Abstract
Rac plays a pivotal role in the assembly of the superoxide-generating NADPH oxidase of phagocytes. In resting cells, Rac is found in the cytosol in complex with Rho GDP dissociation inhibitor (RhoGDI). NADPH oxidase assembly involves dissociation of the Rac·RhoGDI complex and translocation of Rac to the membrane. We reported that liposomes containing high concentrations of monovalent anionic phospholipids cause Rac·RhoGDI complex dissociation (Ugolev, Y., Molshanski-Mor, S., Weinbaum, C., and Pick, E. (2006) J. Biol. Chem.281 ,19204 -19219). We now designed an in vitro model mimicking membrane phospholipid remodeling during phagocyte stimulation in vivo. We showed that liposomes of “resting cell membrane” composition (less than 20 mol % monovalent anionic phospholipids), supplemented with 1 mol % of polyvalent anionic phosphatidylinositol 3,4,5-trisphosphate (PtdIns(3,4,5)P3) in conjunction with constitutively active forms of the guanine nucleotide exchange factors (GEFs) for Rac, Trio, or Tiam1 and a non-hydrolyzable GTP analogue, cause dissociation of Rac1(GDP)·RhoGDI complexes, GDP to GTP exchange on Rac1, and binding of Rac1(GTP) to the liposomes. Complexes were not dissociated in the absence of GEF and GTP, and optimal dissociation required the presence of PtdIns(3,4,5)P3 in the liposomes. Dissociation of Rac1(GDP)·RhoGDI complexes was correlated with the affinity of particular GEF constructs, via the N-terminal pleckstrin homology domain, for PtdIns(3,4,5)P3 and involved GEF-mediated GDP to GTP exchange on Rac1. Phagocyte membranes enriched in PtdIns(3,4,5)P3 responded by NADPH oxidase activation upon exposure in vitro to Rac1(GDP)·RhoGDI complexes, p67phox, GTP, and Rac GEF constructs with affinity for PtdIns(3,4,5)P3 at a level superior to that of native membranes.
The superoxide
(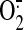 )2-generating
NADPH oxidase of phagocytes (briefly “oxidase”) consists of a
membrane-localized cytochrome b559 and four cytosolic
components, p47phox, p67phox,
p40phox, and the small GTPase Rac (Rac1 or Rac2) (for
reviews, see Refs.
1-4).
The separation between membrane-bound cytochrome b559 and
the cytosolic components is a characteristic of the resting phagocyte. In
response to stimuli acting via cell surface receptors, the cytosolic
components become engaged in protein-protein and protein-membrane lipid
interactions culminating in their translocation to the plasma membrane
(PM).
)2-generating
NADPH oxidase of phagocytes (briefly “oxidase”) consists of a
membrane-localized cytochrome b559 and four cytosolic
components, p47phox, p67phox,
p40phox, and the small GTPase Rac (Rac1 or Rac2) (for
reviews, see Refs.
1-4).
The separation between membrane-bound cytochrome b559 and
the cytosolic components is a characteristic of the resting phagocyte. In
response to stimuli acting via cell surface receptors, the cytosolic
components become engaged in protein-protein and protein-membrane lipid
interactions culminating in their translocation to the plasma membrane
(PM).
Oxidase assembly can be reproduced in vitro by mixing phagocyte membranes with the cytosolic components p47phox, p67phox, and Rac and using an anionic amphiphile as activator (for a review, see Ref. 5). Oxidase assembly can also be achieved in vitro in the absence of an activator in a system consisting of phagocyte membranes, p67phox, and Rac, even in the absence of p47phox, provided that Rac is in the prenylated form (6).
The absolute requirement for Rac, a member of the Rho family of small GTPases, in oxidase function has been confirmed both in vitro and in vivo (7). It is generally assumed that Rho proteins must associate with the PM to trigger a cellular response. A polybasic domain and an isoprenyl (geranylgeranyl) group, both at the C terminus, have been shown to play a critical role in anchoring Rac to the PM (8-11). Polyphosphorylated derivatives of phosphatidylinositol, collectively referred to as polyphosphoinositides (pPIs), which are transiently generated in the cytosolic leaflet of the PM upon phagocyte stimulation (for a review, see Ref. 12), are likely candidates for recruiting the prenylated C-terminal polybasic domain of Rac1 to the PM. Indeed it has been shown that Rac1 selectively interacts with phosphatidylinositol 3,4,5-trisphosphate (PtdIns(3,4,5)P3) and, to a lesser extent, with phosphatidylinositol 3,4-bisphosphate (PtdIns(3,4)P2) (13, 14). Furthermore Heo et al. (14) demonstrated that phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) and PtdIns(3,4,5)P3 target a variety of small GTPases to the PM.
As true of all Rho GTPases, Rac acts as a molecular switch cycling between inactive (GDP-bound) and active (GTP-bound) conformations. This cycle is regulated by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins, which stimulate GTP loading and hydrolysis, respectively (for a review, see Ref. 15). In addition, Rac cycles between cytosolic and membrane-associated forms through the association with and dissociation from the cytosolic regulatory proteins, Rho GDP dissociation inhibitors (RhoGDIs) (for reviews, see Refs. 16 and 17).
The canonical GEFs for Rho GTPases share a common motif designated the Dbl homology (DH) domain and an adjacent pleckstrin homology (PH) domain C-terminal to the DH domain (for a review, see Ref. 18). The DH domain makes extensive contacts with the switch I and II regions of Rho GTPases and contains virtually all the residues required for substrate recognition, binding, and guanine nucleotide exchange (19-22). PH domains of many Dbl family members have been shown to direct membrane localization of their host proteins through binding to PM pPIs (23). For example, it has been shown that the PH domain of Dbl binds to PtdIns(4,5)P2 and PtdIns(3,4,5)P3, and these interactions modulate the nucleotide exchange activity and subcellular localization of Dbl (24). Mutational analysis of the GEF P-Rex1 demonstrated that recruitment to the PM of P-Rex1 mutants missing the PH domain, known to selectively bind to PtdIns(3,4,5)P3, was severely impaired (25). Several groups of investigators provided evidence for the requirement of the N-terminal PH domain of Tiam1 for localization to the PM through its apparent binding preference for PtdIns(3,4,5)P3, PtdIns(3,4)P2, and PtdIns(4,5)P2 (26-29). Furthermore binding of Vav1 (30, 31) and Sos1 (32) to PtdIns(3,4,5)P3 is believed to regulate their targeting to the PM. Very recently, the PH domain of the GEF Asef was shown to bind to PtdIns(3,4,5)P3 and target it to the PM (33).
Dissociation of prenylated Rac from RhoGDI is an obligatory step in the assembly of the oxidase, preceding translocation of Rac from the cytosol to the PM (34-36). A hydrophobic pocket, within the C-terminal immunoglobulin-like domain of RhoGDI, regulates the partitioning of Rac between the cytosol and PM by binding the prenylated polybasic C terminus of Rac (for reviews, see Refs. 16 and 17).
The mechanism(s) responsible for dissociation of Rho GTPases from RhoGDI in general and of Rac from RhoGDI in the context of oxidase assembly in particular remains largely unsolved. Several reports point to the possibility that biologically relevant anionic lipids might serve as RhoGDI displacement factors (for a review, see Ref. 17). In an earlier publication, we demonstrated that Rac·RhoGDI complexes were dissociated by exposure to liposomes containing anionic but not neutral phospholipids, suggesting that the stability of Rac·RhoGDI complexes is regulated by the phospholipid composition of the phagocyte membrane with emphasis on the charge of phospholipids (37).
A large body of evidence supports the idea that generation of PtdIns(3,4,5)P3 in the PM, upon cell stimulation, is functionally related to two events culminating in the formation of GTP-bound and membrane-anchored Rac: the dissociation of the Rac·RhoGDI complex and activation and translocation of a Rac GEF from the cytosol to the PM. In the present study, we set out to explore the relationship between these events by modifying the liposome-based system used earlier (37) by reducing the concentration of monovalent anionic phospholipid to that present in the membranes of resting phagocytes, supplementing the liposomes with PtdIns(3,4,5)P3, and adding a Rac GEF and free GTP. We found the following. 1) Anionic liposomes enriched in PtdIns(3,4,5)P3 combined with DH-PH tandem regions of Rac GEFs cooperatively promoted the dissociation of Rac·RhoGDI complexes. 2) The affinity of specific PH domains for PtdIns(3,4,5)P3 determined the efficacy of Rac·RhoGDI dissociation by PtdIns(3,4,5)P3-enriched liposomes and Rac GEF. 3) GDP to GTP exchange on Rac by GEFs occurred at the level of the liposomes and was critical for Rac·RhoGDI dissociation to become irreversible and be followed by binding of Rac(GTP) to the liposomes. 4) DH-PH tandems of Rac GEFs, with a binding preference for PtdIns(3,4,5)P3, elicited oxidase activation in a cell-free system consisting of PtdIns(3,4,5)P3-enriched phagocyte membranes, Rac1(GDP)·RhoGDI complexes, and GTP in the absence of an amphiphilic activator.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents—The hydrolysis-resistant GTP analogue GMPPNP (tetralithium salt, 83%, <0.2% GTP) was purchased from Roche Applied Science. GTP (lithium salt, 97%) and GDP (sodium salt, type I, 98%) were obtained from Sigma. The fluorescent hydrolysis-resistant GTP analogues mant-GMPPNP and mant-GDP were obtained from Jena Bioscience GmbH. The following synthetic phospholipids (purity > 99%) were purchased from Avanti Polar Lipids: 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC; product 850375P), 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS; sodium salt, product 830035P), PtdIns(3,4,5)P3 (tetra-ammonium salt, product 850156P), phosphatidylinositol 3,5-bisphosphate (PtdIns(3,5)P2; triammonium salt, product 850154P), PtdIns(3,4)P2 (triammonium salt, product 850153P), and PtdIns(4,5)P2 (triammonium salt, product 850155P). Common laboratory chemicals were from Sigma or Merck.
Construction of Tiam1(C1199) Protein—pcDNA 3.0 plasmid carrying the full-length mouse Tiam1 gene (corresponding to amino acids 1-1591) was obtained from J. G. Collard. PCR primers 5′-NdeI-6×His (5′-GTGTGTCATATGCATCACCATCATCATCATGGCTCCAGTACCACCAACAGTGAGAGCC-3′) and 3′-Stop-SalI (5′-CTCTCTGTCGACTCATATCTCCGTGTTCAGTTTCC-3′) were designed to amplify a Tiam1 fragment consisting of amino acids 394-1591 (referred to as Tiam(C1199)). The PCR product was digested with NdeI and SalI restriction enzymes and subcloned into a pET-30a vector (Novagen) to allow expression of His6-tagged Tiam1(C1199) in Escherichia coli. The DNA construct was verified by automated sequencing. Point mutations K452A, K453A, R459A, and R460A within the N-terminal PH (PHn) domain of Tiam1(C1199) (modified from Ref. 29) were generated by PCR-based site-directed mutagenesis and confirmed by DNA sequencing. The mutated protein is referred to as Tiam1(C1199) KK/AA, RR/AA.
Preparation of Macrophage Membrane Liposomes—Membranes were isolated from guinea pig peritoneal macrophages as described previously (38). Membranes, suspended to a concentration of 500 × 106 cell eq/ml, were first solubilized by 40 mm octyl glucoside and then reconstituted into liposomes by dialysis against detergent-free buffer as reported before (39). The specific cytochrome b559 heme content of membrane liposomes was measured by the difference spectrum of sodium dithionite-reduced minus oxidized samples (40).
