to the IFNγ receptor. Thus, GSK3 is intimately associated with the IFNγ receptor signaling to STAT3 in a selective manner. This critical role of GSK3 in the IFNγ receptor intracellular signaling protein complex regulating STAT3 activation may underlie some of the pleiotropic actions of GSK3 on inflammation and cell proliferation and survival (17, 53).
The dependence of STAT3 activation on GSK3 was a widespread regulatory interaction, because it was evident with multiple stimuli and in several types of cells. Thus, IFNγ, IFNα, IL-6, and insulin each activated STAT3 in a GSK3-dependent manner, although each activates a different type of receptor. Taken in conjunction with the association of STAT3 with GSK3, the lack of GSK3 dependence of JAK1, JAK2, and Tyk2 activation, as well as the lack of effect on STAT1 activation and the lack of association of GSK3 with STAT1, these findings suggest that STAT3 association with GSK3 represents the key regulatory interaction to promote STAT3 tyrosine phosphorylation. Stimulation with IFNγ recruited STAT3 to the IFNγ receptor for its activating tyrosine phosphorylation and increased the receptor-associated activity of GSK3, and STAT3 recruitment to the receptor was blocked by inhibiting GSK3. Because inflammatory signals are transduced via transmembrane receptors that serve as docking sites for intracellular signaling proteins, identification of a new partner, GSK3, recruited to this receptor complex provides a novel target for regulating inflammatory signaling. This finding of receptor-associated GSK3 initially may seem surprising considering the dogma that GSK3 is predominantly a cytosolic enzyme. However, GSK3 has recently been shown to be directly associated with a number of other plasma membrane receptors (24, 54–56). GSK3 is characteristically inactivated by external receptor-mediated signals impacting its activity, in contrast to most kinases that are activated by receptor-coupled signal transduction mechanisms (57–59). This GSK3-inactivating mechanism is used by several types of receptors as they generate intracellular signaling cascades, such as growth factor receptors, G-protein-coupled receptors, steroid receptors, and ionotropic glutamatergic GluR1 receptors (24, 60–62). Thus, constitutively active GSK3 is directly or indirectly associated with many types of plasma membrane receptors, and their activation of intracellular signaling often involves inactivation of GSK3. The present study extends the known receptors that GSK3 associates with to the IFNγ receptor and reveals the less common feature that IFNγ receptor stimulation activates GSK3. This effect is not unique, because Wnt stimulation causes activation of GSK3 associated with the LRP6 receptor (56, 63). Thus, as with many other types of receptors, GSK3 is intimately involved in IFNγ-mediated signal transduction mechanisms and may have similar roles in signaling induced by the other stimuli that activate STAT3 in a GSK3-dependent manner.
The GSK3 dependence of STAT3 activation did not extend to all members of the STAT family of transcription factors. Extensive examination of IFNγ-stimulated activation of STAT1 failed to reveal any dependence on GSK3. This is noteworthy because it demonstrates that the regulatory action of GSK3 does not extend to all signals generated by the IFNγ receptor, supporting the conclusion that it is the interaction between GSK3 and STAT3 that is key to regulation, rather than regulation by GSK3 of the receptor or receptor-associated tyrosine kinases. Furthermore, GSK3 did not regulate STAT6 tyrosine phosphorylation induced by IL-4, the most highly documented activator of STAT6 (45). However, GSK3 was found to be required for STAT5 activation, because GSK3 inhibitors reduced STAT5 activation in astrocytes stimulated with IFNγ or IFNα or microglia stimulated by GM-CSF or IFNγ. Knocking down the level of GSK3β in astrocytes further confirmed the dependence of STAT5 activation on GSK3. The mechanism for this regulatory effect of GSK3 on STAT5 activation remains to be studied and is currently under investigation. Overall, these results show that in astrocytes the activation of STAT3 and STAT5, but not STAT1 or STAT6, is dependent on GSK3, demonstrating that GSK3 inhibitors can be utilized to differentially modulate the activation of STAT family members. This selectivity may be a particularly attractive characteristic for application of GSK3 inhibitors because, for example, both STAT3 and STAT5 have been shown to be highly activated in certain types of cancer (35, 47).
