Abstract
Adhesive PfEMP1 proteins are displayed on the surface of malaria-infected red blood cells. They play a critical role in the disease, tethering infected cells away from destruction by the spleen and causing many severe symptoms. A molecular understanding of how these domains maintain their binding properties while evading immune detection will be important in developing therapeutics. In malaria of pregnancy, domains from the var2csa-encoded PfEMP1 protein interact with chondroitin sulfate on the placenta surface. This causes accumulation of infected red blood cells, leading to placental inflammation and block of blood flow to the developing fetus. This is associated with maternal anemia, low birth weight, and premature delivery and can lead to the death of mother and child. Here I present the structure of the chondroitin sulfate-binding DBL3X domain from a var2csa-encoded PfEMP1 protein. The domain adopts a fold similar to malarial invasion proteins, with extensive loop insertions. One loop is flexible in the unliganded structure but observed in the presence of sulfate or disaccharide, where it completes a sulfate-binding site. This loop, and others surrounding this putative carbohydrate-binding site, are flexible and polymorphic, perhaps protecting the binding site from immune detection. This suggests a model for how the domain maintains ligand binding while evading the immune response and will guide future drug and vaccine development.
Malaria is the most deadly parasitic disease affecting humanity, causing 500 million serious cases and 2 million deaths each year (1). People living in endemic countries develop partial immunity after multiple disease episodes, and this immunity correlates with acquisition of strain-specific antibodies that recognize PfEMP12 proteins (2, 3). These proteins are expressed on the surface of infected red blood cells, where they bind to human receptors, tethering the cells away from spleen-mediated destruction (4–6). This protects the parasite and prolongs the infection. These interactions also cause infected cells to accumulate in the brain and on the placenta during cerebral malaria and malaria of pregnancy, leading to some of the most deadly symptoms of the disease (2, 7).
As PfEMP1 proteins must be surface-exposed for their adhesive functions, they are also exposed to the immune system, and to avoid immune detection, they undergo antigenic variation. The genome of the 3D7 strain of Plasmodium falciparum contains 59 genes for PfEMP1 proteins (8). One gene is expressed at a time, and the parasite avoids detection by switching which PfEMP1 is produced (5). PfEMP1 domains also show significant polymorphism, particularly in strains from different locations, allowing them to overcome resistance (9, 10). A molecular understanding of how Duffy binding-like (DBL) domains maintain their ligand binding properties while evading immune detection will be important in development of therapeutics to target severe malaria.
Pregnant women are particularly susceptible to malaria as the placenta provides a new target for the adhesion of infected erythrocytes. These erythrocytes are tethered to the placenta surface through interactions between PfEMP1 domains and chondroitin sulfate A (CSA) (11–13), leading to inflammation and block of blood flow to the developing child (14, 15). This causes the death or underweight birth of many children, killing an estimated 75,000–200,000 fetuses annually (13). Immunity develops following multiple pregnancies and is associated with antibodies that bind to PfEMP1 proteins (16) and block CSA binding (3, 17, 18), suggesting the CSA-PfEMP1 interaction as the principle target for development of therapeutics to target placental malaria.
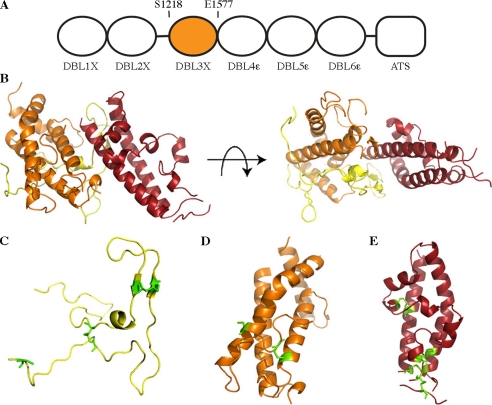
The PfEMP1s are multidomain adhesive proteins (4–6) containing 2–10 discrete extracellular domains that mostly fall into two families, the cysteine-rich interdomain region, and the DBL domains (8). These have important binding properties, interacting with a variety of human receptors (3, 19–21). Although several DBL domains have been shown to interact with CSA, the most likely CSA receptor in placental malaria is the product from the var2csa gene (13, 19). This protein consists of six DBL domains, a single transmembrane helix, and a cytoplasmic acidic terminal sequence (ATS) (see Fig. 1A). Expression of var2csa is closely associated with malaria of pregnancy and three of the DBL domains, including the DBL3X domain, have been shown to bind CSA (9, 22, 23). This study describes the structure of the var2csa DBL3X domain from the A4 strain of P. falciparum.