Preparation of Anionic Phospholipid Liposomes—These were prepared essentially as described in Refs. 37 and 41. Briefly DOPC and DOPS were dissolved at a concentration of 1 mm in the buffer also used for membrane solubilization containing 40 mm octyl glucoside. To facilitate solubilization, the phospholipids were first homogenized by using an ultrasonic processor (Vibra Cell, VCX 400 watts, Sonics and Materials) at 20% amplitude for three 10-s cycles in ice-cooled tubes. Next the mixtures were stirred magnetically in ice-cooled glass vials until a clear solution was obtained. All pPIs (PtdIns(3,4,5)P3, PtdIns(3,4)P2, PtdIns(3,5)P2, and PtdIns(4,5)P2) were also dissolved in the buffer used for membrane solubilization containing 40 mm octyl glucoside at a concentration of 100 μm. As before, the solutions were first subjected to homogenization in an ultrasonic processor at 60% amplitude for three 20min cycles in ice-cooled tubes. This was followed by gentle agitation for 3 h at 4 °C. Liposomes consisting of a mixture of DOPC and DOPS were prepared by mixing 1.48-ml aliquots of a 1 mm solution of DOPC with 0.32-ml aliquots of a 1 mm solution of DOPS and 0.20 ml of solubilization buffer, all containing 40 mm octyl glucoside. The final phospholipid concentrations in DOPC/DOPS liposomes were 0.74 mm DOPC (82.22 mol %) and 0.16 mm DOPS (17.77 mol %). Liposomes consisting of DOPC/DOPS/pPI were prepared by mixing 1.48-ml aliquots of a 1 mm solution of DOPC with 0.32-ml aliquots of a 1 mm solution of DOPS and 0.20-ml aliquots of a 100 μm solution of pPI. The final phospholipid concentrations in DOPC/DOPS/PtdIns liposomes were 0.74 mm DOPC (81.30 mol %), 0.16 mm DOPS (17.58 mol %), and 10 μm PtdIns (1.10 mol %). The mixtures were dialyzed against a 500-fold excess of detergent-free buffer using dialysis membranes with a molecular weight cutoff of 25,000 (Spectrum Laboratories) for 18 h at 4 °C. On gel filtration on a Superose 12 10/300 GL fast protein liquid chromatography (FPLC) column (GE Healthcare), the vesicles elute in the exclusion volume (corresponding to an Mr of ≥2 × 106).
Determination of Membrane Phospholipid Concentration—The concentration of total phospholipids in native macrophage membrane liposomes was measured as described in Ref. 42 following extraction by chloroform as described in Ref. 43. The concentrations varied from 3.2 to 3.5 mm.
Enrichment of Phagocyte Membranes in PtdIns(3,4,5)P3—The lipid composition of macrophage membranes was modified by mixing 1.35 ml of phagocyte membranes, solubilized by 40 mm octyl glucoside (at a concentration above 2100 pmol of cytochrome b559 heme/ml), with 0.15 ml of PtdIns(3,4,5)P3 (at a concentration of 100 μm) also dissolved in buffer containing 40 mm octyl glucoside. The mixtures were reconstituted into liposomes by dialysis against detergent-free buffer as described under “Preparation of Anionic Phospholipid Liposomes.” The final concentration of PtdIns(3,4,5)P3 in the enriched phagocyte membranes was 10 μm, corresponding to 0.28-0.31 mol % in relation to total membrane phospholipids. The concentration of cytochrome b559 was measured after dialysis and was normally found to be close to 1900 pmol of heme/ml. The final concentration of cytochrome b559 heme of PtdIns(3,4,5)P3-enriched membranes in cell-free oxidase activation assays was 5 nm, and the final concentration of PtdIns(3,4,5)P3 in the assay was 25 nm.
Preparation of Recombinant Proteins—Non-prenylated Rac1 and RhoGDI were expressed as glutathione S-transferase (GST) fusion proteins in E. coli BL21-Codon-Plus™ competent cells (Stratagene) and purified by affinity chromatography on glutathione-agarose (Sigma) followed by cleavage by thrombin in situ as described in Ref. 44. Full-length p67phox, Tiam1 (residues 394-1591) (referred to as Tiam1(C1199)), Tiam1(C1199) point mutant in PHn (referred to as Tiam1(C1199) KK/AA, RR/AA), Tiam1 (residues 1033-1406) (referred to as Tiam1(C374)), and Trio (residues 1225-1537) (referred to as TrioN (the N-terminal DH-PH tandem of Trio)) were expressed as N-terminal His6-tagged fusion proteins and purified by metal chelate affinity chromatography. Briefly pET-30a plasmids carrying cDNA of p67phox or Tiam1(C1199) either wild type or point mutant, pProEX HT plasmid carrying cDNA of Tiam1(C374), and pET15b plasmid carrying cDNA of TrioN were introduced into E. coli BL21 (DE3, pRIL)-CodonPlus™ cells. The bacteria were induced with 0.4 mm isopropyl β-d-thiogalactopyranoside at 18 °C for 16 h. The induced cells were suspended in a buffer (buffer A), consisting of 20 mm sodium phosphate buffer, pH 7.4, 500 mm NaCl, and 20 mm imidazole, supplemented with “Complete EDTA-free” protease inhibitor (Roche Applied Science). The bacteria were disrupted by exposure to lysozyme (Sigma) at a concentration of 0.5 mg/ml for 20 min at 4 °C with stirring followed by sonication by a 500-watt sonic disruptor (Vibra Cell, Sonics and Materials) at 20% amplitude for 5 min in the 50% pulse mode in an ice-cooled beaker. The resulting material was supplemented with 1% Triton X-100 (Sigma) and stirred on ice for 15 min. Insoluble material was sedimented by centrifugation at 23,000 × g for 25 min at 4 °C, and the cleared cell-free extract was added to nickel-Sepharose 6 Fast Flow Affinity Resin beads (GE Healthcare). Binding was performed in the batch mode at room temperature for 60 min. Following binding, the beads were washed twice with buffer A and twice with buffer A supplemented with 40 mm imidazole. The His6-tagged proteins were eluted from the resin with buffer A supplemented with 400 mm imidazole. All purified proteins were supplemented with 30% glycerol and stored in small aliquots at -75 °C.
Protein Concentration—This was estimated by the method of Bradford (45) modified for use with 96-well microplates (Technical Bulletin 1177EG, Bio-Rad) using the Bio-Rad protein assay dye reagent concentrate and bovine γ-globulin as standard.
SDS-PAGE and Immunoblotting—These were performed as described in Ref. 46. Molecular mass standards in the 97.4-14.4 kDa range (unstained SDS-PAGE standards, low range) were purchased from Bio-Rad. Rac1 was detected by affinity-purified rabbit polyclonal anti-Rac1 C-terminal peptide antibody C11 (Santa Cruz Biotechnology, product SC-95) used at a dilution of 1: 500. The second antibody was an affinity-purified goat anti-rabbit IgG (heavy + light chains) conjugated with peroxidase (Jackson ImmunoResearch Laboratories, product 111-035-003) used at a dilution of 1:5000. RhoGDI was detected by a mouse monoclonal anti-RhoGDI antibody (Transduction Laboratories, product R26320) used at a dilution of 1:2500. The second antibody was rabbit anti-mouse IgG (heavy + light chains) conjugated with peroxidase (Jackson ImmunoResearch Laboratories, product 315035003) used at dilution of 1:5000. Exposure of blots to the first antibodies was for overnight at 4 °C and to the second antibodies was for 1 h at room temperature. The bands were detected by enhanced chemiluminescence Western blotting substrate (Pierce, product 32106).
Assessment of the Translocation of His6-tagged Proteins to Liposomes by ELISA—Column fractions derived from gel filtration, corresponding to the exclusion volume, were diluted 1:4 in phosphate-buffered saline (PBS; consisting of 137 mm NaCl, 2.7 mm KCl, 4.3 mm Na2HPO4, and 1.4 mm KH2PO4, pH 7.2) and were added in volumes of 100 μl, in duplicates, to 96-well microplates (Dynex Technologies, Immulon 4 HBX, High Binding Extra, product 3855). The proteins were allowed to adhere to the wells for 18 h at 4 °C, and the plates were processed as described in Ref. 47. The wells were washed with PBS supplemented with 0.05% (v/v) Tween 20 (Sigma). Blocking buffer consisted of PBS supplemented with 0.1% (v/v) Tween 20 and 3% (w/v) bovine serum albumin. His6-tagged proteins were detected using a mouse monoclonal anti-polyhistidine antibody conjugated with peroxidase (Sigma, product A7058) diluted 1:4000 in blocking buffer. Peroxidase was quantified with the 3,3′,5,5′-tetramethylbenzidine liquid substrate system (Dako Cytomation, product S1599) followed by stopping the reaction with 1 m H2SO4, and the absorbance at 450 nm was read in a Spectramax 340 microplate reader (Molecular Devices).
Enzymatic Prenylation of Rac1—Recombinant non-prenylated Rac1 was prenylated in vitro by recombinant geranylgeranyltransferase type I as described before (48).
Isolation of Pure Rac1·RhoGDI Complexes in Vitro—These were prepared essentially as described in Ref. 37. Briefly nonprenylated Rac1(GDP) expressed in E. coli was prenylated in vitro and either used in native (=GDP-bound) form or exchanged to the fluorescent GDP analogue mant-GDP. Prenylated Rac1 was mixed with RhoGDI at a ratio of 1:4, and the mixture was incubated in a rotary mixer (Thermomix Comfort, Eppendorf) set at 500 rpm for 20 min at room temperature. Following incubation, the solution, now containing Rac1·RhoGDI complexes, was subjected to ultrafiltration (using Amicon Ultra-4 centrifugal filter units with a pore size of 10,000 Da, Millipore) to remove any unbound fluorescent nucleotide and also to transfer the proteins into the loading buffer for the final purification step by anion exchange chromatography, which was performed as described in Ref. 37.
Assessment of Rac1 Dissociation from RhoGDI by In-line Fluorescence Assay—In experiments aimed to examine the ability of pPI-enriched liposomes to dissociate Rac1·RhoGDI complexes, DOPC/DOPS or DOPC/DOPS/pPI liposomes (90 nmol of total phospholipids) were mixed with Rac1·RhoGDI complexes (1 nmol) in which the native nucleotide on Rac1 (GDP) was exchanged to the fluorescent analogue mant-GDP. In experiments aimed to study the effect of various GEF constructs in cooperation with pPI-enriched liposomes on the stability of Rac1·RhoGDI complexes, Rac1 in the Rac1·RhoGDI complex carried its native nucleotide, GDP. In these experiments, either mant-GDP or mant-GMPPNP (10 nmol) and various GEF constructs (3 nmol) were added to the reaction mixtures containing liposomes (90 nmol of phospholipid) and Rac1·RhoGDI complexes (2 nmol). In both types of experiments, the mixtures were incubated for 12 min in a rotary mixer set to room temperature and 500 rpm and injected immediately into a Superose 12 10/300 GL FPLC gel filtration column. Chromatography was performed with a buffer consisting of 50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, and 150 mm NaCl on an HPLC system (Waters) at a flow rate of 0.2 ml/min at 4 °C, and both absorbance (at 280 nm) and the fluorescent signal (excitation = 361 nm; emission = 440 nm) were measured continuously by passing the column eluate through a diode array detector (MD-1510, Jasco) and a spectrofluorometer (model FP-750, Jasco) fitted with an HPLC flow cell (MFC-132, Jasco). We expected to obtain a fluorescent signal in the exclusion volume if prenylated Rac1 (labeled with fluorescent nucleotide), originating from the dissociation of Rac1·RhoGDI complexes, was bound to the liposomes. The intact Rac1·RhoGDI complex was expected to elute in a volume corresponding to its molecular mass.
