Abstract
The term “oxidative stress” links the production of reactive oxygen species to a variety of metabolic outcomes, including insulin resistance, immune dysfunction, and inflammation. Antioxidant defense systems down-regulated due to disease and/or aging result in oxidatively modified DNA, carbohydrates, proteins, and lipids. Increased production of hydroxyl radical leads to the formation of lipid hydroperoxides that produce a family of α,β-unsaturated aldehydes. Such reactive aldehydes are subject to Michael addition reactions with the side chains of lysine, histidine, and cysteine residues, referred to as “protein carbonylation.” Although not widely appreciated, reactive lipids can accumulate to high levels in cells, resulting in extensive protein modification leading to either loss or gain of function. The use of mass spectrometric methods to identify the site and extent of protein carbonylation on a proteome-wide scale has expanded our view of how oxidative stress can regulate cellular processes.
Chemistry of Reactive Oxygen Species and Production of Reactive Aldehydes
ROS2 are formed as a result of numerous metabolic processes, including oxidation of NADPH by NADPH oxidase, uncoupling of the mitochondrial electron transport chain, and oxidation of xanthine by xanthine oxidase (1). Oxidative stress, which refers to a state of elevated levels of ROS, occurs from a variety of conditions that stimulate either ROS production or a decline in antioxidant defenses (2). In some cases such as stimulation of ROS production by macrophages as an innate immune response to bacterial infection, ROS are protective. However, the dysregulation of ROS levels in a variety of tissues has been linked to a number of inflammatory and age-associated disease states, including macular degeneration, muscular dystrophy, and insulin resistance associated with type 2 diabetes (3–5). During oxidative stress, the oxidation of cellular components results in the modification of DNA, proteins, lipids, and carbohydrates. In the case of proteins, numerous post-translational modifications have been characterized resulting either from direct oxidation of amino acid residues or through the formation of reactive intermediates by the oxidation of other cellular components. The oxidation of methionine to a sulfoxide as well as cysteine to sulfenic, sulfinic, and sulfonic acids has been shown to occur frequently and often can (with the exception of sulfonic acid) be reduced enzymatically. The oxidation of carbohydrates provides intermediates for reactions with proteins in the formation of advanced glycation end products, some of which are reactive toward protein (6). Furthermore, it has been observed that a significant portion of ROS-induced post-translational modifications result in the addition of reactive carbonyl functional groups on proteins, generically termed “protein carbonylation,” with the most reactive and common of these carbonyl groups being in the form of aldehydes.
Direct protein carbonylation can be achieved through a variety of reactions. Oxidation of amino acid side chains with metals and hydrogen peroxide is known to cause the formation of semialdehyde amino acids, with the majority of these reactions occurring with lysine, arginine, and proline residues (7). Alternatively, protein carbonylation can result from an indirect mechanism involving the hydroxyl radical-mediated oxidation of lipids. Polyunsaturated acyl chains of phospholipids or polyunsaturated fatty acids such as arachidonic acid and linoleic acid are highly susceptible to peroxidation and breakdown through non-enzymatic Hock cleavage, forming a variety of lipid-derived aldehydes and ketones (8). Lipid peroxidation products can diffuse across membranes, allowing the reactive aldehyde-containing lipids to covalently modify proteins localized throughout the cell and relatively far away from the initial site of ROS formation. Recent studies have suggested that protein carbonylation formed from lipid-derived aldehydes is more prevalent than that formed via direct amino acid side chain oxidation (9). The cellular metabolism of lipid peroxidation products and the fates of their protein targets are diagramed in Fig. 1.
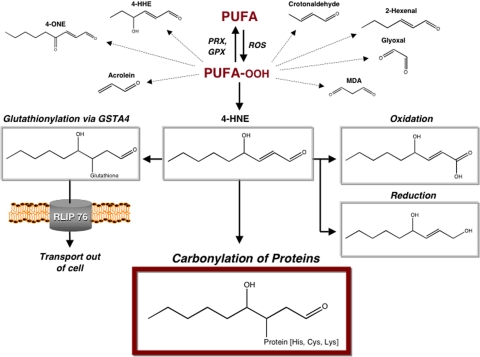
FIGURE 1.
