Abstract
An accurate protein concentration is an essential component of most biochemical experiments. The simplest method to determine a protein concentration is by measuring the A280, using an absorption coefficient (ε), and applying the Beer-Lambert law. For some metalloproteins (including all transferrin family members) difficulties arise because metal binding contributes to the A280 in a non-linear manner. The Edelhoch method is based on the assumption that the ε of a denatured protein in 6 M guanidine-HCl can be calculated from the number of the tryptophan, tyrosine, and cystine residues. We extend this method to derive ε values for both apo- and iron-bound transferrins. The absorbance of an identical amount of iron containing protein is measured in: 1) 6 M guanidine-HCl (denatured, no iron); 2) pH 7.4 buffer (non-denatured with iron); and 3) pH 5.6 (or lower) buffer with a chelator (non-denatured without iron). Since the iron free apo-protein has an identical A280 under non-denaturing conditions, the difference between the reading at pH 7.4 and the lower pH directly reports the contribution of the iron. The method is fast and consumes ~1 mg of sample. The ability to determine accurate ε values for transferrin mutants that bind iron with a wide range of affinities has proven very useful; furthermore a similar approach could easily be followed to determine ε values for other metalloproteins in which metal binding contributes to the A280.
Keywords: Transferrin, molar absorption coefficient, metalloproteins, Edelhoch method, ligand metal charge transfer
The transferrins (TF) are a family of glycoproteins whose members (serum transferrin, ovotransferrin and lactoferrin) are responsible for transporting iron and/or preventing bacterial growth by sequestering iron. Human serum transferrin (hTF) is an 80 kDa glycoprotein comprised of two homologous lobes, termed the N- and C-lobes, each folding to form a cleft in which ferric iron (Fe3+) binds [1]. Diferric hTF delivers iron to cells by binding to a specific transferrin receptor (TFR) and undergoing receptor mediated endocytosis [2]. The reduced pH within the endosome facilitates iron release from hTF (which remains bound to TFR) and is then returned to the blood to acquire more iron. The Fe3+ ion bound in each lobe of hTF is coordinated by one aspartate, two tyrosines, and one histidine residue [3,4]. The coordination sphere around the Fe3+ ion is completed by two oxygen atoms from the synergistic anion, carbonate, which is anchored by an arginine residue. The Fe3+ binding ligands are held in position through an extensive hydrogen bonding network referred to as the “second shell”. As might be expected, mutation of any of the residues involved in the second shell (for example in the N-lobe of hTF, Gly65, Glu83, Tyr85, Arg124, Lys206, Ser248, and Lys296) changes both the iron coordination and binding affinity [5].
We have developed a robust expression system to produce large quantities of both recombinant hTF N-lobe and full length hTF [6–9]. To accurately assess the effect of a mutation, various assays have been developed to measure Fe3+ binding and release, in the presence and absence of the receptor [10–14]. Many of these assays take advantage of the ligand to metal charge transfer (LMCT) band (centered at ~ 470 nm) produced by interaction of the two tyrosine ligands with Fe3+ [15]. A unique and interesting consequence of this LMCT is the disruption of the π to π* transition energy of the liganding tyrosine residues. This disruption results in an increase in the A280 that extends and overlaps the intrinsic tryptophan fluorescence [14,16]. As a result, tryptophan fluorescence in Fe3+-bound hTF is quenched ~70% compared to apo-hTF. Several laboratories, including our own, have monitored the recovery of tryptophan fluorescence to derive rate constants for iron release [10,14,17,18]. To allow valid comparisons of the properties of the various mutants, it is essential have a method to determine accurate concentrations for both the apo- and iron-bound conformations of hTF and hTF mutants. In particular, surface plasmon resonance binding studies and steady-state/time resolved fluorescence measurements require precise knowledge of their concentrations.
The simplest method for determining protein concentration is by measuring the A280 and using the Beer-Lambert law:
| (1) |
where, ε is the molar absorption coefficient (M−1 cm−1), l is the path length (cm), and C is the protein concentration (M). Obviously, this approach can be used only when an accurate ε280 is available. For many years ε280 values were experimentally determined by three techniques: the dry weight method, composition determinations by quantitative amino acid analysis, and Kjeldahl nitrogen determination [19–21]. All three methods are technically challenging and consume large amounts of both time and sample. Additionally, although each technique provided reproducible ε280 values in the hands of skilled practitioners, substantial deviations among them often occurred [22,23].