Assessment of Nucleotide Exchange Activity of GEFs on Liposome-bound Prenylated Rac1 by an In-line Fluorescence Assay—In this set of experiments, prenylated Rac1(GDP) (2 nmol), mant-GDP (10 nmol), and various GEF constructs (3 nmol) were added to DOPC/DOPS liposomes (180 nmol of total phospholipids). The mixtures were incubated for 12 min in a rotary mixer set to room temperature and 500 rpm and injected immediately into a Superose 12 10/300 GL FPLC gel filtration column. Chromatography and measurements of absorbance and fluorescent signal were performed as described in the preceding section. We expected to obtain a fluorescent signal in the exclusion volume consequent to and proportional with the extent of GEF-mediated GDP to mant-GDP exchange having taken place on prenylated Rac1 bound to the liposomes.
Cell-Free NADPH Oxidase Activation Assay—In these
experiments, native phagocyte membranes or phagocyte membranes supplemented
with PtdIns(3,4,5)P3 were used. Oxidase activation was assayed in a
cell-free system (5) consisting
of either native or supplemented phagocyte membranes (equivalent to 5
nm cytochrome b559 heme), free prenylated
Rac1(GDP) or Rac1(GDP)·RhoGDI complexes (both at 100 nm),
p67phox (300 nm), guanine nucleotides (GTP or
GDP) (1 μm), and one of the following GEF constructs: TrioN,
Tiam1(C1199), Tiam1(C1199) KK/AA, RR/AA, or Tiam1(C374) (200 or 400
nm). The order of addition of the components was as follows: a
mixture of phagocyte membrane and p67phox, free prenylated
Rac1(GDP) or Rac1(GDP)·RhoGDI complex, GEF, and guanine nucleotides.
The assay mixtures were incubated for 5 min at room temperature in the absence
of an anionic amphiphile, and
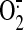 production was initiated
by the addition of NADPH (240 μm).
production was initiated
by the addition of NADPH (240 μm).
RESULTS
Effect of DOPC/DOPS Liposomes Enriched in PtdIns(3,4,5)P3 on the Stability of Rac1(GDP)·RhoGDI Complexes—In an earlier study we demonstrated that Rac1(GDP)·RhoGDI complexes were dissociated by contact with protein-free liposomes containing monovalent anionic phospholipids (having a net charge of -1) serving as “membrane models” (37). However, the amount of anionic phospholipid required for significant dissociation of Rac1·RhoGDI complexes was above that present in the PM of resting and stimulated phagocytes (discussed in Ref. 37). On the other hand, PM pPIs such as PtdIns(4,5)P2 and PtdIns(3,4,5)P3 with high net negative charges of -5 and -7, respectively, were found to promote membrane targeting of Rac1 (14). PtdIns(3,4,5)P3 is present at very low concentrations in quiescent cells but is transiently produced in the PM of stimulated cells (12, 49). We thus reasoned that PtdIns(3,4,5)P3 or PtdIns(3,4)P2 at low concentrations may represent the physiological equivalents of the monovalent anionic phospholipids present at a high concentration in the liposomes used in our earlier study (37). With the purpose of studying the effect of pPIs on the stability of Rac1·RhoGDI complexes, two new types of protein-free membrane models were designed: (a) liposomes containing physiological amounts of the synthetic neutral phospholipid DOPC (82 mol %) and the synthetic anionic phospholipid DOPS (18 mol %), representative of the PM of resting phagocytes, and (b) liposomes containing almost identical amounts of DOPC and DOPS supplemented with various pPIs (1.1 mol %), mimicking the PM of stimulated phagocytes. To study the stability of Rac1·RhoGDI complexes in the presence of liposomes of well defined composition, we utilized an assay based on FPLC gel filtration combined with in-line detection of fluorescent mant-guanine nucleotide-labeled Rac1. The assay allows accurate and easy quantification of both intact Rac1·RhoGDI complexes and of Rac1 dissociated from RhoGDI and bound to liposomes (37).
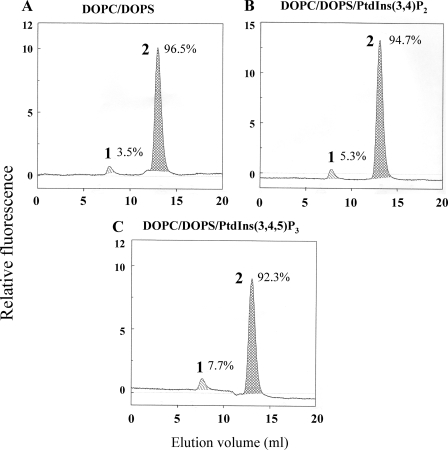
We first assessed the ability of free prenylated Rac1(mant-GDP) to bind to the newly designed liposomes. We found that free prenylated Rac1(mant-GDP) was efficiently targeted to DOPC/DOPS liposomes. We observed no significant increase in Rac1(mant-GDP) binding when such liposomes were enriched with 1.1 mol % PtdIns(3,4,5)P3 (results not shown). Next we created highly purified Rac1(mant-GDP)·RhoGDI complexes in vitro and incubated these with DOPC/DOPS liposomes or with DOPC/DOPS liposomes enriched with either PtdIns(3,4,5)P3 or PtdIns(3,4)P2 (1.1 mol %). As apparent in Fig. 1A, exposure of Rac1(mant-GDP)·RhoGDI complexes to DOPC/DOPS liposomes resulted in only minor dissociation of the complex (3.5%). DOPC/DOPS liposomes enriched with either PtdIns(3,4)P2 or PtdIns(3,4,5)P3 (1.1 mol %) caused only a marginal increase in Rac1·RhoGDI dissociation (5.3 and 7.7%, respectively) (Fig. 1, B and C). These results indicate that despite the reported property of polyvalent pPIs to promote binding of Rac1 to the PM (14) mere enrichment of the liposomes with such pPIs was not sufficient for driving the equilibrium between RhoGDI-bound and membrane-bound Rac toward the latter state.
FIGURE 1.
DOPC/DOPS liposomes enriched in PtdIns(3,4,5)P3 or PtdIns(3,4)P2 cause only minimal dissociation of Rac1(mant-GDP)·RhoGDI complexes. 1 nmol of Rac1(mant-GDP)·RhoGDI complex was mixed with liposomes (90 nmol of total phospholipids) comprising 0.74 mm (82.22 mol %) DOPC and 0.16 mm (17.77 mol %) DOPS (A); 0.74 mm (81.30 mol %) DOPC, 0.16 mm (17.58 mol %) DOPS, and 10 μm (1.10 mol %) PtdIns (3,4)P2 (B); or 0.74 mm (81.30 mol %) DOPC, 0.16 mm (17.58 mol %) DOPS, and 10 μm (1.10 mol %) (PtdIns(3,4,5)P3 (C). In all experiments the Rac1(mant-GDP)·RhoGDI complex was preincubated with phospholipid liposomes for 12 min at room temperature. The mixtures were subjected to gel filtration on a Superose 12 FPLC column, and the eluates were monitored in line for the fluorescent signal of Rac1(mant-GDP). In each panel, peak 1 represents Rac1(mant-GDP) dissociated from the Rac1(mant-GDP)·RhoGDI complex and bound to the liposomes eluting in the exclusion volume, and peak 2 represents the non-dissociated Rac1(mant-GDP)·RhoGDI complex. The numbers next to peaks 1 and 2 express the amount of Rac1(mant-GDP) dissociated from the complex and left in the complex, respectively, expressed as a percentage of the total amount of Rac1(mant-GDP) present in the complex injected into the column. Calculations were based on the integration of the relevant peaks. The panels illustrate representative individual experiments of at least two to three experiments performed for each combination of components.
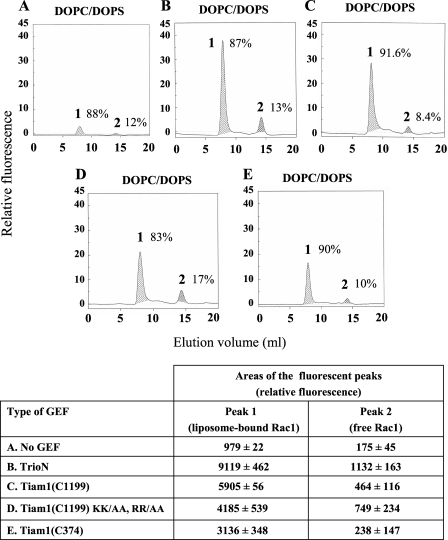
DOPC/DOPS Liposomes Enriched in PtdIns(3,4,5)P3 Combined with TrioN and Free GMPPNP Promote Rac1·RhoGDI Dissociation—Upon cell stimulation, the generation of PtdIns(3,4,5)P3 at the PM occurs coincidentally with the translocation of Rac1 and of Rac-specific GEFs to the PM. Given the large body of evidence showing an important role of PM pPIs in the recruitment of GEFs to the PM (24-33), we examined the effect of Dbl-like GEFs on the stability of Rac1(GDP)·RhoGDI complexes in the presence of PtdIns(3,4,5)P3-enriched liposomes. N-terminal truncation of many Dbl-like GEFs enhances their nucleotide exchange activity in vitro and promotes their association with the PM in vivo (26, 50). Because it has been shown that certain DH-PH tandems of GEFs are capable of independent membrane translocation and can act as constitutively active GEFs in vitro and in vivo (24, 25, 51-54), we decided to investigate the effect of constitutively active DH-PH tandems and free guanine nucleotides on the dissociation of Rac1·RhoGDI complexes in a controlled liposome environment. The schematic representations of the N-terminally truncated Rac GEFs used in this study are shown in Fig. 2. We first tested the ability of various DH-PH domains to catalyze nucleotide exchange on prenylated Rac1(GDP) bound to DOPC/DOPS liposomes. Prenylated Rac1(GDP) was incubated with DOPC/DOPS liposomes in the presence of mant-GDP and various GEF constructs. As shown in Fig. 3, all GEF constructs exhibited the ability to catalyze nucleotide exchange on Rac1 in the presence of DOPC/DOPS liposomes; the C-terminal DH-PH tandem of Tiam1 (Tiam1(C374)) was the least efficient. Most significantly, all GEF constructs preferentially catalyzed guanine nucleotide exchange on prenylated Rac1 bound to DOPC/DOPS compared with free prenylated Rac1 (Fig. 3). This result shows that liposome-associated prenylated Rac1 is a much better substrate for Dbl-like GEFs than free prenylated Rac1, confirming earlier findings by Antonny and co-workers (55). It is also apparent in Fig. 3 that mutating essential residues in the N-terminal PH domain of an N-terminally truncated form of Tiam1 (Tiam1(C1199) KK/AA, RR/AA) (29) led to only a moderate reduction in nucleotide exchange ability.