Cellular fates of α,β-unsaturated aldehydes and carbonylated proteins. ROS stimulate peroxidation of polyunsaturated fatty acids (PUFA), an oxidative event that is reversible through reduction by peroxiredoxin (PRX) and glutathione peroxidase (GPX) enzymes. The lipid hydroperoxides (PUFA-OOH) generated are unstable and lead to a variety of reactive aldehydes. The lipid peroxidation products generated include the α,β-unsaturated aldehydes 4-HNE, 4-ONE, 4-hydroxy-(2E)-hexanal (4-HHE), (2E)-hexenal, crotonaldehyde, and acrolein as well as the dialdehydes glyoxal and malondialdehyde (MDA). GSTA4 catalyzes the conjugation of the highly reactive α,β-unsaturated aldehydes to glutathione, leading to their efflux from the cell by the glutathione conjugate transporter RLIP76. In addition, oxidation by aldehyde dehydrogenase or reduction by alcohol dehydrogenase, aldehyde reductase, or aldose reductase converts free aldehydes into less toxic molecules. The α,β-unsaturated aldehydes that escape cellular metabolism serve as electrophiles in the covalent modification of proteins via non-enzymatic Michael addition. The resulting aliphatic carbonyl adducts on cysteine, histidine, or lysine residues may alter the activity of protein targets or cause them to become degraded by the proteasome.
The most reactive of the aldehydes generated from polyunsaturated fatty acid oxidation are α,β-unsaturated aldehydes, including 4-HNE, 4-ONE, and acrolein (for an excellent recent review on the chemistry of lipid peroxidation, see Ref. 10). Because of the presence of electron-withdrawing functional groups, the double bond of 4-HNE or 4-ONE serves as a site for Michael addition with the sulfur atom of cysteine, the imidizole nitrogen of histidine, and, to a lesser extent, the amine nitrogen of lysine. There is some evidence for reaction with arginine as well, albeit to a lesser extent than with lysine. Although 4-HNE has historically been the most well studied lipid peroxidation product (11), 4-ONE is also highly reactive (12). After forming Michael adducts, the aldehyde moiety may in some cases undergo Schiff base formation with amines of adjacent lysines, producing intra- and/or intermolecular cross-linked amino acids (13, 14). In the case of 4-ONE, modification of lysine residues through 1,2-addition (Schiff base formation and the addition of water to the double bond) can also result in ketoamide adducts (15). In model systems, mass spectrometric analysis has demonstrated that >99% of proteins modified in vitro by 4-HNE retain a free carbonyl group (16).
A variety of antioxidant enzymes and proteins function to eliminate reactive lipid peroxidation products (17). Lipid hydroperoxides can be reduced via peroxiredoxins and glutathione peroxidases, thereby preventing reactive aldehyde formation. Although aldehyde dehydrogenase converts (4R)-HNE into a carboxylic acid and alcohol dehydrogenase, and aldehyde reductase and aldose reductase convert the aldehyde into the corresponding alcohol (18–20), leading to markedly reduced reactivity of the lipid, a major route of detoxification is via glutathionylation by GST. Of the various GST isoforms, GSTA4 exhibits the greatest specificity for 4-HNE (21, 22). Once 4-HNE/glutathione adducts have been formed, they are removed from the cell by the 76-kDa Ral-binding GTPase-activating protein RLIP76. Although the quenching of 4-HNE occurs largely via GSTA4-controlled metabolism, GSTA4 efficiently recognizes only the S-stereoisomer of 4-HNE (23). Finally, cytoplasmic FABP can scavenge both the R- and S-stereoisomers via covalent protein adduction (24).
Functional Outcomes of Protein Modification
Lipid peroxidation products have been shown to have a wide variety of effects on cells in vitro depending upon the concentration utilized, and as such, interpretation of experimental results must be considered cautiously. For example, treatment of epithelial and smooth muscle cells with physiological concentrations of 4-HNE (∼0.1–1 μm) promoted cellular proliferation, whereas treatment of erythroleukemic cells with supraphysiological concentrations (20–40 μm) induced cell cycle arrest, erythrocyte differentiation, and apoptosis (25, 26).
Because the side chains of Cys, His, and Lys are often used in catalysis, the most common effect of protein carbonylation is enzyme inactivation. The inactivation of several membrane transporters, including the Na+-K+-ATPase and glucose (GLUT3) transporters, by lipid-derived aldehydes in the brain has been linked to neurodegenerative disorders (27, 28). The modification of the adipocyte FABP (as well as the epithelial isoform) by 4-HNE occurs on a conserved cysteine residue (Cys117), decreases the protein's affinity for fatty acids, and may contribute to obesity-linked insulin resistance (24, 29). Cytosolic and mitochondrial NADP+-dependent isocitrate dehydrogenase isoforms are inactivated by lipid peroxidation products, which may lead to the dysregulation of NADPH levels (30). The inactivation of thioredoxin and thioredoxin reductase through modification of their active-site cysteine and selenocysteine residues by 4-HNE and acrolein has been linked to dysregulation of cellular redox status and stress signaling (31–33). Likewise, the inactivation of glutathione peroxidase by modification with methylglyoxal on an arginine residue at its glutathione-binding site amplifies oxidative stress by increasing peroxide levels in the cell (34). In addition, Hsp90 and protein-disulfide isomerase have also recently been shown to be inactivated by modification with reactive aldehydes (35, 36).