Examination of isoform-specific effects of GSK3 revealed that GSK3β predominately promotes STAT3 activation at the receptor in the plasma membrane, whereas the effect of GSK3α was less clear. STATs activated by tyrosine phosphorylation accumulate in the nucleus bound to DNA (64), following which they are dephosphorylated and exported with an active half-life estimated to be 15 min for STAT1 (8). This short active half-life requires that STATs are exported from the nucleus to re-associate with the activated receptor-tyrosine kinase complex for reactivation. Both GSK3 (65) and STAT3 (34) contain nuclear localization sequences and undergo rapid nucleocytoplasmic shuttling under basal conditions. Because in the nucleus GSK3α was predominantly associated with STAT3, and siRNA-mediated knockdown of GSK3α increased nuclear levels of phospho-Tyr705-STAT3, GSK3α may play a role in promoting the inactivating dephosphorylation of Tyr705-STAT3 that leads to nuclear export, but this will require further investigation. This action would be consistent with a previous report showing that GSK3 promotes Dd-STATa export from the nucleus in Dictyostelium (66). These findings add to previous reports of isoform-specific regulatory actions of GSK3 on other transcription factors, such as NF-κB (20, 67) and Smad3/4 (67).
GSK3 is recognized as an integrator of many signaling pathways and the subsequent regulation of a large number of transcription factor effectors (17), and the results reported here extend this function of GSK3 to STAT3, a crucial component of inflammatory signaling. This adds STAT3 to other transcription factors involved in inflammatory signaling that are known to be regulated by GSK3, including NF-κB, CREB, and AP-1 (14, 15, 20, 21). These transcription factors act both independently to control the expression of inflammatory molecules and also interactively regulate each other. Thus, in contrast to interventions targeting individual transcription factors, inhibiting GSK3 enables simultaneous regulation of multiple transcription factors involved in inflammatory signaling. Regulation of inflammation continues to be a pressing problem because of the widespread association of excessive inflammation with many diseases. This is particularly true of neuroinflammation, which contributes to a broad range of neurodegenerative diseases (1–3). The present results show that inhibitors of GSK3 hold promise for therapeutic intervention of neuroinflammation based on the findings that GSK3 promotes STAT3 activation in primary astrocytes and microglia, a therapeutic potential strengthened by the rapidly growing armament of GSK3 inhibitors (32) that includes lithium, a drug already used therapeutically in psychiatric diseases (68).
Acknowledgments
We thank Dr. S. M. Michalek for the LPS, Dr. E. Benveniste for the RAW264.7 cells and for comments on the project, and Drs. M. Ozaki and C. Jobin for providing the Ad5STAT3C adenovirus.
This work was supported, in whole or in part, by National Institutes of Health Grants AG021045, MH38752, and NS37768. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: JAK, Janus kinase; CREB, cAMP response element-binding protein; GFAP, glial fibrillary acidic protein; GM-CSF, granulocyte-macrophage colony-stimulating factor; GSK3, glycogen synthase kinase-3; IL-4, interleukin-4; IFN, interferon; LPS, lipopolysaccharide; NF-κB, nuclear factor κB; STAT, signal transducer and activator of transcription; TLR, toll-like receptor; siRNA, small interference RNA; BIO, 6-bromoindirubin-3′-oxime; TDZD-8, 4-benzyl-2-methyl-1,2,4-thiadiazolidine-3,5-dione.