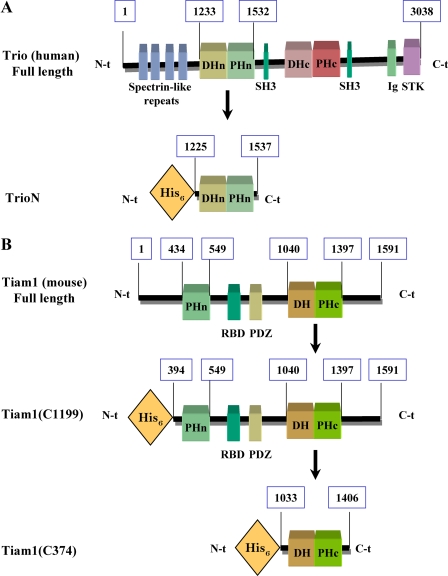
FIGURE 2.
Schematic representation of GEF constructs used in this study. A, domain architecture of full-length Trio and His6-tagged TrioN. Numbers indicate the position of amino acids in the full-length protein. DHn and DHc stand for the N- and C-terminal Dbl homology domains, respectively; SH3 stands for Src homology 3 domain; Ig stands for Ig-like domain, and STK stands for serine-threonine kinase domain. B, domain architecture of full-length Tiam1 and its His6-tagged derivatives Tiam1(C1199) (residues 394-1591) and Tiam1(C374) (residues 1033-1406). Numbers indicate the position of amino acids in the full-length protein. RBD stands for Ras-binding domain, and PDZ stands for PSD-95 Discs large/ZO-1 homology domain.
FIGURE 3.
All GEF constructs display nucleotide exchange activity on prenylated Rac1 bound to DOPC/DOPS liposomes. DOPC/DOPS liposomes (180 nmol of total phospholipids) were mixed with 2 nmol of prenylated Rac1(GDP), 10 nmol of mant-GDP, and one of the following components: buffer (A), 3 nmol of TrioN (B), 3 nmol of Tiam1(C1199) (C), 3 nmol of Tiam1(C1199) KK/AA, RR/AA (D), or 3 nmol of Tiam1(C374) (E). Reaction mixtures were incubated for 12 min at room temperature. The mixtures were subjected to gel filtration on a Superose 12 FPLC column, and the eluates were monitored in line for the fluorescent signal of Rac1(mant-GDP). In each panel, peak 1 represents liposome-bound prenylated Rac1 labeled with mant-GDP eluting in the exclusion volume, and peak 2 represents free prenylated Rac1 labeled with mant-GDP eluting in a volume corresponding to its molecular mass. The numbers next to peaks 1 and 2 express the percentage of the total amount of prenylated Rac1 labeled with mant-GDP associated with the respective peaks. Calculations were based on the integration of the areas of peaks 1 and 2 in the five experimental situations (A, B, C, D, and E), which are listed in the table at the bottom of the figure. The panels illustrate representative individual experiments of three experiments performed for each combination of components. The numerical results in the table are means ± S.E. of three experiments.
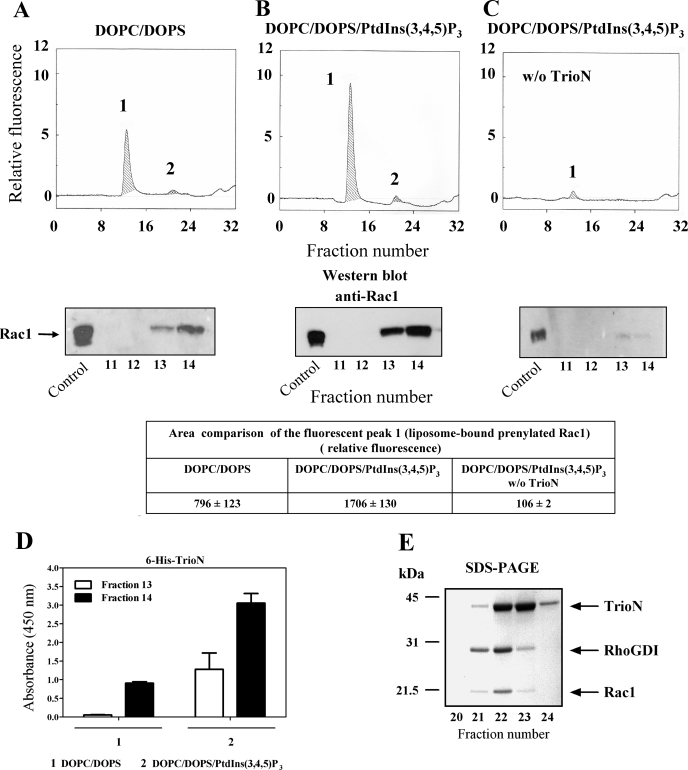
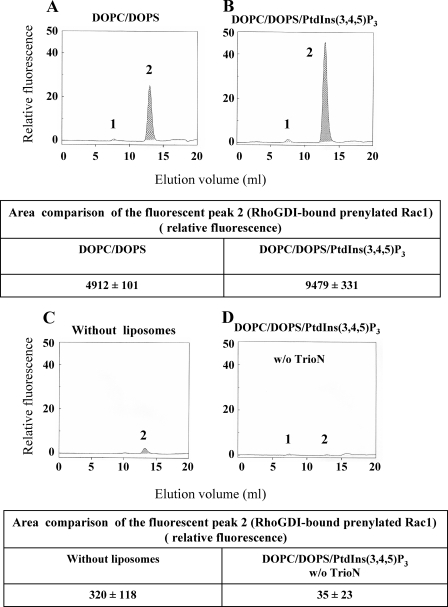
To explore the possible destabilizing effect of various GEFs on Rac1·RhoGDI complexes, we incubated Rac1(GDP)·RhoGDI complexes with DOPC/DOPS or PtdIns(3,4,5)P3-enriched DOPC/DOPS liposomes, TrioN, and mant-GMMPNP. As apparent in Fig. 4A, exposure of Rac1(GDP)·RhoGDI complexes to DOPC/DOPS liposomes, TrioN, and mant-GMPPNP resulted in a fluorescent peak in the exclusion volume (peak 1), indicating that both dissociation of Rac1(GDP)·RhoGDI and exchange of GDP to mant-GMPPNP on Rac1 took place. Enrichment of DOPC/DOPS liposomes with just 1.1 mol % PtdIns(3,4,5)P3 led to a 2-fold increase in the level of fluorescence in peak 1 (Fig. 4, B and associated table), demonstrating an enhancing effect of liposome-incorporated PtdIns(3,4,5)P3 on the ability of TrioN to promote Rac1·RhoGDI dissociation and GDP to mant-GMPPNP exchange on Rac1. The exposure of Rac1·RhoGDI complexes to PtdIns(3,4,5)P3-enriched liposomes and mant-GMPPNP in the absence of TrioN did not result in the appearance of a fluorescent peak in the exclusion volume (Fig. 4C), confirming that the dissociation of Rac1·RhoGDI complexes and GDP to mant-GMPPNP exchange were GEF-dependent. Enhanced dissociation of Rac1·RhoGDI complexes, similar in intensity to that found with DOPC/DOPS liposomes enriched with PtdIns(3,4,5)P3, was also obtained with PtdIns(3,4)P2-, PtdIns(3,5)P2-, and PtdIns(4,5)P2-enriched DOPC/DOPS liposomes in the presence of TrioN and mant-GMPPNP (data not shown). No effect of TrioN and mant-GMPPNP on the stability of Rac1·RhoGDI complexes in the absence of liposomes was observed (data not shown).
FIGURE 4.
Exposure of Rac1(GDP)·RhoGDI complexes to anionic liposomes, mant-GMMPNP, and TrioN causes dissociation of complexes, GDP to mant-GMPPNP exchange on Rac1, and binding of Rac1-GMPPNP to liposomes. 2 nmol of Rac1(GDP)·RhoGDI complex were mixed with 10 nmol of mant-GMPPNP and DOPC/DOPS liposomes (90 nmol of total phospholipids) and 3 nmol of TrioN (A), DOPC/DOPS/PtdIns(3,4,5)P3 liposomes (90 nmol of total phospholipids) and 3 nmol of TrioN (B), or DOPC/DOPS/PtdIns(3,4,5)P3 liposomes (90 nmol of total phospholipids) and buffer (C). In all experiments, the reaction mixtures were incubated for 12 min at room temperature. The mixtures were subjected to gel filtration on a Superose 12 FPLC column, and the eluates were monitored in line for the fluorescent signal of Rac1(mant-GMPPNP) (upper panels). In each panel, peak 1 represents Rac1 dissociated from the Rac1(GDP)·RhoGDI complex, labeled with mant-GMPPNP, and bound to the liposomes eluting in the exclusion volume, and peak 2 represents the nondissociated Rac1(GDP)·RhoGDI complex. The areas of the fluorescent peak 1 in the three experimental situations (A, B, and C) are listed in the table associated with the figure and represent means ± S.E. of three experiments. The panels illustrate representative individual experiments of at least three experiments performed for each combination of components. The dissociation and translocation of Rac1 to liposomes was confirmed by Western blot analysis (lower panels). 35 μl of each fraction (0.6 ml), corresponding to peak 1 (fractions 11 to 14), were subjected to SDS-PAGE followed by immunoblotting with anti-Rac1 antibody as described under “Experimental Procedures.” Rac1·RhoGDI complex, applied to the first lane at the left, served as a control for the ability of anti-Rac1 antibody to detect Rac1. The figure illustrates representative individual experiments of two performed with Rac1(GDP)·RhoGDI complexes mixed with liposomes in the three experimental conditions (A, B, and C). D, detection of liposome-bound His6-tagged TrioN by ELISA. 25-μl aliquots of fractions 13 and 14, corresponding to peak 1, were used to coat the wells of 96-well microplates. His6-tagged TrioN was detected using anti-polyhistidine antibody as described under “Experimental Procedures”; results are means ± S.E. of three experiments. Error bars represent S.E. E, SDS-PAGE analysis of peak 2. 35 μl of each fraction corresponding to peak 2 (fractions 20-24) were analyzed by SDS-PAGE. D and E illustrate representative individual experiments of at least two to three experiments performed. w/o, without.
The quantitative and temporal parameters chosen for the performance of these experiments were derived from preliminary work. This involved varying the concentration of PtdIns(3,4,5)P3 in the DOPC/DOPS liposomes from 0.55 to 2.2 mol %. This range was chosen based on the knowledge that pPIs phosphorylated at position 3 on the inositol ring represent less than 0.025 mol % of total membrane lipids in the resting state and that their concentration increases markedly upon cell stimulation (56). We opted for working with 1.1 mol % PtdIns(3,4,5)P3, taking into consideration that dissociation of the Rac1·RhoGDI complex increased by only 54.9 ± 4.1% when the concentration of PtdIns(3,4,5)P3 was increased 4-fold from 0.55 to 2.2 mol %. The duration of incubation of the Rac1·RhoGDI complex with liposomes and supplements was chosen as 12 min based on the preliminary finding that dissociation of the Rac1·RhoGDI complex at 1-min incubation was 71.4 ± 2.5% of the plateau value, which was reached at 5 min; we chose 12 min to be certain that we were recording results at a state of equilibrium. These time measurements did not include the time elapsed from the injection of the mixture in the gel filtration column to the emergence of the liposomes with bound Rac (40 min).