To circumvent these difficulties, Edelhoch developed a spectroscopic method to accurately predict the number of tryptophan and tyrosine residues in a protein of unknown composition by comparison to model compounds in a denaturant. Wetlaufer, Edelhoch and others had determined that these two residues, along with cystines, are the only amino acids that contribute to protein absorbance above 275 nm [24–26]. It was found that denaturing globular proteins in 6 M guanidine-HCl (GdHCl) provided a reliable method to determine ε280 [27,28]. With the assumption that the ε280 of a denatured protein in 6 M GdHCl can be calculated from the number of the Trp, Tyr, and cystine residues using the ε280 of appropriate model peptides or derivatived amino acids in 6 M GdHCl, the Edelhoch method was born.
With the enormous increase in the availability of DNA sequence information to provide exact amino acid compositions, much subsequent effort has been devoted to improving and substantiating the validity of predicting accurate ε280 values from the composition alone [29–31]. In the most recent update, Pace et al. [23] evaluated 116 ε280 values for 80 proteins. Based on this analysis the ε280 of a folded protein in water can be predicted by the following equation:
| (2) |
Although this calculation provides a reasonable estimate of ε280 for many proteins, it cannot be used for any protein containing a metal ligand or a prosthetic group that absorbs in the near UV.
A common approach for obtaining ε280 values for members of the TF family involves titration of apo samples with ferric iron [6,32–35]. This is a lengthy and tedious process because it involves the displacement of a chelator (needed to keep Fe3+ in solution) by the TF (see below). Additionally, since the visible signal produced is relatively weak, this approach is inherently rather insensitive and prone to error. More recently, cobalt was substituted for Fe3+ because Co3+: 1) binds to TF rapidly and tightly, 2) forms a stable complex, and 3) produces a stronger LMCT signal (centered at 410 nm) [36]. At a saturating concentration of cobalt, a break-point is reached corresponding to the stoichiometric binding of Co3+ to TF. The concentration of Co3+ (determined by atomic absorbance analysis) at the break-point allows an accurate determination of the TF concentration. The major limitation of any titration method is that only mutants that bind metal tightly yield a sharp break-point. In addition, although Co3+ does not result in the destruction of the protein, the binding is essentially irreversible and, due to low sensitivity, ~ 6–10 mg of protein is required for triplicate determinations.
We recognized that a faster and a more sensitive method was needed to determine accurate ε280 values for our many recombinant hTF mutants in which metal binding contributes variably to the UV spectrum [5]. We report here an extension of the Edelhoch method to determine the ε280 of apo- and iron-bound wild-type (WT) and mutant constructs of hTF N-lobe, hTF and ovotransferrin (oTF). A comparison between previously reported ε280 values and the ε280 values obtained demonstrates the utility, accuracy and simplicity of this approach. Furthermore, it could easily be adapted to determine accurate ε280 values of other metalloproteins with minimal expenditure of sample.
Materials and methods
Expression and purification of hTF, oTF, and hTF N-lobe constructs
All recombinant proteins including full length hTF, oTF, hTF N-lobe and mutants thereof, were expressed in baby hamster kidney cells containing the relevant cDNA in the pNUT vector and purified as previously described [7–9,37,38].