References
- 1.Campbell, I. L. (2005) Brain Res. Brain Res. Rev. 48 166–177 [DOI] [PubMed] [Google Scholar]
- 2.Maragakis, N. J., and Rothstein, J. D. (2006) Nat. Clin. Pract. Neurol. 2 679–689 [DOI] [PubMed] [Google Scholar]
- 3.Zipp, F., and Aktas, O. (2006) Trends Neurosci. 29 518–527 [DOI] [PubMed] [Google Scholar]
- 4.Levy, D. E., and Darnell, J. E., Jr. (2002) Nat. Rev. Mol. Cell. Biol. 3 651–662 [DOI] [PubMed] [Google Scholar]
- 5.Brierley, M. M., and Fish, E. N. (2005) J. Interferon Cytokine Res. 25 733–744 [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharya, S., and Schindler, C. (2003) J. Clin. Invest. 111 553–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lodige, I., Marg, A., Wiesner, B., Malecova, B., Oelgeschlager, T., and Vinkemeier, U. (2005) J. Biol. Chem. 280 43087–43099 [DOI] [PubMed] [Google Scholar]
- 8.Lerner, L., Henriksen, M. A., Zhang, X., and Darnell, J. E., Jr. (2003) Genes Dev. 17 2564–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonni, A., Sun, Y., Nadal-Vicens, M., Bhatt, A., Frank, D. A., Rozovsky, I., Stahl, N., Yancopoulos, G. D., and Greenberg, M. E. (1997) Science 278 477–483 [DOI] [PubMed] [Google Scholar]
- 10.He, F., Ge, W., Martinowich, K., Becker-Catania, S., Coskun, V., Zhu, W., Wu, H., Castro, D., Guillemot, F., Fan, G., de Vellis, J., and Sun, Y. E. (2005) Nat. Neurosci. 8 616–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jee, Y., Kim, G., Tanuma, N., and Matsumoto, Y. (2001) J. Neuroimmunol. 114 40–47 [DOI] [PubMed] [Google Scholar]
- 12.Justicia, C., Gabriel, C., and Planas, A. M. (2000) Glia 30 253–270 [DOI] [PubMed] [Google Scholar]
- 13.Sriram, K., Benkovic, S. A., Hebert, M. A., Miller, D. B., and O'Callaghan, J. P. (2004) J. Biol. Chem. 279 19936–19947 [DOI] [PubMed] [Google Scholar]
- 14.Martin, M., Rehani, K., Jope, R. S., and Michalek, S. M. (2005) Nat. Immunol. 6 777–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu, X., Paik, P. K., Chen, J., Yarilina, A., Kockeritz, L., Lu, T. T., Woodgett, J. R., and Ivashkiv, L. B. (2006) Immunity 24 563–574 [DOI] [PubMed] [Google Scholar]
- 16.Woodgett, J. R. (1990) EMBO J. 9 2431–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jope, R. S., and Johnson, G. V. (2004) Trends Biochem. Sci. 29 95–102 [DOI] [PubMed] [Google Scholar]
- 18.Jope, R. S., Yuskaitis, C. J., and Beurel, E. (2007) Neurochem. Res. 32 577–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshimura, A. (2006) Cancer Sci. 97 439–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoeflich, K. P., Luo, J., Rubie, E. A., Tsao, M. S., Jin, O., and Woodgett, J. R. (2000) Nature 406 86–90 [DOI] [PubMed] [Google Scholar]
- 21.Steinbrecher, K. A., Wilson, W., 3rd, Cogswell, P. C., and Baldwin, A. S. (2005) Mol. Cell. Biol. 25 8444–8455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirschfeld, M., Ma, Y., Weis, J. H., Vogel, S. N., and Weis, J. J. (2000) J. Immunol. 165 618–622 [DOI] [PubMed] [Google Scholar]
- 23.McCarthy, K. D., and de Vellis, J. (1980) J. Cell Biol. 85 890–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peineau, S., Taghibiglou, C., Bradley, C., Wong, T. P., Liu, L., Lu, J., Lo, E., Wu, D., Saule, E., Bouschet, T., Matthews, P., Isaac, J. T., Bortolotto, Z. A., Wang, Y. T., and Collingridge, G. L. (2007) Neuron 53 703–717 [DOI] [PubMed] [Google Scholar]