To confirm the fact that the fluorescent signal associated with material eluting in the exclusion volume represented liposome-bound Rac1 no longer associated with RhoGDI, we analyzed these fractions by immunoblotting with anti-Rac1 (Fig. 4, A, B, and C, middle panels) and anti-RhoGDI antibodies. It is apparent that the amounts of Rac1 translocated to the liposomes as detected by immunoblotting are in good correlation with the levels of fluorescence found in peak 1. No RhoGDI was detected by immunoblotting in any of the fractions derived from peak 1 (results not shown).
We next analyzed fractions containing liposome-bound Rac1 for TrioN co-localization. As seen in Fig. 4D, TrioN was detected by ELISA in fractions also containing liposome-bound Rac1(mant-GMP-PNP). Higher amounts of TrioN were found in fractions containing PtdIns(3,4,5)P3-enriched DOPC/DOPS liposomes, demonstrating a higher affinity of the N-terminal DH-PH module of Trio for PtdIns(3,4,5)P3-enriched DOPC/DOPS liposomes versus DOPC/DOPS liposomes.
Fractions 21-24, corresponding to peak 2, were analyzed by SDS-PAGE, and typical results are illustrated in Fig. 4E. They were found to contain the proteins initially present in the reaction mixture injected into the gel filtration column not bound to the liposomes and were recovered at the elution volumes corresponding to their molecular mass. These were undissociated Rac1(GDP)·RhoGDI complexes, TrioN, and probably some free RhoGDI (dissociated from Rac1 and eluting as a homodimer (37)).
Replacement of GMPPNP by GDP Abrogates the Dissociation of Rac1·RhoGDI Complexes by PtdIns(3,4,5)P3-enriched DOPC/DOPS Liposomes and TrioN—To further characterize the mechanism of Rac1·RhoGDI dissociation by anionic liposomes and GEF, we examined whether the nature of the guanine nucleotide added to the reaction mixture was a determining factor in the dissociation of the complex. For this purpose, we exposed Rac1(GDP)·RhoGDI complexes to DOPC/DOPS or DOPC/DOPS/PtdIns(3,4,5)P3 liposomes in the presence of TrioN and mant-GDP (instead of mant-GMPPNP). As seen in Fig. 5, A and B, no fluorescence was detected in the exclusion volume (peak 1), indicating that replacement of mant-GMP-PNP by mant-GDP completely abrogated the dissociation of the Rac1(GDP)·RhoGDI complex. Surprisingly peak 2, corresponding to the elution volume of the undissociated Rac1·RhoGDI complex, became labeled with mant-GDP, indicating that an exchange of native GDP, bound to Rac1, for exogenous mant-GDP took place. An ∼2-fold increase in the fluorescent signal of peak 2 was observed when PtdIns(3,4,5)P3-enriched DOPC/DOPS liposomes were used compared with DOPC/DOPS liposomes (see table associated with Fig. 5, A and B). Only a minor fluorescent signal was obtained in peak 2 when Rac1(GDP)·RhoGDI was incubated with mant-GDP and TrioN but in the absence of liposomes (Fig. 5C and associated table). This result indicates that GDP to mant-GDP exchange on Rac1 occurred exclusively at the liposome level. Incubation of Rac1(GDP)·RhoGDI with PtdIns(3,4,5)P3-enriched DOPC/DOPS liposomes and mant-GDP but in the absence of TrioN prevented the appearance of fluorescent peak 2, proving that the exchange of GDP to mant-GDP was TrioN-dependent (Fig. 5D and associated table).
FIGURE 5.
Replacement of mant-GMPPNP by mant-GDP eliminates the dissociation of Rac1·RhoGDI complexes by anionic liposomes and TrioN, but nucleotide exchange on Rac1 in complex with RhoGDI is conserved. 2 nmol of Rac1(GDP)·RhoGDI complex and 10 nmol of mant-GDP were mixed with DOPC/DOPS liposomes (90 nmol of total phospholipids) and 3 nmol of TrioN (A), DOPC/DOPS/PtdIns(3,4,5)P3 liposomes (90 nmol of total phospholipids) and 3 nmol of TrioN (B), 3 nmol of TrioN without liposomes (C), and DOPC/DOPS/PtdIns(3,4,5)P3 liposomes (90 nmol of total phospholipids) without TrioN (D). In all experiments, the reaction mixtures were incubated for 12 min at room temperature. The mixtures were subjected to gel filtration on a Superose 12 FPLC column, and the eluates were monitored in line for the fluorescent signal of Rac1(mant-GDP). In each panel, peak 1 represents Rac1 dissociated from the Rac1(GDP)·RhoGDI complex, labeled with mant-GDP, and bound to the liposomes eluting in the exclusion volume, and peak 2 represents the non-dissociated Rac1(mant-GDP)·RhoGDI complex. The areas of fluorescent peak 2 appear in the tables below the upper and lower panels and represent means ± S.E. of three experiments. The panels illustrate representative individual experiments of three experiments performed for each combination of components. w/o, without.
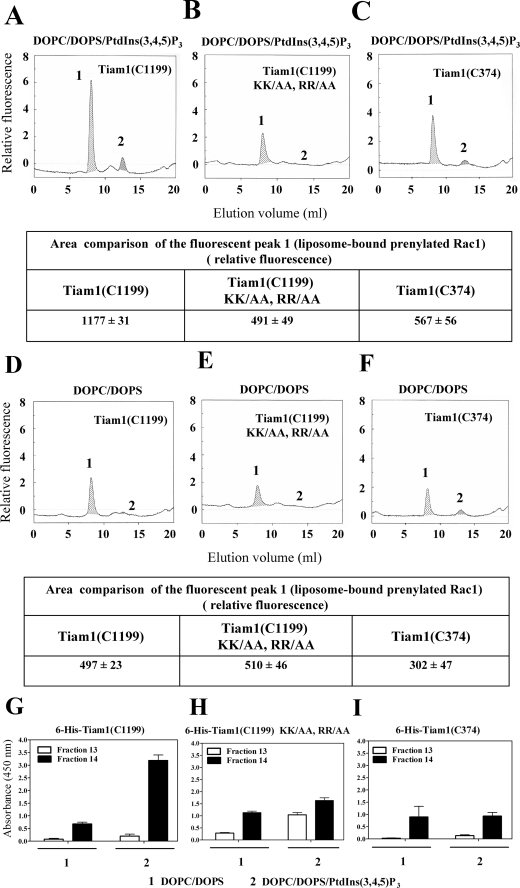
Affinity of the PH Domains of GEFs for PtdIns(3,4,5)P3 Determines the Efficacy of Rac1·RhoGDI Dissociation by PtdIns(3,4,5)P3-enriched Liposomes, GMPPNP, and GEF—Tiam1, a specific GEF for Rac, is a unique Dbl family member that contains a PHn domain in addition to the C-terminal PH (PHc) domain present in the DH-PH tandem. The PHn domain of Tiam1 was shown to possess a higher affinity for PtdIns than the PHc domain with a preference for PtdIns(3,4,5)P3 over PtdIns(3,4)P2 and PtdIns(4,5)P2 (26-29). This is in contrast to the PHc domain, which was shown to interact only with phosphatidylinositol 3-phosphate (57). The enhanced Rac1-(GDP)·RhoGDI dissociation by PtdIns(3,4,5)P3-enriched liposomes versus DOPC/DOPS liposomes (Fig. 4, B versus A) led us to hypothesize that the affinity of certain PH domains for specific pPIs determines the efficacy of a particular GEF in causing Rac1(GDP)·RhoGDI dissociation. To test this assumption, we took advantage of the unique properties of Tiam1 PH domains and made use of the Tiam1 constructs illustrated in Fig. 2B and of a Tiam1 construct with mutations in the PHn domain.
It has been demonstrated that positively charged residues in the β1-β2 loop of PH domains are critical for high affinity binding of phosphoinositide 3-kinase products (58) and that substitution of arginines 459 and 460 with alanines within the N-terminal PH domain of Tiam1 significantly reduced its binding affinity for PtdIns(3,4,5)P3 (29). To impair the ability of Tiam1(C1199) to bind PtdIns(3,4,5)P3 we substituted Lys-452, Lys-453, Arg-459, and Arg-460 with alanines. Tiam1(C1199) KK/AA, RR/AA retained most of its ability to catalyze nucleotide exchange on liposome-bound prenylated Rac1(GDP) (Fig. 3D). Exposure of Rac1(GDP)·RhoGDI complexes to PtdIns(3,4,5)P3-enriched DOPC/DOPS liposomes, mant-GMPPNP, and the Tiam1(C1199) construct (containing both the C-terminal DH-PH tandem and the PHn domain) led to a 2-fold increase in the fluorescent signal in the exclusion volume (peak 1) compared with DOPC/DOPS liposomes (Fig. 6, A versus D and associated tables). On the other hand, incubation of Rac1(GDP)·RhoGDI complexes with PtdIns(3,4,5)P3-enriched DOPC/DOPS liposomes, mant-GMPPNP, and the Tiam1(C1199) KK/AA, RR/AA construct led to a moderate fluorescent signal in the exclusion volume (peak 1) very similar to that observed in the presence of DOPC/DOPS liposomes (Fig. 6, B versus E and associated tables). Exposure of Rac1(GDP)·RhoGDI complexes to PtdIns(3,4,5)P3-enriched DOPC/DOPS liposomes, mant-GMPPNP, and the Tiam1(C374) construct (containing only the C-terminal DH-PH tandem) resulted in a moderate dissociating effect, reflecting the lower affinity of the PHc domain of Tiam1 for PtdIns(3,4,5)P3 (Fig. 6C and associated table). Exposure of Rac1(GDP)·RhoGDI complexes to DOPC/DOPS liposomes, mant-GMPPNP, and Tiam1(C374) resulted in an even lesser dissociating effect (Fig. 6F and associated table). The binding of Tiam1(C1199), Tiam1(C1199) KK/AA, RR/AA, and Tiam1(C374) proteins to liposomes, as assessed by ELISA, revealed a good correlation with the results obtained in the dissociation assays based on in-line recording of fluorescence (Fig. 6, G-I). These results support the hypothesis that the higher the affinity of a PH domain for PtdIns(3,4,5)P3 the more efficient the GEF harboring this domain is in assisting dissociation of Rac1(GDP)·RhoGDI complexes by PtdIns(3,4,5)P3-enriched DOPC/DOPS liposomes.
FIGURE 6.