FIGURE 1.
Structure of a chondroitin sulfate A-binding DBL3X domain. A, the var2csa protein consists of six extracellular DBL domains followed by a single transmembrane helix and a cytoplasmic acidic terminal sequence (ATS). Residues 1218–1577 form the DBL3X domain. B, orthogonal views of the DBL3X domain with subdomain 1 in yellow, subdomain 2 in orange, and subdomain 3 in red. C–E, subdomains 1, 2, and 3, respectively, with residues involved in disulfide bonds highlighted in green.
EXPERIMENTAL PROCEDURES
Crystallization and Data Collection—The DBL3X domain of var2csa (accession codes AY372123 and AAQ73926) was cloned from A4 strain genomic DNA, expressed in the Origami B strain of Escherichia coli, and purified and crystallized as described in Ref. 24. The crystals that diffracted to the highest resolution were grown in 25% PEG 4000, 20 mm Tris, pH 8.0. They were cryoprotected by transfer into 25% PEG 4000, 20 mm Tris, pH 8.0, 25% glycerol and flash-frozen in liquid nitrogen. Diffraction data from native crystals were collected at 100 K using a copper anode fitted on a Rigaku rotating anode generator. Data were processed using MOSFLM (25) and SCALA (26) from the CCP4 suite (27).
Phase Determination, Model Building, and Refinement—Phase information was determined using single wavelength anomalous scattering from a xenon derivative. Crystals were labeled with xenon by transfer into a cryoprotectant mixture (25% PEG 4000, 0.1 m MES, pH 6.0, 25% glycerol) and were mounted in a cryoloop (Hampton). The loop was placed into an Xcell xenon pressure chamber (Oxford cryosystems) and was exposed to xenon (BOC) at a pressure of 8 megapascals for 15 min before flash freezing in liquid nitrogen. Data were collected on a Rigaku generator fitted with a chromium anode (2.29 Å wavelength) and an R-AXIS IV detector with a helium path and processed using MOSFLM (25) and SCALA (26).
The structure was determined using single isomorphous replacement with anomalous scattering (SIRAS) phasing with data from a xenon derivative (180° of data collected at 3.0 Å resolution) and a native crystal (90° of data at 2.2 Å resolution). A single xenon site was found and refined using autoSHARP (28). SIRAS phases, following density modification, were used to calculate an initial map, allowing model building in Coot (29). The structure was refined using refmac (30) and CNS (31) with 5% of reflections kept aside for evaluation of Rfree. Data collection and refinement statistics are outlined in supplemental Tables 1 and 2.
Preparation of Chondroitin Sulfate Fragments—Chondroitin sulfate A (Sigma) was dissolved to 10 mg/ml in 5 mm sodium phosphate, pH 7.4, 0.2 mm NaCl and digested by the addition of 0.5 units of chondroitinase ABC (Sigma). The reaction was followed by measuring the absorption at 232 nm and disaccharide was purified using a BioGel P-10 column (16 × 900 mm) eluted with 0.2 m ammonium bicarbonate. Purified disaccharide was desalted into water using a PD10 column and dried before use.
Structure Determination in the Presence of Sulfate or Disaccharide—Crystals were soaked with ammonium sulfate (to a final concentration of 0.3 m) or CSA disaccharide (to a final concentration of 10 mm). Ligands were prepared as concentrated stocks and added to the crystallization droplet. Crystals were soaked in ligand overnight before freezing. Diffraction data from sulfate-soaked crystals were collected at 100 K on beamline id14.4 at the European Synchrotron Radiation Facility (ESRF), whereas data from disaccharide-soaked crystals were collected at 100 K on beam line I04 at the Diamond light source. The structures of the sulfate- and disaccharide-soaked proteins were determined by refinement in refmac (30) using the model for the apo structure.