- 25.Zhou, W., Grandis, J. R., and Wells, A. (2006) Br. J. Cancer 95 164–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schroder, K., Sweet, M. J., and Hume, D. A. (2006) Immunobiology 211 511–524 [DOI] [PubMed] [Google Scholar]
- 27.Schroder, K., Hertzog, P. J., Ravasi, T., and Hume, D. A. (2004) J. Leukocyte Biol. 75 163–189 [DOI] [PubMed] [Google Scholar]
- 28.Qing, Y., and Stark, G. R. (2004) J. Biol. Chem. 279 41679–41685 [DOI] [PubMed] [Google Scholar]
- 29.Klein, P. S., and Melton, D. A. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 8455–8459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coghlan, M. P., Culbert, A. A., Cross, D. A., Corcoran, S. L., Yates, J. W., Pearce, N. J., Rausch, O. L., Murphy, G. J., Carter, P. S., Roxbee Cox, L., Mills, D., Brown, M. J., Haigh, D., Ward, R. W., Smith, D. G., Murray, K. J., Reith, A. D., and Holder, J. C. (2000) Chem. Biol. 7 793–803 [DOI] [PubMed] [Google Scholar]
- 31.Leclerc, S., Garnier, M., Hoessel, R., Marko, D., Bibb, J. A., Snyder, G. L., Greengard, P., Biernat, J., Wu, Y. Z., Mandelkow, E. M., Eisenbrand, G., and Meijer, L. (2001) J. Biol. Chem. 276 251–260 [DOI] [PubMed] [Google Scholar]
- 32.Martinez, A., Alonso, M., Castro, A., Perez, C., and Moreno, F. J. (2002) J. Med. Chem. 45 1292–1299 [DOI] [PubMed] [Google Scholar]
- 33.Stambolic, V., Ruel, L., and Woodgett, J. R. (1996) Curr. Biol. 6 1664–1668 [DOI] [PubMed] [Google Scholar]
- 34.Ma, J., Zhang, T., Novotny-Diermayr, V., Tan, A. L., and Cao, X. (2003) J. Biol. Chem. 278 29252–29260 [DOI] [PubMed] [Google Scholar]
- 35.Bromberg, J. F., Wrzeszczynska, M. H., Devgan, G., Zhao, Y., Pestell, R. G., Albanese, C., and Darnell, J. E., Jr. (1999) Cell 98 295–303 [DOI] [PubMed] [Google Scholar]
- 36.Li, L., and Shaw, P. E. (2006) J. Biol. Chem. 281 33172–33181 [DOI] [PubMed] [Google Scholar]
- 37.Brocke-Heidrich, K., Ge, B., Cvijic, H., Pfeifer, G., Loffler, D., Henze, C., McKeithan, T. W., and Horn, F. (2006) Oncogene 25 7297–7304 [DOI] [PubMed] [Google Scholar]
- 38.Hirano, T., Ishihara, K., and Hibi, M. (2000) Oncogene 19 2548–2556 [DOI] [PubMed] [Google Scholar]
- 39.Nakashima, K., Wiese, S., Yanagisawa, M., Arakawa, H., Kimura, N., Hisatsune, T., Yoshida, K., Kishimoto, T., Sendtner, M., and Taga, T. (1999) J. Neurosci. 19 5429–5434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams, L. M., Sarma, U., Willets, K., Smallie, T., Brennan, F., and Foxwell, B. M. (2007) J. Biol. Chem. 282 6965–6975 [DOI] [PubMed] [Google Scholar]
- 41.Blaskovich, M. A., Sun, J., Cantor, A., Turkson, J., Jove, R., and Sebti, S. M. (2003) Cancer Res. 63 1270–1279 [PubMed] [Google Scholar]
- 42.Stahl, N., Farruggella, T. J., Boulton, T. G., Zhong, Z., Darnell, J. E., Jr., and Yancopoulos, G. D. (1995) Science 267 1349–1353 [DOI] [PubMed] [Google Scholar]
- 43.Platanias, L. C. (2005) Nat. Rev. Immunol. 5 375–386 [DOI] [PubMed] [Google Scholar]
- 44.Coffer, P. J., van Puijenbroek, A., Burgering, B. M., Klop-de Jonge, M., Koenderman, L., Bos, J. L., and Kruijer, W. (1997) Oncogene 15 2529–2539 [DOI] [PubMed] [Google Scholar]