The N-terminal PH domain of Tiam1 is essential for promoting the dissociation of the Rac1(GDP)·RhoGDI complex in the presence of PtdIns(3,4,5)P3-enriched liposomes and mant-GMPPNP. 2 nmol of Rac1(GDP)·RhoGDI complex and 10 nmol of mant-GMPPNP were mixed with DOPC/DOPS/PtdIns(3,4,5)P3 liposomes (90 nmol of total phospholipids) and 3 nmol of Tiam1(C1199) (A), DOPC/DOPS/PtdIns(3,4,5)P3 liposomes (90 nmol of total phospholipids) and 3 nmol of Tiam1(C1199) KK/AA, RR/AA (B), DOPC/DOPS/PtdIns(3,4,5)P3 liposomes (90 nmol of total phospholipids) and 3 nmol of Tiam1(C374) (C), DOPC/DOPS liposomes (90 nmol of total phospholipids) and 3 nmol of Tiam1(C1199) (D), DOPC/DOPS liposomes (90 nmol of total phospholipids) and 3 nmol of Tiam1(C1199) KK/AA, RR/AA (E), and DOPC/DOPS liposomes (90 nmol of total phospholipids) and 3 nmol of Tiam1(C374) (F). In all experiments, the reaction mixtures were incubated for 12 min at room temperature. The mixtures were subjected to gel filtration on a Superose 12 FPLC column, and the eluates were monitored in line for the fluorescent signal of Rac1(mant-GMPPNP). In each panel, peak 1 represents Rac1 dissociated from the Rac1(GDP)·RhoGDI complex, labeled with mant-GMPPNP, and bound to the liposomes eluting in the exclusion volume, and peak 2 represents the non-dissociated Rac1(GDP)·RhoGDI complex. The areas of fluorescent peak 1 in the six experimental situations are presented in the tables beneath the panels illustrating the in-line recording of fluorescence (A, B, C, D, E, and F) and represent means ± S.E. of three experiments. The panels illustrate representative individual experiments of at least three experiments performed for each combination of components. G, H, and I, detection of liposome-bound His6-tagged Tiam1 constructs by ELISA. 25-μl aliquots of fractions 13 and 14, corresponding to peak 1, derived from the experiments illustrated in A-F were used to coat the wells of 96-well microplates. The His6-tagged Tiam1 proteins were detected using anti-polyhistidine antibody as described under “Experimental Procedures.” Results are means ± S.E. of three experiments. Error bars represent S.E.
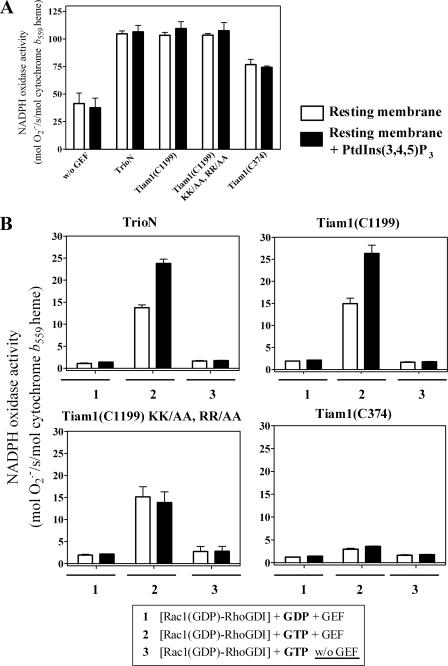
Rac GEFs Harboring PH Domains with High Affinity for
PtdIns(3,4,5)P3 Support Oxidase Activation in Vitro—We
previously reported that oxidase activation can be achieved in vitro
by mixtures of phagocyte membrane liposomes, p67phox, and
prenylated Rac1(GTP) in the absence of an amphiphilic activator and of
p47phox
(6). This was followed by the
reconstitution of oxidase activity in a cell-free system consisting of
membrane liposomes, p67phox, prenylated Rac1(GDP), the
GEFs TrioN or Tiam1(C374), and GTP
(44). We reasoned that
exposure of Rac1(GDP)·RhoGDI complexes to
PtdIns(3,4,5)P3-enriched phagocyte membranes in the presence of a
GEF and GTP will mimic the destabilizing effect on Rac1·RhoGDI
complexes of protein-free anionic liposomes, under the same circumstances,
with the ensuing translocation of prenylated Rac1 to the phagocyte membrane
leading to oxidase activation. We posit that such a system represents a more
physiologic validation of that recently described by us in which oxidase
activation was elicited by Rac1·RhoGDI complexes added to membrane
liposomes enriched with high concentrations of monovalent anionic
phospholipids (37). A
cell-free oxidase activation system was constructed consisting of native or
PtdIns(3,4,5)P3-enriched membrane liposomes, purified Rac1
(GDP)·RhoGDI complexes, p67phox, DH-PH tandems of
various GEFs, and guanine nucleotides. Assays were performed in the absence of
an anionic amphiphilic activator. First we tested whether the enrichment of
phagocyte membrane with PtdIns(3,4,5)P3 will influence the ability
of DH-PH tandems of various GEFs to support oxidase activation in the presence
of free prenylated Rac1(GDP) and GTP. We found that both native phagocyte
membrane and membrane supplemented with PtdIns(3,4,5)P3 were
equally efficient in supporting high levels of
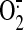 production
(Fig. 7A). The
relative abilities of DH-PH tandems of various GEFs to support oxidase
activation was as follows: TrioN = Tiam1(C1199) = Tiam1(C1199) KK/AA, RR/AA
> Tiam1(C374). When, however, free prenylated Rac1(GDP) was replaced by
Rac1(GDP)·RhoGDI complex, PtdIns(3,4,5)P3-enriched membrane
was more active in
production
(Fig. 7A). The
relative abilities of DH-PH tandems of various GEFs to support oxidase
activation was as follows: TrioN = Tiam1(C1199) = Tiam1(C1199) KK/AA, RR/AA
> Tiam1(C374). When, however, free prenylated Rac1(GDP) was replaced by
Rac1(GDP)·RhoGDI complex, PtdIns(3,4,5)P3-enriched membrane
was more active in 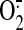 production than was native membrane in the presence of TrioN or Tiam1(C1199)
and GTP (Fig. 7B). No
advantage of PtdIns(3,4,5)P3-enriched membrane over native membrane
was phagocyte membrane supplemented with PtdIns(3,4,5)P3 is
superior to native membrane only when prenylated Rac1(GDP) is found in complex
with RhoGDI and a GEF harboring a PH domain with preference for
PtdIns(3,4,5)P3 is present in the reaction. No oxidase activation
was found with either native or supplemented membrane when GTP was replaced by
GDP.
production than was native membrane in the presence of TrioN or Tiam1(C1199)
and GTP (Fig. 7B). No
advantage of PtdIns(3,4,5)P3-enriched membrane over native membrane
was phagocyte membrane supplemented with PtdIns(3,4,5)P3 is
superior to native membrane only when prenylated Rac1(GDP) is found in complex
with RhoGDI and a GEF harboring a PH domain with preference for
PtdIns(3,4,5)P3 is present in the reaction. No oxidase activation
was found with either native or supplemented membrane when GTP was replaced by
GDP.
FIGURE 7.
Amphiphile-independent oxidase activation in native versus
PtdIns(3,4,5)P3-enriched phagocyte membranes by
Rac1(GDP)·RhoGDI complexes, guanine nucleotides, and GEFs.
In these experiments, native phagocyte membranes or membranes supplemented
with 10 μm PtdIns(3,4,5)P3 were prepared as described
under “Experimental Procedures.” Oxidase activation was assayed in
a cell-free system consisting of either native or supplemented membranes
(equivalent to 5 nm cytochrome b559 heme) mixed
with prenylated Rac1(GDP) (100 nm), p67phox
(300 nm), one of the GEF constructs (200 nm), and GTP (1
μm) (A) or with Rac1(GDP)·RhoGDI complex (100
nm), p67phox (300 nm), one of the
GEF constructs (400 nm), and either GDP or GTP (1 μm)
(B). Assay mixtures were incubated for 5 min at room temperature in
the absence of an anionic amphiphile, and
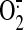 production was initiated
by the addition of NADPH (240 μm). The results are means
± S.E. of three experiments. Error bars represent S.E.
w/o, without.
production was initiated
by the addition of NADPH (240 μm). The results are means
± S.E. of three experiments. Error bars represent S.E.
w/o, without.
DISCUSSION
The small GTPase Rac is required for the assembly and activation of the phagocyte oxidase complex. In resting cells, Rac is found in the cytosol as a prenylated protein, in the GDP-bound form, associated with RhoGDI. Upon cell stimulation, the Rac·RhoGDI complex dissociates, and Rac translocates to the PM. This event is probably preceded by the generation of PtdIns(3,4,5)P3 in the PM and the movement of a Rac-specific GEF to the PM.
In the present work, we establish a connection between Rac·RhoGDI dissociation, the generation of PtdIns(3,4,5)P3, and GEF-mediated GDP to GTP exchange on Rac, all occurring at the level of the PM. To support this contention, we designed an in vitro system, which is meant to reproduce the events in the intact cell, consisting of liposomes of controlled phospholipid composition, purified Rac·RhoGDI complexes, constitutively active Rac GEF constructs, and guanine nucleotides.
The PM of the resting mammalian cell contains less than 20 mol % monovalent anionic phospholipids, mainly phosphatidylserine, in its inner leaflet (56, 59, 60). We reported previously that exposing Rac(GDP)·RhoGDI complexes to liposomes containing negatively charged monovalent phospholipids at concentrations well in excess of those present in the PM of resting cells caused dissociation of RacGDP from RhoGDI and translocation of Rac to liposomes (37). Upon cell activation, the concentration of anionic phospholipids in the inner leaflet of the PM increases sharply as exemplified by the rapid generation of PtdIns(3,4,5)P3 from the constitutive membrane component PtdIns(4,5)P2 by type I phosphoinositide 3-kinase (for a review, see Ref. 49). Thus, we designed an in vitro system based on liposomes with a phospholipid composition imitating that of the PM of either resting or activated phagocytes. We found that exposure of Rac1(GDP)·RhoGDI complexes to DOPC/DOPS liposomes (82.22/17.77 mol %), representing the PM of resting cells, caused only minimal dissociation of the complex. This result is in agreement with our earlier finding that only marginal levels of dissociation were obtained upon exposure of Rac1(GDP)·RhoGDI complexes to native membranes derived from resting phagocytes (37). Enrichment of DOPC/DOPS liposomes with 1.1 mol % PtdIns(3,4,5)P3 or PtdIns(3,4)P2, representing an attempt to mimic the PM of stimulated cells, caused only minimal increases in the level of Rac1·RhoGDI dissociation compared with DOPC/DOPS liposomes.
In the light of these results, we reasoned that a proper model of a stimulated phagocyte should also include the changes in the subcellular localization of Rac GEFs. The majority of Dbl family GEFs are located in the cytosol of resting cells, and membrane association is generally accepted to be a prerequisite for nucleotide exchange on Rac. In the case of GEFs homologous to Dbl, the isolated DH-PH tandem alone is sufficient for PM localization and biological activity in vivo (24, 25, 51-54). Therefore, to investigate the possible role of Dbl-related GEFs in Rac1(GDP)·RhoGDI dissociation, we designed a system consisting of DOPC/DOPS or PtdIns(3,4,5)P3-enriched DOPC/DOPS liposomes (representing the PM of resting and stimulated phagocytes, respectively), DH-PH tandems of GEF, Rac1(GDP)·RhoGDI complexes, and guanine nucleotides. We found indeed that three constitutively active Rac GEF constructs, together with mant-GMPPNP, caused various degrees of Rac1·RhoGDI dissociation and association of mant-GMPPNP-labeled Rac1 with the two liposome prototypes; the liposomes enriched in PtdIns(3,4,5)P3 were more effective. The efficacy of Rac1·RhoGDI dissociation by PtdIns(3,4,5)P3-enriched liposomes appears to be correlated with the presence and affinity of the N-terminal PH domain of a particular GEF for PtdIns(3,4,5)P3. The essential role of DH-PH tandems of GEFs in the dissociation of Rac1·RhoGDI complexes is in good agreement with findings that transgenic expression of the constitutively active, N-terminally truncated Rac GEFs Vav1, Vav2 and Tiam1 causes membrane targeting and activation of endogenous Rac1 (61, 62).