Targeted degradation of carbonylated proteins occurs via at least two different mechanisms. The 20 S ubiquitin-independent proteasome, which degrades misfolded proteins based on its ability to detect exposed hydrophobic residues, is responsible for the degradation of many carbonylated proteins (37). However, the 26 S proteasome has also been demonstrated to have a role in the degradation of modified proteins after they have undergone ubiquitination, as is the case for alcohol dehydrogenase (38). Although targeted degradation minimizes the amount of carbonylated proteins during conditions of mild oxidative stress, the 4-HNE modification and inhibition of the proteasome machinery itself amplify the accumulation of modified and misfolded proteins during conditions of increased ROS (39).
Although carbonylation most typically inactivates protein targets, such modification can also result in a gain of function for certain metabolic signaling systems. For example, transcriptional activation of antioxidant-response genes is up-regulated by protein carbonylation. Several genes containing antioxidant-responsive elements are activated by 4-HNE-linked processes. Nrf2 (NF-E2-related factor 2) is a central transcription factor involved in the regulation of antioxidant-responsive element-containing genes that are often activated in response to oxidative stress. The alkylation of the cytoplasmic inhibitor of Nrf2, Keap1 (Kelch-like ECH-associated protein 1), by 4-HNE and other electrophiles results in the dissociation of the Keap1-Nrf2 complex. Once freed from inhibition by Keap1, Nrf2 translocates to the nucleus and activates the expression of antioxidant-responsive element-containing genes, increasing antioxidant defenses (40).
The ability of α,β-unsaturated aldehydes to regulate inflammation, apoptosis, and other cellular signaling (41) has largely been attributed to regulation of critical signaling kinases through loss- and gain-of-function modifications, frequently on activation loop cysteine or histidine residues in the case of inhibitory modifications. The 4-HNE modification of extracellular signal-regulated kinase (ERK) on His178 inhibits its ability to become phosphorylated and, as a result, decreases its kinase activity (42). Similarly, the modification of AMP kinase kinase (LKB1/STK11) with 4-HNE, 4-ONE, and a variety of other reactive lipids on activation loop residue Cys120 inhibits its kinase activity and attenuates downstream AMP kinase signaling (43). In addition, IKKβ is inactivated by 4-HNE modification, although the site has yet to be determined (44). It will be interesting to determine whether IKKβ inactivation by 4-HNE is due to modification on Cys179, as inhibition of IKKβ with cyclopentenone prostaglandins, which are also α,β-unsaturated carbonyl-containing lipids that form Michael addition adducts (45), has been mapped to this thiol. Reactive aldehydes have also been reported to stimulate the activities of certain kinases. For example, the JNK upstream kinase ASK1 is activated by 4-HNE, leading to stimulation of the ASK1-SEK1-JNK pathway linking oxidative stress to inflammation (46). In addition, the epidermal growth factor receptor is activated by 4-HNE in the absence of ligand binding by inducing clustering and autophosphorylation (47). However, the specific amino acid targets and mechanisms responsible for both epidermal growth factor receptor and JNK activation by 4-HNE are still unclear and warrant future investigation.
Mass Spectrometric Methods for Detecting and Quantifying Protein Carbonylation
The emergence of two modern ionization technologies, matrix-assisted laser desorption ionization and electrospray ionization, has enabled the direct structural analysis of aldehyde modification in single-protein models as reviewed previously (10, 48). Although these techniques have proven useful for investigating protein carbonylation in simple in vitro model systems, studies of these proteins do not necessarily reflect accurately their endogenous modification state. Therefore, to better understand the role of carbonyl modifications in vivo, more advanced methods are required to characterize directly modified proteins isolated from complex biological systems.
For the study of proteins containing endogenous reactive carbonyl modifications, large-scale mass spectrometry-based proteomic methods offer the potential to discover new protein targets susceptible to these modifications, to quantitatively profile changes in these modifications with disease state or aging, and to characterize the exact amino acid site and type of carbonyl modification via MS/MS analysis and sequence data base searching (49). Similar to the proteomic study of other post-translational modifications to proteins, reactive carbonyl-modified components generally make up a relatively small proportion of the total proteins within a complex biological sample. Therefore, methods to enrich for this subset of modified proteins prior to MS analysis are generally necessary. Proteomic methods that have been described for carbonylated protein analysis in complex systems are summarized in supplemental Fig. 1 and discussed below.