Determination of molar absorption coefficient
Samples of TF saturated with Fe3+ (~ 0.3 A280 units, 1–10 µL of stock solution) are added to a 1.0 mL quartz cuvette with a 1 cm path length containing 6 M GdHCl (final volume of 500 µL), mixed thoroughly and equilibrated for 10 minutes at 25°C to ensure complete denaturation. To determine the λmax of the denatured protein, absorbance scans (from 240 – 340 nm) are recorded at 25°C on a Varian Cary 100 spectrophotometer in dual beam mode using a reference cuvette containing only 6 M GdHCl. The ε in 6 M GdHCl is calculated at the absorbance maximum using the equation:
| (3) |
where, #Trp, #Tyr, and #cystine are the number of each type of residue in the protein and the ελ values at the λmax for Trp, Tyr, and cystine (in 6 M GdHCl) come from Pace et al. [23]. The protein concentration in 6 M GdHCl is then calculated by recording the absorbance value at λmax and using Equation 3 below:
| (3) |
An identical amount of Fe3+ sample is then added to 100 mM HEPES, pH 7.4, and either 100 mM MES, pH 5.6 with a chelator (4 mM EDTA) or 100 mM acetate buffer, pH 4.0 with 4 mM EDTA (final volume 500 µL) and equilibrated for ~ 10 minutes. The choice of MES or acetate buffer (to remove the iron yielding the apo protein) is dictated by the binding affinity (see Results). Since the amount of protein added to each buffer is identical to the amount added to 6 M GdHCl, the concentration of protein in buffer (C(buffer)) is identical to the concentration determined in 6 M GdHCl (Cλmax (6 M GdHCl)). The absorbance at 280 nm of native protein is recorded by scanning between 240 and 340 nm and the ε280 calculated using the relationship:
| (4) |
As described by Pace et al [23], the contribution of light scattering to the A λmax in GdHCl is determined by multiplying the absorbance at 329 nm by 2 and corrected by subtracting that value from the A λmax [23]. (All TF samples had a λmax in GdHCl of 276 nm). Likewise for the apo samples in each buffer the contribution from light scattering at 280 nm is corrected for by multiplying the absorbance at 333 nm by 2 and subtracting that value from the value at A280 [23]. We note that the presence of the LMCT precludes correction for the contribution of light scattering in any of the iron bound samples. Following this protocol, the ε280 of an individual protein can be calculated in less than 1 h resulting in the consumption of ~1 mg of protein (with determinations in triplicate for each buffer). Obviously, the proteins from the determinations under non-denaturing conditions (HEPES, MES and acetate) can easily be recovered.
Results and discussion
Since it was first described, the Edelhoch method has provided a simple and accurate method for experimentally determining ε280 values for many different proteins. The basic tenet of the method is that denaturation of any protein removes all interactions of Trp, Tyr and cystine residues with nearby residues which influence their spectral properties. In any unfolded protein, the Trp, Tyr and cystine residues are thus “normalized” and their ε values are equal to model compounds in 6 M GdHCl. The ε for the unfolded protein is calculated based on the number of each residue, providing an accurate estimate of the protein concentration. The ε280 of the native protein is then determined by placement of an equal amount of protein solution into a suitable buffer. In the present study an extension of the Edelhoch method has been used to determine the ε280 of apo- and Fe3+-bound TF samples. For iron-bound TF, placement into 6 M GdHCl must result in the loss of the LMCT between Fe3+ and tyrosines such that the two liganding tyrosines will have normalized ε values. To document that this is the case, the spectra of the Fe3+ bound hTF N-lobe in HEPES, MES (with chelator), and 6 M GdHCl are shown in Figure 1. As expected there is a hypochromic shift and a decrease in the absorbance above 300 nm when Fe3+-bound N-lobe is placed in the lower pH buffer with chelator, signifying generation of apo-N-lobe. The spectrum of the Fe3+-bound N-lobe in 6 M GdHCl is nearly identical to the spectrum of apo-N-lobe indicating that denaturation does result in the complete loss of Fe3+ (and validating the method). Similar results were found for all TF samples that were analyzed.
Figure 1.
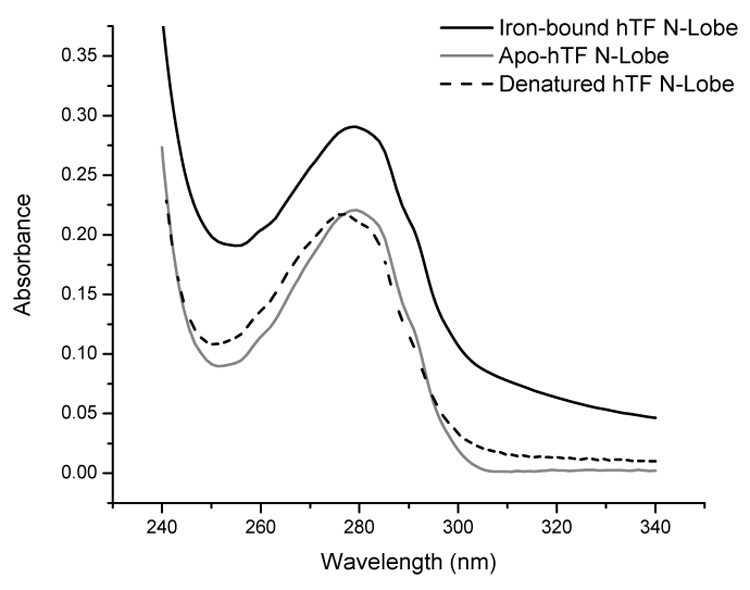
Baseline corrected absorbance scans of iron-bound hTF N-lobe (100 mM HEPES, pH 7.4), apo-hTF N-lobe (100 mM MES, pH 5.6 and 4 mM EDTA) and denatured hTF N-lobe (6 M GdHCl). Samples were equilibrated in buffer for ~ 10 minutes. Spectra were collected by scanning between 240–340 nm at 25°C and baseline corrected.