RESULTS
The Structure of a Chondroitin Sulfate-binding DBL Domain—The structure of the DBL3X domain (Fig. 1B) was determined from a xenon derivative using single-wavelength dispersion data collected using chromium Kα radiation and was refined to 1.8 Å resolution. The domain adopts a similar basic fold to the DBL domains of malarial invasion protein EBA-175 (32) and the Pkα-DBL domain from the Duffy antigen/receptor for chemokines (DARC)-binding protein (33). A predominantly “boomerang”-shaped α-helical core contains 44% of residues, whereas extensive loops form the remainder. Loop extensions not seen in other DBL domains, including a loop that becomes ordered only in the presence of sulfate ions, mainly occupy the concave surface (Figs. 2 and 3). Eight disulfide bonds stabilize the structure, with one holding the N terminus in place and three stabilizing the C terminus.
FIGURE 2.
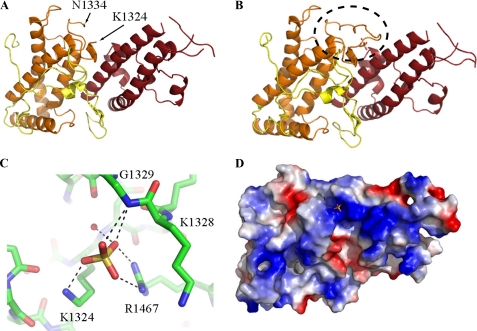
Sulfate binding induces formation of an ordered loop. A and B, the structure of the DBL3X domain in the absence (A) and in the presence (B) of sulfate. In the absence of sulfate, residues 1325–1333 are disordered. The binding of sulfate stabilizes a single conformation of a previously disordered loop, highlighted with a dashed ellipse. C, the sulfate ion interacts with the side chains of residues Lys-1324 and Arg-1467 and the main chain amine of Gly-1329. The side chain of Lys-1328 interacts with Arg-1467 to stabilize the conformation of the sulfate-binding loop. D, an electrostatic surface representation shows that the sulfate-binding loop completes a positively charged patch on the protein surrounding the sulfate-binding site. Positively charged residues (+1.8) are blue, whereas negatively charged residues (–1.8) are red.
FIGURE 3.
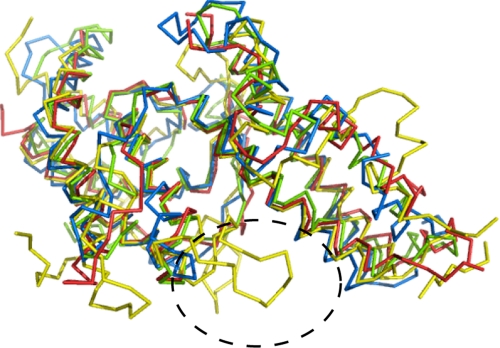
Alignment of DBL domain structures. Alignment of the structure of the DBL3X domain in the presence of sulfate (yellow) with those of DBL domains from EBA-175 F1 (green), EBA-175 F2 (blue), and Pkα-DBL (red) is shown. The loop extensions on the concave surface are indicated by a dashed ellipse.
Subdomain 1 (Fig. 1C) is formed predominantly from random coil and is longer than those found in invasion DBL domains. Residues 1236–1246 form a loop insertion that extends into the concave face, and a 12-residue N-terminal extension folds round the back of subdomain 2. This is held in place by a disulfide bond between Cys-1219 and Cys-1418, determining the direction of the preceding linker. This disulfide is not essential for the domain as both Cys-1219 and Cys-1418 are mutated in some variants (10). Two other disulfides stabilize the subdomain and are also found in invasion DBL domains.
Subdomain 2 (residues 1292–1443) is formed from a bundle of four α-helices with extensive loop insertions (Fig. 1D) and is stabilized by a single disulfide bond that is also found in the invasion DBL domains. Residues 1325–1333 and 1388–1396 are disordered in the structure, with 1325–1333 becoming ordered on binding to a sulfate ion (Fig. 2).
Subdomain 3 (Fig. 1E) contains two 40 Å long α-helices. A third strand completes the bundle and is linked to the first helix by a single disulfide. Three further disulfide bonds stabilize the distal part of the bundle, stapling the end of the structure together. Two of these are also observed in invasion DBL domains, whereas a third holds together a unique “hammerhead” loop structure, the extremities of which are poorly ordered in the crystal.