- 45.Wurster, A. L., Tanaka, T., and Grusby, M. J. (2000) Oncogene 19 2577–2584 [DOI] [PubMed] [Google Scholar]
- 46.Mui, A. L., Wakao, H., O'Farrell, A. M., Harada, N., and Miyajima, A. (1995) EMBO J. 14 1166–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bowman, T., Garcia, R., Turkson, J., and Jove, R. (2000) Oncogene 19 2474–2488 [DOI] [PubMed] [Google Scholar]
- 48.Murray, P. J. (2007) J. Immunol. 178 2623–2629 [DOI] [PubMed] [Google Scholar]
- 49.Bowman, C. C., Rasley, A., Tranguch, S. L., and Marriott, I. (2003) Glia 43 281–291 [DOI] [PubMed] [Google Scholar]
- 50.Carpentier, P. A., Begolka, W. S., Olson, J. K., Elhofy, A., Karpus, W. J., and Miller, S. D. (2005) Glia 49 360–374 [DOI] [PubMed] [Google Scholar]
- 51.Wang, Y., and Zhou, C. F. (2005) Glia 50 56–65 [DOI] [PubMed] [Google Scholar]
- 52.Ahmed, C. M., Burkhart, M. A., Mujtaba, M. G., Subramaniam, P. S., and Johnson, H. M. (2003) J. Cell Sci. 116 3089–3098 [DOI] [PubMed] [Google Scholar]
- 53.Doble, B. W., and Woodgett, J. R. (2003) J. Cell Sci. 116 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Espinosa, L., Ingles-Esteve, J., Aquilera, C., and Bigas, A. (2003) J. Biol. Chem. 278 32227–32235 [DOI] [PubMed] [Google Scholar]
- 55.Plotnikov, A., Li, Y., Tran, T. H., Tang, W., Palazzo, J. P., Rui, H., and Fuchs, S. Y. (2008) Cancer Res. 68 1354–1361 [DOI] [PubMed] [Google Scholar]
- 56.Zeng, X., Tamai, K., Doble, B., Li, S., Huang, H., Habas, R., Okamura, H., Woodgett, J., and He, X. (2005) Nature 438 873–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grimes, C. A., and Jope, R. S. (2001) Prog. Neurobiol. 65 391–426 [DOI] [PubMed] [Google Scholar]
- 58.Manoukian, A. S., and Woodgett, J. R. (2002) Adv. Cancer Res. 84 203–229 [DOI] [PubMed] [Google Scholar]
- 59.Woodgett, J. R. (2003) Curr. Drug Targets Immune Endocr. Metabol. Disord. 3 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beaulieu, J. M., Sotnikova, T. D., Yao, W. D., Kockeritz, L., Woodgett, J. R., Gainetdinov, R. R., and Caron, M. G. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 5099–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cardona-Gomez, P., Perez, M., Avila, J., Garcia-Segura, L. M., and Wandosell, F. (2004) Mol. Cell Neurosci. 25 363–373 [DOI] [PubMed] [Google Scholar]
- 62.Cross, D. A., Alessi, D. R., Cohen, P., Andjelkovich, M., and Hemmings, B. A. (1995) Nature 378 785–789 [DOI] [PubMed] [Google Scholar]
- 63.Wei, Q., Yokota, C., Semenov, M. V., Doble, B., Woodgett, J., and He, X. (2007) J. Biol. Chem. 282 15903–15911 [DOI] [PubMed] [Google Scholar]
- 64.Reich, N. C., and Liu, L. (2006) Nat. Rev. Immunol. 6 602–612 [DOI] [PubMed] [Google Scholar]
- 65.Meares, G. P., and Jope, R. R. (2007) J. Biol. Chem. 282 16989–17001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ginger, R. S., Dalton, E. C., Ryves, W. J., Fukuzawa, M., Williams, J. G., and Harwood, A. J. (2000) EMBO J. 19 5483–5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liang, M. H., and Chuang, D. M. (2006) J. Biol. Chem. 281 30479–30484 [DOI] [PubMed] [Google Scholar]
- 68.Jope, R. S. (1999) Mol. Psychiatry 4 117–128 [DOI] [PubMed] [Google Scholar]