The experimentally derived apo- and Fe3+-bound ε280/1000 (mM absorption coefficient) values for a variety of hTF N-lobe mutants are reported in Table 1. Critical to the interpretation of the results, the mM ε280 value of apo-hTF N-lobe placed into either HEPES or MES buffer is identical. WT apo-hTF N-lobe has a molecular weight of 37,151 Da and contains 3 Trp, 14 Tyr and 16 cysteine residues (forming 8 cystines). As shown in Table 1, we were able to obtain mM ε280 values for the apo form of most of the mutants. In addition to the experimentally derived values, the calculated mM ε280 values for each apo sample (using Equation 2) are also presented in Table 1. It is significant that the percent deviation for the experimental apo-samples from the calculated values is very small (standard deviation of 1.6%), indicating that the calculated values provide reasonable estimates of the concentration. We note that the mM ε280 value for the apo form of the K206E mutant could not be determined because iron was not completely removed in a reasonable period of time under either the MES or acetate buffer conditions. Due to the excellent agreement between the experimental and calculated values, the calculated mM ε280 value for the apo-form of the K206E mutant would provide a satisfactory estimate of the concentration while the ε value for the Fe3+ form are accurately determined experimentally.
Table 1. Millimolar absorption coefficients (ε) at 280 nm for hTF N-lobe and various mutants.
Iron samples were assayed either in 100 mM HEPES, pH 7.4 (Iron) or 100 mM MES, pH 5.6 and 4 mM EDTA (Apo) to derive experimental ε280 as indicated below.
| hTF N-lobe | Calculated ε280 (Apo)a | Experimental ε280 (Apo)b | Experimental ε280 (Iron)b | % Increase due to iron c | % Difference calc/expd |
|---|---|---|---|---|---|
| Wild Type | 38.4 | 38.1 ± 0.2 | 50.1 ± 0.4 | 31.5 | −0.8 |
| F94S | 38.4 | 37.6 ± 0.5 | 50.2 ± 0.4 | 33.5 | −2.1 |
| K206E | 38.4 | - | 50.0 ± 0.1 | 30.2 | − |
| D63E | 38.4 | 39.0 ± 0.3 | 48.5 ± 0.3 | 24.4 | 1.5 |
| H119Q | 38.4 | 39.9 ± 0.9 | 49.0 ± 0.4 | 22.8 | 3.8 |
| H207E | 38.4 | 37.9 ± 0.3 | 45.5 ± 0.3 | 20.1 | −1.3 |
| W8Y | 34.4 | 34.8 ± 0.6 | 41.3 ± 0.2 | 18.7 | 1.1 |
| W264Y | 34.4 | 34.8 ± 0.4 | 41.3 ± 0.2 | 18.7 | 1.1 |
| W128Y | 34.4 | 33.9 ± 0.7 | 40.2 ± 0.3 | 18.6 | −1.5 |
| L66E | 38.4 | 38.9 ± 0.5 | 46.0 ± 0.2 | 18.3 | 1.3 |
| L66W | 43.9 | 45.3 ± 0.5 | 52.4 ± 0.3 | 15.7 | 3.1 |
| Y85F | 36.9 | 37.8 ± 0.2 | 42.2 ± 0.2 | 11.6 | 2.4 |
| D63S | 38.4 | 39.0 ± 0.2 | 43.0 ± 0.3 | 10.3 | 1.5 |
| Y95F | 36.9 | 37.5 ± 0.2 | 40.4 ± 0.3 | 7.7 | 1.6 |
| E83A | 38.4 | 38.5 ± 0.4 | 41.3 ± 0.5 | 7.3 | 0.3 |
| Y188F | 36.9 | 37.1 ± 0.1 | - | - | 0.5 |
Calculated from Eq. 2.