Our study provides additional proof for observations made in the past that membrane-bound prenylated GTPases are better substrates for GEFs (55, 63). We found indeed that the GEF constructs used in our study preferentially catalyzed guanine nucleotide exchange on prenylated Rac1 bound to DOPC/DOPS liposomes as opposed to free prenylated Rac1. Thus, release of Rac1(GDP) from RhoGDI and translocation to the membrane must precede its interaction with GEF. This conclusion has to be reconciled with our finding that replacement of mant-GMPPNP by mant-GDP completely abolished Rac1(GDP)·RhoGDI dissociation and liposome targeting of Rac1 but did not prevent TrioN-dependent exchange of endogenous GDP for exogenous mant-GDP on Rac1. The persistence of GEF-mediated nucleotide exchange on Rac1(GDP) in complex with RhoGDI in the absence of significant complex dissociation and binding of Rac1 to liposomes is best explained by the occurrence of some degree of dissociation or partial opening (64) of the complex (upon random contact with PtdIns(3,4,5)P3-containing liposomes) followed by the transient association of small amounts of Rac1(GDP) with the liposomes. Activated GEF, also bound to liposomes, catalyzes the exchange of Rac1-bound GDP for mant(GDP), and Rac1(mant-GDP) reassociates with RhoGDI due to the higher affinity of Rac1(GDP) for RhoGDI (37, 65).
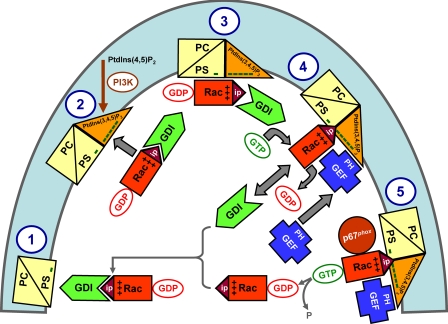
On the basis of our findings, we propose a model for Rac(GDP)·RhoGDI dissociation in the intact cell by the cooperative action of membrane-localized PtdIns(3,4,5)P3, a Rac GEF, and free cytosolic GTP (see Fig. 8 and legend). The dissociation of the Rac(GDP)·RhoGDI complex can be visualized in the form of a “two-step” or “one-step” mechanism. In both, what drives the reaction toward dissociation of the complex is the higher affinity of Rac(GTP) for PtdIns(3,4,5)P3, at the cytosolic aspect of the membrane, than for RhoGDI. In the two-step mechanism, limited spontaneous dissociation of the complex is the initiator of the process followed by the GEF-induced conversion of Rac(GDP) to Rac(GTP), enhanced complex dissociation, and binding of Rac(GTP) to the membrane. In the one-step mechanism, complex dissociation is, from the start, dependent on the GEF-induced GDP to GTP exchange on Rac. We favor the two-step mechanism but are aware of the fact that our methodology only permits measurement of Rac binding to liposomes at equilibrium (=infinite time) and is inappropriate for distinguishing between the two mechanisms.
FIGURE 8.
Extrapolation of the in vitro mechanism of Rac·RhoGDI dissociation by the cooperative action of PtdIns(3,4,5)P3-containing liposomes, GTP, and GEF to events hypothesized to occur in the course of oxidase activation in the intact phagocyte. The proposed sequence of events is as follows. 1, in the PM of the resting cell, represented by a phospholipid composition of less than 20% anionic lipids (phosphatidylcholine (PC)/phosphatidylserine (PS)), the Rac·RhoGDI complexes are in the cytosol. 2 and 3, upon phagocyte stimulation, PtdIns(3,4,5)P3 is generated on the cytosolic aspect of the PM resulting in a marked increase in negative charge. A small proportion of the Rac·RhoGDI complexes dissociates spontaneously, and Rac(GDP) translocates to the PtdIns(3,4,5)P3-enriched PM. 4, a marked enhancement of the dissociation of Rac·RhoGDI complexes takes place upon the translocation of a Rac-specific GEF to the PM by virtue of the affinity of the PH domain of GEF for PtdIns(3,4,5)P3. This leads to GDP to GTP exchange on Rac, also bound to the PM, preventing reassociation with RhoGDI due to the lower affinity of the latter for Rac(GTP). 5, membrane-associated Rac(GTP) interacts with downstream effectors, exemplified by p67phox. Intrinsic or GTPase-activating protein-enhanced GTPase activity leads to the conversion of Rac(GTP) to Rac(GDP), which reassociates with RhoGDI and is returned to the cytosol. “Minus” symbols represent the negative charge of the phospholipids on the cytosolic aspect of the PM, “plus” symbols represent the positive charge of the polybasic C terminus of Rac, and “ip” stands for isoprenyl. PI3K, phosphoinositide 3-kinase. Some elements in this scheme are inspired by a model proposed by Bokoch et al. (see Fig. 7 in Ref. 67).
Recent reports have established the importance of polyvalent anionic pPIs for the targeting of proteins containing polybasic prenylated motifs to the PM. Thus, Yeung and Grinstein (66) demonstrated that membrane-associated cationic molecules, such as K-Ras and Rac1, were released from membrane subdomains following hydrolysis of pPIs and displacement of phosphatidylserine, resulting in a decrease in negative charge. Furthermore Heo et al. (14) reported that the majority of PM-localized small GTPases possess polybasic clusters and bind to the PM by electrostatic interaction with the negatively charged PtdIns(3,4,5)P3 and PtdIns(4,5)P2. Our findings provide a good illustration of the concept emphasizing the contribution of anionic phospholipids, at the inner aspect of the PM, to the attraction, anchoring, and release of cationic cytosolic proteins to and from the membrane (for a review, see Ref. 56). At a more particular level, we provide the first definite experimental demonstration of the involvement of a membrane-bound GEF in the dissociation of the Rac(GDP)·RhoGDI complex and the membrane association of Rac(GTP) as first proposed in a landmark study by Bokoch et al. (67).
Acknowledgments
We thank J. G. Collard (The Netherlands Cancer Institute) for the gift of pcDNA 3.0 plasmid carrying the full-length mouse Tiam1, J. Sondek (University of North Carolina at Chapel Hill) for the gift of mouse Tiam1 DH-PH (residues 1033-1406) expression plasmid, Y. Zheng (Children's Hospital Research Foundation, Cincinnati, OH) for the gift of human TrioN expression plasmid (residues 1225-1537), F. Wientjes (University College, London, UK) for the gift of the GST-p67phox expression plasmid, D. Leonard (Cornell University) for the gift of GST-RhoGDI expression plasmid, and S. Molshanski-Mor for assistance with computer graphics.
This work was supported, in whole or in part, by National Institutes of Health Grant GM46372 (to C. W.). This work was also supported by the Julius Friedrich Cohnheim-Minerva Center for Phagocyte Research, the Ela Kodesz Institute of Host Defense against Infectious Diseases, Israel Science Foundation Grant 19/05, and the Roberts-Guthman Chair in Immunopharmacology (to E. P.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are:
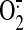 , superoxide; PM, plasma
membrane; DH, Dbl homology; PH, pleckstrin homology; GDI, GDP dissociation
inhibitor; GEF, guanine nucleotide exchange factor; GMPPNP, guanylyl
imidodiphosphate; mant-GMPPNP, 2′-(or
3′)-O-(N-methylanthraniloyl)-guanylyl
imidodiphosphate; mant-GDP, 2′-(or
3′)-O-(N-methylanthraniloyl)-guanosine
5′-diphosphate; FPLC, fast protein liquid chromatography; HPLC, high
pressure liquid chromatography; DOPC,
1,2-dioleoyl-sn-glycero-3-phosphocholine; DOPS,
1,2-dioleoyl-sn-glycero-3-phospho-l-serine; pPI(s),
polyphosphoinositide(s); PtdIns(3,4,5)P3, phosphatidylinositol
3,4,5-trisphosphate; PtdIns(3,4)P2, phosphatidylinositol
3,4-bisphosphate; PtdIns(4,5)P2, phosphatidylinositol
4,5-bisphosphate; PtdIns(3,5)P2, phosphatidylinositol
3,5-bisphosphate; PHn, N-terminal PH; TrioN, N-terminal DH-PH tandem of Trio;
ELISA, enzyme-linked immunosorbent assay; PHc, C-terminal PH.
, superoxide; PM, plasma
membrane; DH, Dbl homology; PH, pleckstrin homology; GDI, GDP dissociation
inhibitor; GEF, guanine nucleotide exchange factor; GMPPNP, guanylyl
imidodiphosphate; mant-GMPPNP, 2′-(or
3′)-O-(N-methylanthraniloyl)-guanylyl
imidodiphosphate; mant-GDP, 2′-(or
3′)-O-(N-methylanthraniloyl)-guanosine
5′-diphosphate; FPLC, fast protein liquid chromatography; HPLC, high
pressure liquid chromatography; DOPC,
1,2-dioleoyl-sn-glycero-3-phosphocholine; DOPS,
1,2-dioleoyl-sn-glycero-3-phospho-l-serine; pPI(s),
polyphosphoinositide(s); PtdIns(3,4,5)P3, phosphatidylinositol
3,4,5-trisphosphate; PtdIns(3,4)P2, phosphatidylinositol
3,4-bisphosphate; PtdIns(4,5)P2, phosphatidylinositol
4,5-bisphosphate; PtdIns(3,5)P2, phosphatidylinositol
3,5-bisphosphate; PHn, N-terminal PH; TrioN, N-terminal DH-PH tandem of Trio;
ELISA, enzyme-linked immunosorbent assay; PHc, C-terminal PH.