Two-dimensional Gel-based Approaches
2DGE has frequently been used for the identification of carbonylated proteins in complex mixtures. Specific detection of gel-separated reactive carbonyl-containing proteins is then achieved via immunoblotting, in some cases using antibodies that directly recognize the carbonyl modification on the protein (e.g. anti-4-HNE antibodies). In other cases, reactive carbonyls are labeled covalently with nucleophilic hydrazide- or hydrazine-based probes. These groups have highly specific reactivity with aldehydes (and to a far lesser extent, other carbonyls such as ketones), forming a covalent Schiff base that can then be reduced to a highly stable carbon–nitrogen single bond. For example, labeling of reactive carbonyl-containing proteins prior to 2DGE with DNPH, followed by immunoblotting with anti-DNPH antibodies (commercially marketed as OxyBlot™), has been commonly used. Many times, two gels are run in parallel, and one is immunoblotted to visualize the migration pattern of carbonylated proteins, whereas the other is stained for total protein so that spots corresponding to the locations of the carbonylated proteins can be excised, in gel-digested with trypsin, and identified by mass spectrometry.
2DGE coupled with immunostaining and MS analysis has been used in a variety of studies. Epithelial FABP has been shown to be carbonylated in vivo by 4-HNE modification using a 2DGE approach (29). Ferrington and Kapphahn (39) identified subunits of rat liver 20 S proteasome containing HNE adducts by a similar antibody-based detection method, providing important details regarding the catalytic site-specific inhibition of the proteasome by 4-HNE. This approach was also used in the identification of in vivo 4-HNE-modified retinal proteins from young and old rat eyes, cultured ARPE19 cells, and human donor eyes (50). Alternatively, the OxyBlot™ detection strategy was used to identify six carbonylated proteins from Alzheimer disease brain (51, 52), providing a step forward in understanding the relationship between oxidative modifications and neuronal death. An alternative method for visualizing oxidized proteins, relying on similar hydrazide chemistry, was also developed using biotinylation and avidin-fluorescein isothiocyanate affinity staining (53). A limitation of all hydrazide-based labeling methods described above is the general reactivity of hydrazide to all reactive carbonyl modifications, precluding the identification of the exact type of carbonyl modification (e.g. lipid-derived reactive aldehyde or the result of direct amino acid side chain oxidation).
Gel-free Proteomic Approaches
Despite the contributions of 2DGE-based methods to proteomic studies of reactive carbonyl-modified proteins, 2DGE has a number of well described (54) limitations as a general platform for large-scale proteomic studies, including analysis of membrane and low-abundance proteins. Therefore, “gel free”-based proteomic methods have begun to emerge. For these methods, carbonylated proteins are enriched from complex mixtures using affinity methods (outlined in blue boxes in supplemental Fig. 1) (55–57). The enriched proteins are then digested by peptides, and the putatively carbonyl-modified proteins are identified via LC-MS/MS analysis.
This general gel-free method has been used in a number of different studies. One of the first descriptions identified proteins in aged mouse brain homogenates (55). Carbonylation was found on several low-abundance receptor proteins, mitochondrial proteins, and tyrosine phosphatases known to be associated with insulin and insulin-like growth factor metabolism and cell signaling pathways. Another study used multidimensional LC-MS/MS of enriched protein digests to identify targets of amyloid β-induced oxidative stress in cultured primary cortical mouse neurons (58), identifying the carbonylation of the Golgi-resident enzyme glucuronyltransferase. Proteins susceptible to reactive carbonyl modification in rat liver homogenates from nitropropane-treated animals have also been identified using this method (56). In work investigating the possible linkage between obesity and oxidative protein damage, such technology was utilized to identify aldehyde-modified proteins from adipose tissue of lean insulin-sensitive and obese insulin-resistant C57BL/6J mice (24). Adipocyte FABP, a protein implicated in the regulation of insulin resistance, was found to be a target of 4-HNE modification in vivo. An extension of this technology utilizes stable isotope labeling of enriched biotin hydrazide-labeled protein digests prior to μLC-MS/MS analysis with iTRAQ™ reagent (59). Applying such analysis to carbonylated proteins from rat muscle mitochondria (60) distinguished carbonylated proteins from non-carbonylated background proteins that may nonspecifically bind the avidin column.