An Ordered Loop Completes a Sulfate-binding Site and Positively Charged Patch—A loop in the concave surface (residues 1325–1333) is not observed in the structure of the apo protein (Fig. 2A) but becomes ordered when crystals are soaked with sulfate ions (Fig. 2B). This is one of three loop insertions that distinguish the domain from those found in the invasion DBLs. The ordered loop forms part of a binding site for a single sulfate ion that is coordinated by the side chains of Lys-1324 and Arg-1467 and the backbone amine of Gly-1329 (Fig. 2C). The side chains of Lys-1324 and Arg-1467 are observed in electron density of the apo protein, and sulfate binding leads to only subtle changes in the placement of the lysine amine. This suggests that these residues act as a preformed partial sulfate-binding site and that the presence of a sulfate stabilizes the flexible loop in a conformation that completes the binding pocket. This sulfate-binding pocket forms part of the largest patch of positive charge on the surface of the domain (Fig. 2D).
In crystals soaked with CSA disaccharide, the loop is in a similar conformation, and a sulfate ion is seen with the same coordination. Other parts of the disaccharide are not observed clearly in the electron density.
DISCUSSION
Comparison of DBL Domain Structures—Here I present the first structure of a DBL domain from a PfEMP1 protein. Alignment of this structure with those of DBL domains from the malarial invasion proteins, EBA-175 (32), and the DARC-binding protein (33) reveals the degree of structural similarity of the molecular scaffold of the domain (Fig. 3). Despite low sequence conservation, the four α-helices of subdomain 2 and the two long α-helices of subdomain 3 overlap with an root mean squared deviation of 1.5, 2.2, or 1.7 Å when compared with the DARC-binding protein and the first and second DBL domains of EBA-175, respectively. However, subdomain 1, the loops of subdomain 2, and the third strand and loops of subdomain 3 vary significantly, with insertions in DBL3X when compared with the invasion DBL domains. The PfEMP1 protein DBL domains are exposed to the immune system for longer than DBL domains present in invasion proteins, and variation in loop insertions may contribute to immune evasion.
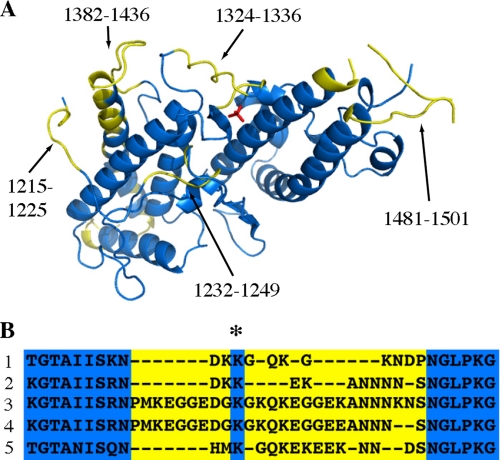
The Arrangement of Polymorphic Residues on the Domain Surface—Sequence analysis of the DBL3X domains of parasites taken from pregnant women show the degree of variability of this domain. There are three significant regions of increased polymorphism that show evidence of diversifying selection pressure (Fig. 4A) (9, 10). These correspond to the sulfate-binding loop (residues 1324–1336), the exposed face of the final helix of subdomain 2 together with the associated loop (residues 1382–1436), and the loops that form the hammerhead structure at the distal end of subdomain 3 (residues 1481–1501). The N terminus also displays polymorphism in residues 1215–1225 and 1232–1249 (10). These polymorphic loops are the principle targets of antibodies from resistant woman (9) and will act as epitopes for vaccine development.
FIGURE 4.
Polymorphic residues are found predominantly on one side of the DBL3X domain and include the sulfate-binding loop. A, highly polymorphic regions (shown in yellow) consist of residues 1215–1225, 1232–1249, 1324–1336, 1382–1436, and 1481–1501 and include the sulfate-binding loop. B, alignment of DBL3X domain sequences showing residues involved in the sulfate-binding loop, with polymorphic sequences in yellow. The conserved side chain of residue Lys-1328 is marked (*). 1 is residues 1317–1342 from the structure presented in this report, DBL3X from AAQ73926; 2 is from AAQ73930; 3 is from DQ995592; 4 is from DQ995593; and 5 is from DQ995594.