References
- 1.Nauseef, W. M. (2004) Histochem. Cell Biol. 122277 -291 [DOI] [PubMed] [Google Scholar]
- 2.Groemping, Y., and Rittinger, K. (2005) Biochem. J. 386401 -416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quinn, M. T., and Gauss, K. A. (2004) J. Leukoc. Biol. 76760 -781 [DOI] [PubMed] [Google Scholar]
- 4.Perisic, O., Wilson, M. I., Karathanassis, D., Bravo, J., Pacold, M. E., Ellson, C. D., Hawkins, P. T., Stephens, L., and Williams, R. L. (2004) Adv. Enzyme Regul. 44 279-298 [DOI] [PubMed] [Google Scholar]
- 5.Molshanski-Mor, S., Mizrahi, A., Ugolev, Y., Dahan, I., Berdichevsky, Y., and Pick, E. (2007) in Neutrophil Methods and Protocols (Quinn, M. T., DeLeo, F. R., and Bokoch, G. M., eds) pp. 385-428, Humana Press, Totowa, NJ
- 6.Gorzalczany, Y., Sigal, N., Itan, M., Lotan, O., and Pick, E. (2000) J. Biol. Chem. 27540073 -40081 [DOI] [PubMed] [Google Scholar]
- 7.Bokoch, G. M., and Zhao, T. (2006) Antioxid. Redox Signal. 81533 -1548 [DOI] [PubMed] [Google Scholar]
- 8.Joseph, G., Gorzalczany, Y., Koshkin, V., and Pick, E. (1994) J. Biol. Chem. 26929024 -29031 [PubMed] [Google Scholar]
- 9.Kreck, M. L., Freeman, J. L., Abo, A., and Lambeth, J. D. (1996) Biochemistry 3515683 -15692 [DOI] [PubMed] [Google Scholar]
- 10.Sarfstein, R., Gorzalczany, Y., Mizrahi, A., Berdichevsky, Y., Molshanski-Mor, S., Weinbaum, C., Hirshberg, M., Dagher, M. C., and Pick, E. (2004) J. Biol. Chem. 27916007 -16016 [DOI] [PubMed] [Google Scholar]
- 11.Ueyama, T., Eto, M., Kami, K., Tatsuno, T., Kobayashi, T., Shirai, Y., Lennartz, M. R., Takeya, R., Sumimoto, H., and Saito, N. (2005) J. Immunol. 1752381 -2390 [DOI] [PubMed] [Google Scholar]
- 12.Stephens, L. R., Jackson, T. R., and Hawkins, P. T. (1993) Biochim. Biophys. Acta 1179 27-75 [DOI] [PubMed] [Google Scholar]
- 13.Missy, K., Van Poucke, V., Raynal, P., Viala, C., Mauco, G., Plantavid, M., Chap, H., and Payrastre, B. (1998) J. Biol. Chem. 27330279 -30286 [DOI] [PubMed] [Google Scholar]
- 14.Heo, W. D., Inoue, T., Park, W. S., Kim, M. L., Park, B. O., Wandless, T. J., and Meyer, T. (2006) Science 3141458 -1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bokoch, G. M. (2005) Trends Cell Biol. 15163 -171 [DOI] [PubMed] [Google Scholar]
- 16.Dovas, A., and Couchman, J. R. (2005) Biochem. J. 3901 -9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DerMardirossian, C., and Bokoch, G. M. (2005) Trends Cell Biol. 15356 -363 [DOI] [PubMed] [Google Scholar]
- 18.Rossman, K. L., Der, C. J., and Sondek, J. (2005) Nat. Rev. Mol. Cell Biol. 6 167-180 [DOI] [PubMed] [Google Scholar]
- 19.Rossman, K. L., Worthylake, D. K., Snyder, J. T., Siderovski, D. P., Campbell, S. L., and Sondek, J. (2002) EMBO J. 211315 -1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snyder, J. T., Worthylake, D. K., Rossman, K. L., Betts, L., Pruitt, W. M., Siderovski, D. P., Der, C. J., and Sondek, J. (2002) Nat. Struct. Biol. 9 468-475 [DOI] [PubMed] [Google Scholar]
- 21.Worthylake, D. K., Rossman, K. L., and Sondek, J. (2000) Nature 408682 -688 [DOI] [PubMed] [Google Scholar]
- 22.Gao, Y., Xing, J., Streuli, M., Leto, T. L., and Zheng, Y. (2001) J. Biol. Chem. 27647530 -47541 [DOI] [PubMed] [Google Scholar]
- 23.Lemmon, M. A. (2003) Traffic 4 201-213 [DOI] [PubMed] [Google Scholar]
- 24.Russo, C., Gao, Y., Mancini, P., Vanni, C., Porotto, M., Falasca, M., Torrisi, M. R., Zheng, Y., and Eva, A. (2001) J. Biol. Chem. 27619524 -19531 [DOI] [PubMed] [Google Scholar]
- 25.Barber, M. A., Donald, S., Thelen, S., Anderson, K. E., Thelen, M., and Welch, H. C. (2007) J. Biol. Chem. 28229967 -29976 [DOI] [PubMed] [Google Scholar]
- 26.Michiels, F., Stam, J. C., Hordijk, P. L., van der Kammen, R. A., Ruuls-Van Stalle, L., Feltkamp, C. A., and Collard, J. G. (1997) J. Cell Biol. 137387 -398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rameh, L. E., Arvidsson, A., Carraway, K. L., III, Couvillon, A. D., Rathbun, G., Crompton, A., VanRenterghem, B., Czech, M. P., Ravichandran, K. S., Burakoff, S. J., Wang, D. S., Chen, C. S., and Cantley, L. C. (1997) J. Biol. Chem. 27222059 -22066 [DOI] [PubMed] [Google Scholar]
- 28.Fleming, I. N., Gray, A., and Downes, C. P. (2000) Biochem. J. 351173 -182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ceccarelli, D. F., Blasutig, I. M., Goudreault, M., Li, Z., Ruston, J., Pawson, T., and Sicheri, F. (2007) J. Biol. Chem. 28213864 -13874 [DOI] [PubMed] [Google Scholar]
- 30.Han, J., Luby-Phelps, K., Das, B., Shu, X., Xia, Y., Mosteller, R. D., Krishna, U. M., Falck, J. R., White, M. A., and Broek, D. (1998) Science 279558 -560 [DOI] [PubMed] [Google Scholar]
- 31.Das, B., Shu, X., Day, G. J., Han, J., Krishna, U. M., Falck, J. R., and Broek, D. (2000) J. Biol. Chem. 27515074 -15081 [DOI] [PubMed] [Google Scholar]
- 32.Nimnual, A. S., Yatsula, B. A., and Bar-Sagi, D. (1998) Science 279560 -563 [DOI] [PubMed] [Google Scholar]
- 33.Muroya, K., Kawasaki, Y., Hayashi, T., Ohwada, S., and Akiyama, T. (2007) Biochem. Biophys. Res. Commun. 355 85-88 [DOI] [PubMed] [Google Scholar]
- 34.Pick, E., Gorzalczany, Y., and Engel, S. (1993) Eur. J. Biochem. 217441 -455 [DOI] [PubMed] [Google Scholar]
- 35.Abo, A., Webb, M. R., Grogan, A., and Segal, A. W. (1994) Biochem. J. 298585 -591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chuang, T. H., Bohl, B. P., and Bokoch, G. M. (1993) J. Biol. Chem. 26826206 -26211 [PubMed] [Google Scholar]
- 37.Ugolev, Y., Molshanski-Mor, S., Weinbaum, C., and Pick, E. (2006) J. Biol. Chem. 28119204 -19219 [DOI] [PubMed] [Google Scholar]
- 38.Bromberg, Y., and Pick, E. (1984) Cell. Immunol. 88213 -221 [DOI] [PubMed] [Google Scholar]
- 39.Shpungin, S., Dotan, I., Abo, A., and Pick, E. (1989) J. Biol. Chem. 2649195 -9203 [PubMed] [Google Scholar]
- 40.Pick, E., Bromberg, Y., Shpungin, S., and Gadba, R. (1987) J. Biol. Chem. 26216476 -16483 [PubMed] [Google Scholar]
- 41.Sigal, N., Gorzalczany, Y., and Pick, E. (2003) Inflammation 27147 -159 [DOI] [PubMed] [Google Scholar]
- 42.Stewart, J. C. (1980) Anal. Biochem. 10410 -14 [DOI] [PubMed] [Google Scholar]
- 43.Bligh, E. G., and Dyer, W. J. (1959) Can. J. Biochem. Physiol. 37911 -917 [DOI] [PubMed] [Google Scholar]
- 44.Mizrahi, A., Molshanski-Mor, S., Weinbaum, C., Zheng, Y., Hirshberg, M., and Pick, E. (2005) J. Biol. Chem. 2803802 -3811 [DOI] [PubMed] [Google Scholar]
- 45.Bradford, M. M. (1976) Anal. Biochem. 72248 -254 [DOI] [PubMed] [Google Scholar]
- 46.Abo, A., and Pick, E. (1991) J. Biol. Chem. 26623577 -23585 [PubMed] [Google Scholar]
- 47.Jouan, A., Pilloud-Dagher, M. C., Fuchs, A., and Vignais, P. V. (1993) Anal. Biochem. 214252 -259 [DOI] [PubMed] [Google Scholar]
- 48.Gorzalczany, Y., Alloul, N., Sigal, N., Weinbaum, C., and Pick, E. (2002) J. Biol. Chem. 27718605 -18610 [DOI] [PubMed] [Google Scholar]
- 49.Welch, H. C., Coadwell, W. J., Stephens, L. R., and Hawkins, P. T. (2003) FEBS Lett. 546 93-97 [DOI] [PubMed] [Google Scholar]
- 50.Aghazadeh, B., Lowry, W. E., Huang, X. Y., and Rosen, M. K. (2000) Cell 102625 -633 [DOI] [PubMed] [Google Scholar]
- 51.Estrach, S., Schmidt, S., Diriong, S., Penna, A., Blangy, A., Fort, P., and Debant, A. (2002) Curr. Biol. 12 307-312 [DOI] [PubMed] [Google Scholar]
- 52.Bellanger, J. M., Estrach, S., Schmidt, S., Briancon-Marjollet, A., Zugasti, O., Fromont, S., and Debant, A. (2003) Biol. Cell 95625 -634 [DOI] [PubMed] [Google Scholar]
- 53.Baumeister, M. A., Rossman, K. L., Sondek, J., and Lemmon, M. A. (2006) Biochem. J. 400563 -572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hill, K., Krugmann, S., Andrews, S. R., Coadwell, W. J., Finan, P., Welch, H. C., Hawkins, P. T., and Stephens, L. R. (2005) J. Biol. Chem. 2804166 -4173 [DOI] [PubMed] [Google Scholar]
- 55.Robbe, K., Otto-Bruc, A., Chardin, P., and Antonny, B. (2003) J. Biol. Chem. 2784756 -4762 [DOI] [PubMed] [Google Scholar]
- 56.Yeung, T., Terebiznik, M., Yu, L., Silvius, J., Abidi, W. M., Philips, M., Levine, T., Kapus, A., and Grinstein, S. (2006) Science 313347 -351 [DOI] [PubMed] [Google Scholar]
- 57.Snyder, J. T., Rossman, K. L., Baumeister, M. A., Pruitt, W. M., Siderovski, D. P., Der, C. J., Lemmon, M. A., and Sondek, J. (2001) J. Biol. Chem. 27645868 -45875 [DOI] [PubMed] [Google Scholar]
- 58.Isakoff, S. J., Cardozo, T., Andreev, J., Li, Z., Ferguson, K. M., Abagyan, R., Lemmon, M. A., Aronheim, A., and Skolnik, E. Y. (1998) EMBO J. 175374 -5387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quinn, P. J. (2002) Subcell. Biochem. 3639 -60 [DOI] [PubMed] [Google Scholar]
- 60.Mason, R. J., Stossel, T. P., and Vaughan, M. (1972) J. Clin. Investig. 512399 -2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Price, M. O., Atkinson, S. J., Knaus, U. G., and Dinauer, M. C. (2002) J. Biol. Chem. 27719220 -19228 [DOI] [PubMed] [Google Scholar]
- 62.Moissoglu, K., Slepchenko, B. M., Meller, N., Horwitz, A. F., and Schwartz, M. A. (2006) Mol. Biol. Cell 172770 -2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pechlivanis, M., Ringel, R., Popkirova, B., and Kuhlmann, J. (2007) Biochemistry 465341 -5348 [DOI] [PubMed] [Google Scholar]
- 64.Faure, J., Vignais, P. V., and Dagher, M. C. (1999) Eur. J. Biochem. 262879 -889 [DOI] [PubMed] [Google Scholar]
- 65.Sasaki, T., Kato, M., and Takai, Y. (1993) J. Biol. Chem. 26823959 -23963 [PubMed] [Google Scholar]
- 66.Yeung, T., and Grinstein, S. (2007) Immunol. Rev. 21917 -36 [DOI] [PubMed] [Google Scholar]
- 67.Bokoch, G. M., Bohl, B. P., and Chuang, T. H. (1994) J. Biol. Chem. 26931674 -31679 [PubMed] [Google Scholar]