Strategies for Identifying the Site and Type of Carbonyl Modification
Despite the insights provided by the studies above, the methods employed have generally lacked the ability to directly identify the exact amino acid site and type of carbonyl modification. This information is necessary for a deeper understanding of the oxidative mechanisms leading to the protein modification as well as for providing information needed for follow-up studies to assess the functional effects that modification of specific amino acid sites may have on the protein.
Therefore, numerous proteomic methods for the direct identification of reactive carbonyl-modified amino acid sites within peptides have begun to emerge. The use of DNPH as a reactive matrix has been shown to increase the sensitivity for carbonyl-modified peptides when analyzing digested proteins after separation by 2DGE (61). A number of gel-free methods have been developed that use enrichment of carbonylated peptides, followed by direct μLC-MS/MS analysis (see right side of supplemental Fig. 1). One such method (62) labels and enriches carbonylated peptides with Girard reagent P. We have developed a solid-phase “capture and release” strategy utilizing reversible hydrazide chemistry for enriching carbonylated peptides (63). This method, followed by LC-electrospray ionization-MS/MS, was applied to identify 4-HNE modification sites in a digest of a yeast lysate treated with 4-HNE. An alternative method used N′-aminooxymethylcarbonylhydro-d-biotin (64) to label in vitro 4-HNE-modified proteins in rat cardiac mitochondria, followed by tryptic digestion, avidin column enrichment of the modified peptides, and MS/MS analysis. Although these advances are promising, none have been shown to reliably identify carbonylated amino acid sites in low abundance on endogenously modified proteins from complex mixtures, highlighting the need for continued development of more sensitive methods.
Strategies for Quantifying Modification with Reactive Carbonyls
As an alternative to the Levine-Stadtman procedure (DNPH assay) (65) for measuring bulk levels of carbonylated proteins from biological samples, gel-free quantitative proteomic methods have begun to emerge. These methods couple stable isotope labeling of peptides derived from enriched carbonylated proteins or peptides and LC-MS/MS analysis. Multiplexed labeling of digests of enriched carbonylated proteins from yeast lysates with heavy and light isotope-coded Girard reagent P, followed by LC-MS/MS analysis, is one such promising method (66), along with the recently described hydrazide-functionalized isotope-coded affinity tag approach (67). Parallel approaches have used multiplexed iTRAQ™ reagent to label digests of enriched carbonylated proteins from rat muscle mitochondria with stable isotopes, followed by μLC-MS/MS analysis (60).
Challenges and Future Directions
It has become increasingly clear that modification of proteins by reactive aldehydes leads to loss and/or gain of function of target proteins linked to disease states, signaling systems, and age-related conditions. However, despite this growing body of knowledge, a main question still remains as to whether these modifications are causative or consequential factors in their observed association with disease and other conditions. Mass spectrometry-based proteomic methods have begun to provide the tools to help answer this pivotal question, although further advances are still necessary. These include more sensitive and reliable methods to directly identify the site and type of endogenous carbonyl protein modifications and characterization of the stoichiometry of in vivo protein carbonylation, both essential for understanding the possible functional effects of these modifications.
Acknowledgments
We thank Drs. Edgar Arriaga and Deborah Ferrington for critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant AG25371 (to T. J. G.), Grant T32 HL07741 from NHLBI (to P. A. G.), and Grant DK053189 (to D. A. B.). This work was also supported by an award from Eli Lilly and Co. (to T. J. G.) and by the Minnesota Agricultural Experiment Station and the Minnesota Obesity Center (to D. A. B.). This minireview will be reprinted in the 2008 Minireview Compendium, which will be available in January, 2009.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: ROS, reactive oxygen species; 4-HNE, 4-hydroxy-(2E)-nonenal; 4-ONE, 4-oxo-(2E)-nonenal; GST, glutathione S-transferase; FABP, fatty acid-binding protein; IKKβ, IκB kinase β; JNK, c-Jun N-terminal kinase; MS/MS, tandem mass spectrometry; 2DGE, two-dimensional PAGE; DNPH, dinitrophenylhydrazine; LC, liquid chromatography; μLC, microcapillary reversed-phase LC.