It is notable that large areas of the domain surface are not polymorphic. This supports the suggestion from modeling studies and the analysis of antibody binding (34) that much of the DBL domain is not exposed to the immune system, perhaps through the formation of higher order structure when part of intact PfEMP1 proteins.
A Putative Carbohydrate-binding Pocket—Comparison of the structure of the DBL3X domain, with and without sulfate or disaccharide, suggests a putative location for a carbohydrate-binding site. In the presence of sulfate or disaccharide, residues 1325–1333 become ordered, completing a sulfate-binding pocket (Fig. 2). This lies in the center of the major patch of positive charge on the surface of the domain. Indeed, binding studies with another CSA-binding DBL domain have suggested that the interaction with CSA has a major electrostatic component (35). In addition, the close proximity of polymorphic and antibody-accessible regions of the protein shows that this region is surface-exposed, making it a likely carbohydrate-binding surface.
Comparison of the sialic acid-binding site of EBA-175 with the DBL3X domain reveals considerable diversity in the mechanism of carbohydrate binding between these two proteins. EBA-175 domains form a dimer with a glycan-binding site in a positively charged patch on the convex surface. Dimerization is essential for this interaction, and each glycan molecule forms contacts with residues from both molecules of the dimer (32). Thereisnoareaofpositivechargeinthe corresponding region of the DBL3X domain, and DBL3X shows no evidence of dimerization, either in the crystal or in solution. Instead, the CSA-binding site is predicted to lie in a positively charged patch in the concave face of the domain. Therefore EBA-175 and DBL3X each have different multimerization properties and use widely different binding surfaces. This shows the adaptability of DBL domains in the development of different binding phenotypes on a conserved molecular scaffold.
Conservation of the Sulfate-binding Pocket and Positively Charged Patch—Although the sulfate-binding loop is polymorphic in both length and sequence, a single side chain, that of Lys-1328, is conserved (Fig. 4B) (9, 10). In the sulfate-bound form of the protein, this side chain makes hydrogen bonds to Arg-1467, stabilizing the conformation of the loop and positioning the amine group of the main chain of residue 1329 correctly to form hydrogen bonds with the sulfate ion.
In addition, the side chains of Lys-1324 and Arg-1467, both of which form hydrogen bonds with the sulfate ion, have limited variability. Lys-1324 can be replaced by arginine or glutamine, whereas Arg-1467 can be replaced by glutamine. Any of these residues will retain the ability to form hydrogen bonds with the sulfate ion and will maintain the integrity of the binding pocket. The residues that form the positively charged pocket (Lys-1504, Lys-1507, Lys-1509, Lys-1510, Lys-1515, and Lys-1516) are also conserved. The conservation of the chemical nature of these surface residues, including some residues within a polymorphic region, suggests the importance of the formation of the sulfate-binding site and positively charged patch for the function of the domain.
A Model for Carbohydrate Binding and Polymorphism—The structure of the DBL3X domain, in the presence and absence of sulfate or disaccharide, suggests a model for how the module maintains its binding properties while avoiding immune detection. Proteins from pathogenic organisms frequently acquire polymorphic loops that immediately surround regions of functional importance as these surfaces are accessible to both ligand and immune system (36–38). Here the sulfate-binding loop is polymorphic and is flexible in the absence of ligand. It will cover the partial putative CSA-binding surface underneath, protecting its conserved features from the immune detection. In the presence of a sulfate ion, alone or as part of a polysaccharide, the loop folds with the conserved side chain of Lys-1328 interacting with Arg-1467 and the backbone amine of residue 1329 forming hydrogen bonds to the sulfate ion. This completes a positively charged patch that can form electrostatic interactions with other parts of the CSA molecule, generating the binding site.
This provides the first structural insight into one of the PfEMP1 DBL domains from P. falciparum. Comparison with the structures of DBL domains from invasion proteins shows that, despite low sequence identity, the DBL domains maintain a conserved molecular architecture. However, significant variability in the length and structure of loops suggests that individual DBL domain structures, in the presence and absence of ligands, will be needed to understand the binding properties of these domains. The structure also gives clues into the nature of polymorphism in the DBL domain and suggests a likely CSA-binding surface. This will guide future drug and vaccine development work.