References
- 1.Sayre, L. M., Smith, M. A., and Perry, G. (2001) Curr. Med. Chem. 8 721–738 [DOI] [PubMed] [Google Scholar]
- 2.Schafer, F. Q., and Buettner, G. R. (2001) Free Radic. Biol. Med. 30 1191–1212 [DOI] [PubMed] [Google Scholar]
- 3.Beatty, S., Koh, H., Phil, M., Henson, D., and Boulton, M. (2000) Surv. Ophthalmol. 45 115–134 [DOI] [PubMed] [Google Scholar]
- 4.Tidball, J. G., and Wehling-Henricks, M. (2007) J. Appl. Physiol. 102 1677–1686 [DOI] [PubMed] [Google Scholar]
- 5.Eriksson, J. W. (2007) FEBS Lett. 581 3734–3742 [DOI] [PubMed] [Google Scholar]
- 6.Stadtman, E. R. (2006) Free Radic. Res. 40 1250–1258 [DOI] [PubMed] [Google Scholar]
- 7.Stadtman, E. R., and Berlett, B. S. (1991) J. Biol. Chem. 266 17201–17211 [PubMed] [Google Scholar]
- 8.Schneider, C., Tallman, K. A., Porter, N. A., and Brash, A. R. (2001) J. Biol. Chem. 276 20831–20838 [DOI] [PubMed] [Google Scholar]
- 9.Yuan, Q., Zhu, X., and Sayre, L. M. (2007) Chem. Res. Toxicol. 20 129–139 [DOI] [PubMed] [Google Scholar]
- 10.Sayre, L. M., Lin, D., Yuan, Q., Zhu, X., and Tang, X. (2006) Drug Metab. Rev. 38 651–675 [DOI] [PubMed] [Google Scholar]
- 11.Esterbauer, H., Schaur, R. J., and Zollner, H. (1991) Free Radic. Biol. Med. 11 81–128 [DOI] [PubMed] [Google Scholar]
- 12.Lin, D., Lee, H. G., Liu, Q., Perry, G., Smith, M. A., and Sayre, L. M. (2005) Chem. Res. Toxicol. 18 1219–1231 [DOI] [PubMed] [Google Scholar]
- 13.Oe, T., Arora, J. S., Lee, S. H., and Blair, I. A. (2003) J. Biol. Chem. 278 42098–42105 [DOI] [PubMed] [Google Scholar]
- 14.Zhang, W. H., Liu, J., Xu, G., Yuan, Q., and Sayre, L. M. (2003) Chem. Res. Toxicol. 16 512–523 [DOI] [PubMed] [Google Scholar]
- 15.Williams, M. V., Wishnok, J. S., and Tannenbaum, S. R. (2007) Chem. Res. Toxicol. 20 767–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruenner, B. A., Jones, A. D., and German, J. B. (1995) Chem. Res. Toxicol. 8 552–559 [DOI] [PubMed] [Google Scholar]
- 17.Yang, Y., Sharma, R., Sharma, A., Awasthi, S., and Awasthi, Y. C. (2003) Acta Biochim. Pol. 50 319–336 [PubMed] [Google Scholar]
- 18.Honzatko, A., Brichac, J., Murphy, T. C., Reberg, A., Kubatova, A., Smoliakova, I. P., and Picklo, M. J., Sr. (2005) Free Radic. Biol. Med. 39 913–924 [DOI] [PubMed] [Google Scholar]
- 19.Canuto, R. A., Ferro, M., Muzio, G., Bassi, A. M., Leonarduzzi, G., Maggiora, M., Adamo, D., Poli, G., and Lindahl, R. (1994) Carcinogenesis 15 1359–1364 [DOI] [PubMed] [Google Scholar]
- 20.Poli, G., Schaur, R. J., Siems, W. G., and Leonarduzzi, G. (2008) Med. Res. Rev. 28 569–631 [DOI] [PubMed] [Google Scholar]
- 21.Engle, M. R., Singh, S. P., Czernik, P. J., Gaddy, D., Montague, D. C., Ceci, J. D., Yang, Y., Awasthi, S., Awasthi, Y. C., and Zimniak, P. (2004) Toxicol. Appl. Pharmacol. 194 296–308 [DOI] [PubMed] [Google Scholar]
- 22.Bruns, C. M., Hubatsch, I., Ridderstrom, M., Mannervik, B., and Tainer, J. A. (1999) J. Mol. Biol. 288 427–439 [DOI] [PubMed] [Google Scholar]
- 23.Hiratsuka, A., Tobita, K., Saito, H., Sakamoto, Y., Nakano, H., Ogura, K., Nishiyama, T., and Watabe, T. (2001) Biochem. J. 355 237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimsrud, P. A., Picklo, M. J., Sr., Griffin, T. J., and Bernlohr, D. A. (2007) Mol. Cell. Proteomics 6 624–637 [DOI] [PubMed] [Google Scholar]
- 25.Awasthi, Y. C., Sharma, R., Cheng, J. Z., Yang, Y., Sharma, A., Singhal, S. S., and Awasthi, S. (2003) Mol. Aspects Med. 24 219–230 [DOI] [PubMed] [Google Scholar]
- 26.Fazio, V. M., Rinaldi, M., Ciafre, S., Barrera, G., and Farace, M. G. (1993) Mol. Aspects Med. 14 217–228 [DOI] [PubMed] [Google Scholar]
- 27.Miyake, H., Kadoya, A., and Ohyashiki, T. (2003) Biol. Pharm. Bull. 26 1652–1656 [DOI] [PubMed] [Google Scholar]
- 28.Mark, R. J., Pang, Z., Geddes, J. W., Uchida, K., and Mattson, M. P. (1997) J. Neurosci. 17 1046–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennaars-Eiden, A., Higgins, L., Hertzel, A. V., Kapphahn, R. J., Ferrington, D. A., and Bernlohr, D. A. (2002) J. Biol. Chem. 277 50693–50702 [DOI] [PubMed] [Google Scholar]
- 30.Yang, J. H., Yang, E. S., and Park, J. W. (2004) Free Radic. Res. 38 241–249 [DOI] [PubMed] [Google Scholar]
- 31.Fang, J., and Holmgren, A. (2006) J. Am. Chem. Soc. 128 1879–1885 [DOI] [PubMed] [Google Scholar]
- 32.Cassidy, P. B., Edes, K., Nelson, C. C., Parsawar, K., Fitzpatrick, F. A., and Moos, P. J. (2006) Carcinogenesis 27 2538–2549 [DOI] [PubMed] [Google Scholar]
- 33.Park, Y. S., Misonou, Y., Fujiwara, N., Takahashi, M., Miyamoto, Y., Koh, Y. H., Suzuki, K., and Taniguchi, N. (2005) Biochem. Biophys. Res. Commun. 327 1058–1065 [DOI] [PubMed] [Google Scholar]
- 34.Park, Y. S., Koh, Y. H., Takahashi, M., Miyamoto, Y., Suzuki, K., Dohmae, N., Takio, K., Honke, K., and Taniguchi, N. (2003) Free Radic. Res. 37 205–211 [DOI] [PubMed] [Google Scholar]
- 35.Carbone, D. L., Doorn, J. A., Kiebler, Z., Ickes, B. R., and Petersen, D. R. (2005) J. Pharmacol. Exp. Ther. 315 8–15 [DOI] [PubMed] [Google Scholar]
- 36.Carbone, D. L., Doorn, J. A., Kiebler, Z., and Petersen, D. R. (2005) Chem. Res. Toxicol. 18 1324–1331 [DOI] [PubMed] [Google Scholar]
- 37.Grune, T., and Davies, K. J. (2003) Mol. Aspects Med. 24 195–204 [DOI] [PubMed] [Google Scholar]
- 38.Carbone, D. L., Doorn, J. A., and Petersen, D. R. (2004) Free Radic. Biol. Med. 37 1430–1439 [DOI] [PubMed] [Google Scholar]
- 39.Ferrington, D. A., and Kapphahn, R. J. (2004) FEBS Lett. 578 217–223 [DOI] [PubMed] [Google Scholar]
- 40.Levonen, A. L., Landar, A., Ramachandran, A., Ceaser, E. K., Dickinson, D. A., Zanoni, G., Morrow, J. D., and Darley-Usmar, V. M. (2004) Biochem. J. 378 373–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leonarduzzi, G., Robbesyn, F., and Poli, G. (2004) Free Radic. Biol. Med. 37 1694–1702 [DOI] [PubMed] [Google Scholar]
- 42.Sampey, B. P., Carbone, D. L., Doorn, J. A., Drechsel, D. A., and Petersen, D. R. (2007) Mol. Pharmacol. 71 871–883 [DOI] [PubMed] [Google Scholar]
- 43.Wagner, T. M., Mullally, J. E., and Fitzpatrick, F. A. (2006) J. Biol. Chem. 281 2598–2604 [DOI] [PubMed] [Google Scholar]
- 44.Ji, C., Kozak, K. R., and Marnett, L. J. (2001) J. Biol. Chem. 276 18223–18228 [DOI] [PubMed] [Google Scholar]
- 45.Rossi, A., Kapahi, P., Natoli, G., Takahashi, T., Chen, Y., Karin, M., and Santoro, M. G. (2000) Nature 403 103–108 [DOI] [PubMed] [Google Scholar]