Supplementary Material
Acknowledgments
Many thanks to Tom Blundell, Mark Carrington, Ben Luisi, Dima Chirgadze, and Marko Hyvonen for advice and support and to Liz Dukes, Gwyndaf Evans, and Ralf Flaig at beamline I04 at the Diamond light source and Didier Nurizzo at beamline id23.1 at the ESRF for assistance in data collection.
Author's Choice—Final version full access.
The atomic coordinates and structure factors (codes 3BQI, 3BQK, 3BQL) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by grants from the Royal Society and the Wellcome trust. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental figures.
Footnotes
The abbreviations used are: PfEMP1, plasmodium falciparum erythrocyte membrane protein 1; DBL domain, Duffy binding-like domain; CSA, chondroitin sulfate A; PEG, polyethylene glycol; MES, 4-morpholineethanesulfonic acid.
References
- 1.Breman, J. G., Egan, A., and Keusch, G. T. (2001) Am. J. Trop. Med. Hyg. 64 iv–vii [DOI] [PubMed] [Google Scholar]
- 2.Miller, L. M., Baruch, D. I., Marsh, K., and Doumbo, O. K. (2002) Nature 415 673–679 [DOI] [PubMed] [Google Scholar]
- 3.Fried, M., Nosten, F., Brockman, A., Brabin, B. J., and Duffy, P. E. (1998) Nature 395 851–852 [DOI] [PubMed] [Google Scholar]
- 4.Kraemer, S. M., and Smith, J. D. (2006) Curr. Opin. Microbiol. 9 374–378 [DOI] [PubMed] [Google Scholar]
- 5.Su, X., Heatwole, V. M., Wertheimer, S. P., Guinet, F., Herrfeldt, F. A., Peterson, D. S., Ravetch, J. A., and Wellems, T. E. (1995) Cell 82 89–100 [DOI] [PubMed] [Google Scholar]
- 6.Smith, J. D., Chitnis, C. E., Craig, A. G., Roberts, D. J., Hudson-Taylor, D. E., Peterson, D. E., Pinches, R., Newbold, C. I., and Miller, L. H. (1995) Cell 82 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duffy, P. E. (2007) Parasitology 134 1877–1881 [DOI] [PubMed] [Google Scholar]
- 8.Gardner, M. J., Hall, N., Fung, E., White, O., Berrimanm, M., Hymanm, R. W., Carlton, J. M., Pain, A., Nelson, K. E., Bowman, S., et al. (2002) Nature 419 498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahlbäck, M., Rask, T. S., Andersen, P. H., Nielsen, M. A., Ndam, N. T., Resende, M., Turner, L., Deloron, P., Hviid, L., Lund, O., Pedersen, A. G., Theander, T. G., and Salanti, A. (2006) PLoS Pathog. 2 e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bockhorst, J., Lu, F., Janes, J. H., Keebler, J., Gamain, B., Awadalla, P., Su, X. Z., Samudrala, R., Jojic, N., and Smith, J. D. (2007) Mol. Biochem. Parasitol. 155 103–112 [DOI] [PubMed] [Google Scholar]
- 11.Fried, M., and Duffy, P. E. (1996) Science 272 1502–1504 [DOI] [PubMed] [Google Scholar]
- 12.Fried, M., Domingo, G. J., Gowda, C. D., Mutabingwa, T. K., and Duffy, P. E. (2006) Exp. Parasitol. 113 36–42 [DOI] [PubMed] [Google Scholar]
- 13.Hviid, L., and Salanti, A. (2007) Parasitology 134 1871–1876 [DOI] [PubMed] [Google Scholar]
- 14.Dorman, E. K., Shulman, C. E., Kingdom, J., Bulmer, J. N., Mwendwa, J., Peshu, N., and Marsh, K. (2002) Ultrasound Obstet. Gynecol. 19 165–170 [DOI] [PubMed] [Google Scholar]
- 15.Suguitan, A. L., Jr., Leke, R. G., Fouda, G., Zhou, A., Thuita, L., Metenou, S., Fogako, J., Megnekou, R., and Taylor, D. W. (2003) J. Infect. Dis. 188 1074–1082 [DOI] [PubMed] [Google Scholar]