- 46.Soh, Y., Jeong, K. S., Lee, I. J., Bae, M. A., Kim, Y. C., and Song, B. J. (2000) Mol. Pharmacol. 58 535–541 [DOI] [PubMed] [Google Scholar]
- 47.Liu, W., Akhand, A. A., Kato, M., Yokoyama, I., Miyata, T., Kurokawa, K., Uchida, K., and Nakashima, I. (1999) J. Cell Sci. 112 2409–2417 [DOI] [PubMed] [Google Scholar]
- 48.Carini, M., Aldini, G., and Facino, R. M. (2004) Mass Spectrom. Rev. 23 281–305 [DOI] [PubMed] [Google Scholar]
- 49.Sadygov, R. G., Cociorva, D., and Yates, J. R., III (2004) Nat. Methods 1 195–202 [DOI] [PubMed] [Google Scholar]
- 50.Kapphahn, R. J., Giwa, B. M., Berg, K. M., Roehrich, H., Feng, X., Olsen, T. W., and Ferrington, D. A. (2006) Exp. Eye Res. 83 165–175 [DOI] [PubMed] [Google Scholar]
- 51.Castegna, A., Aksenov, M., Thongboonkerd, V., Klein, J. B., Pierce, W. M., Booze, R., Markesbery, W. R., and Butterfield, D. A. (2002) J. Neurochem. 82 1524–1532 [DOI] [PubMed] [Google Scholar]
- 52.Castegna, A., Aksenov, M., Aksenova, M., Thongboonkerd, V., Klein, J. B., Pierce, W. M., Booze, R., Markesbery, W. R., and Butterfield, D. A. (2002) Free Radic. Biol. Med. 33 562–571 [DOI] [PubMed] [Google Scholar]
- 53.Yoo, B. S., and Regnier, F. E. (2004) Electrophoresis 25 1334–1341 [DOI] [PubMed] [Google Scholar]
- 54.Roe, M. R., and Griffin, T. J. (2006) Proteomics 6 4678–4687 [DOI] [PubMed] [Google Scholar]
- 55.Soreghan, B. A., Yang, F., Thomas, S. N., Hsu, J., and Yang, A. J. (2003) Pharm. Res. (N. Y.) 20 1713–1720 [DOI] [PubMed] [Google Scholar]
- 56.Mirzaei, H., and Regnier, F. (2005) Anal. Chem. 77 2386–2392 [DOI] [PubMed] [Google Scholar]
- 57.Mirzaei, H., and Regnier, F. (2007) J. Chromatogr. A 1141 22–31 [DOI] [PubMed] [Google Scholar]
- 58.Thomas, S. N., Soreghan, B. A., Nistor, M., Sarsoza, F., Head, E., and Yang, A. J. (2005) J. Neurochem. 92 705–717 [DOI] [PubMed] [Google Scholar]
- 59.Ross, P. L., Huang, Y. N., Marchese, J. N., Williamson, B., Parker, K., Hattan, S., Khainovski, N., Pillai, S., Dey, S., Daniels, S., Purkayastha, S., Juhasz, P., Martin, S., Bartlet-Jones, M., He, F., Jacobson, A., and Pappin, D. J. (2004) Mol. Cell. Proteomics 3 1154–1169 [DOI] [PubMed] [Google Scholar]
- 60.Meany, D. L., Xie, H., Thompson, L. V., Arriaga, E. A., and Griffin, T. J. (2007) Proteomics 7 1150–1163 [DOI] [PubMed] [Google Scholar]
- 61.Fenaille, F., Tabet, J. C., and Guy, P. A. (2004) Anal. Chem. 76 867–873 [DOI] [PubMed] [Google Scholar]
- 62.Mirzaei, H., and Regnier, F. (2006) Anal. Chem. 78 770–778 [DOI] [PubMed] [Google Scholar]
- 63.Roe, M. R., Xie, H., Bandhakavi, S., and Griffin, T. J. (2007) Anal. Chem. 79 3747–3756 [DOI] [PubMed] [Google Scholar]
- 64.Chavez, J., Wu, J., Han, B., Chung, W. G., and Maier, C. S. (2006) Anal. Chem. 78 6847–6854 [DOI] [PubMed] [Google Scholar]
- 65.Levine, R. L., Williams, J. A., Stadtman, E. R., and Shacter, E. (1994) Methods Enzymol. 233 346–357 [DOI] [PubMed] [Google Scholar]
- 66.Mirzaei, H., and Regnier, F. (2006) J. Chromatogr. A 1134 122–133 [DOI] [PubMed] [Google Scholar]
- 67.Han, B., Stevens, J. F., and Maier, C. S. (2007) Anal. Chem. 79 3342–3354 [DOI] [PubMed] [Google Scholar]