- 16.Salanti, A., Dahlback, M., Turner, L., Nielsen, M. A., Barfod, L., Magistrado, P., Jensen, A. T., Lavstsen, T., Ofori, M. F., Marsh, K., Hviid, L., and Theander, T. G. (2004) J. Exp. Med. 200 1197–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ricke, C. H., Staalsoe, T., Koram, K., Akanmori, B. D., Riley, E. M., Theander, T. G., and Hviid, L. (2001) J. Immunol. 165 3309–3316 [DOI] [PubMed] [Google Scholar]
- 18.O'Neil-Dunne, I., Achur, R. N., Agbor-Enoh, S. T., Valiyaveettil, M., Naik, R. S., Ockenhouse, C. F., Zhou, A., Megnekou, R., Leke, R., Taylor, D. W., and Gowda, D. C. (2001) Infect. Immun. 69 7487–7492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowe, J. A., and Kyes, S. (2004) Mol. Microbiol. 53 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith, J. D., Kyes, S., Craig, A. G., Fagan, T., Hudson-Taylor, D., Miller, L. H., Baruch, D. I., and Newbold, C. I. (1998) Mol. Biochem. Parasitol. 97 133–148 [DOI] [PubMed] [Google Scholar]
- 21.Smith, J. D., Craig, A. G., Kriek, N., Hudson-Taylor, D., Kyes, S., Fagan, T., Pinches, R., Baruch, D. I., Newbold, C. I., and Miller, L. H. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 1766–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gamain, B., Trimnell, A. R., Scheidig, C., Scherf, A., Miller, L. H., and Smith, J. D. (2005) J. Infect. Disease 191 1010–1013 [DOI] [PubMed] [Google Scholar]
- 23.Viebig, N. K., Gamain, B., Scheidig, C., Lepolard, C., Przyborzki, J., Lanzer, M., Gysin, J., and Scherf, A. (2005) EMBO Rep. 6 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins, M. K. (2008) Acta Crystallogr. F Struct. Biol. Crystalliz. Comm. 64 221–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leslie, A. G. (2006) Acta Crystallogr. Sect. D Biol. Crystallogr. 62 48–57 [DOI] [PubMed] [Google Scholar]
- 26.Collaborative Computational project, Number 4 (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760–76315299374 [Google Scholar]
- 27.Evans, P. R. (1993) Proceedings of the CCP4 Study Weekend: Data Collection and Processing January 29–30, 1993 (Sawyer, L., Isaacs, N., and Bailey, S., eds), pp. 114–133, Daresbury Laboratory, Warrington, UK
- 28.Vonrhein, C., Blanc, E., Roversi, P., and Bricogne, G. (2007) Methods Mol. Biol. 364 215–230 [DOI] [PubMed] [Google Scholar]
- 29.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53 240–2525 [DOI] [PubMed] [Google Scholar]
- 31.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905–921 [DOI] [PubMed] [Google Scholar]
- 32.Tolia, N. H., Enemark, E. J., Sim, B. K., and Joshua-Tor, L. (2005) Cell 122 183–193 [DOI] [PubMed] [Google Scholar]
- 33.Singh, S. K., Hora, R., Belrhali, H., Chitnis, C. E., and Sharma, A. (2006) Nature 439 741–744 [DOI] [PubMed] [Google Scholar]
- 34.Andersen, P., Nielsen, M. A., Resende, M., Rask, T. S., Dahlbäck, M., Theander, T., Lund, O., and Salanti, A. (2008) PLoS Pathog. 4 e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Badaut, C., Faure, G., Tuikue-Ndam, N. G., Bertin, G., Chaffotte, A., Khattab, A., Klinkert, M. Q., Deloron, P., and Bentley, G. A. (2007) Mol. Biochem. Parasitol. 151 89–99 [DOI] [PubMed] [Google Scholar]
- 36.Bai, T., Becker, M., Gupta, A., Strike, P., Murphy, V. J., Anders, R. F., and Batchelor, A. H. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 12736–12741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen, B., Vogan, E. M., Gong, H., Skehel, J. J., Wiley, D. C., and Harrison, S. C. (2005) Nature 433 834–841 [DOI] [PubMed] [Google Scholar]
- 38.Skehel, J. J., and Wiley, D. C. (2000) Annu. Rev. Biochem. 69 531–569 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.