Values are means ± STD of at least three determinations and in the case of the apo samples corrected for light scattering.
Percent increase is calculated as 100 * [ε(Iron) - ε(Apo)]/ ε(Apo).
Percent difference is calculated as 100 *[ε(Apo)- ε(Calc)]/ ε(Apo).
Interestingly, the apo-form of the H119Q mutant shows the largest deviation between the calculated and experimental value (3.8 %). As previously observed, His119 strongly quenches the signal from Trp128 which is ~ 7 Å away [39]. Obviously, in the H119Q mutant this quenching effect is ablated with a predictable effect on the A280.
The importance of the present work is that it provides a protocol to easily obtain mM ε280 values for the Fe3+ form of each of the N-lobe mutants (see Table 1- Column entitled “% increase due to iron”). As described earlier, titrations with either iron or cobalt are tedious, time and sample consuming, and, especially if the binding is weak, not accurate. When a new mutant is produced characterization routinely involves determination of its spectral parameters and rate constants for iron release to assess the effect of the mutation on iron coordination. Examination of the change in mM ε280 as a result of iron binding shows that the various mutants roughly segregate into three groups (Table 1). Those with release rates equal to or slower than WT N-lobe (including F94S and K206E), show the largest change (~30%) in mM ε280 as a result of the presence of iron. Mutants with moderate changes in their spectral properties and intermediate rate constants of iron release show a smaller increase (17–25%). As might be expected, mutations which disrupt the iron binding ligands (D63S and Y95F) or second shell residues which weaken binding (E83A and Y85F) have the smallest increase (7–12%) and the fastest rate constants for iron release. The changes in the mM ε280 values for the Fe3+ N-lobe samples compared to the apo-samples thus correlate very well with the properties of each mutant (Table 1) [5].
The experimental and calculated mM ε280 values for apo - and Fe3+ full length hTF (8 Trp, 26 Tyr and 19 cystine residues) and oTF (10 Trp, 21 Tyr and 15 cystine residues) are given in Table 2. In order to completely remove Fe3+ from these samples within 10 minutes, it was necessary to use pH 4.0 acetate buffer (with chelator) in place of MES buffer. Similar to the results with the hTF N-lobe, the calculated apo-values did not appreciably deviate from the experimental values. Additionally, both hTF and oTF show a similar increase in mM ε280 as a result of iron coordination (24%), consistent with fact that both contain identical iron binding ligands in each lobe. Interestingly, the two monoferric hTF samples have a nearly identical increase in the mM ε280 as a result of iron binding despite the fact that there is a 9 nm difference between their LMCT in the visible region [9].
Table 2. Millimolar absorption coefficients (ε) at 280 nm for full length hTF and oTF.
All samples were assayed in 100 mM HEPES, pH 7.4 (iron-bound) or 100 mM Acetate, pH 4.0 and 4 mM EDTA (Apo).
| Protein | Calculated ε280 (Apo)a | Experimental ε280 (Apo)b | Experimental ε280 (Iron)b | % Increase due to ironc | % Difference Calc/Expc |
|---|---|---|---|---|---|
| Diferric hTF | 85.1 | 84.0 ± 0.2 | 103.9 ± 0.2 | 23.7 | −1.3 |
| Monoferric-N hTFd | 82.1 | 81.4 ± 0.3 | 92.5 ± 0.3 | 13.6 | −0.9 |
| Monoferric-C hTFe | 82.1 | 81.5 ± 0.2 | 92.1 ± 0.2 | 13.0 | −0.7 |
| Apo hTFf | 79.2 | 80.1 ± 0.4 | - | - | 1.1 |
| Diferric oTF | 88.2 | 87.9 ± 0.1 | 109.3 ± 0.2 | 24.3 | −0.3 |
Calculated from Eq. 2.
Values are means ± STD of at least three determinations.
Percent increase and difference are calculated as in Table 1.
Recombinant non-glycosylated His tagged hTF with Y426F and Y517F mutations inhibiting its ability to coordinate iron in the C-lobe.
Recombinant non-glycosylated His tagged hTF with Y95F and Y188F mutations inhibiting its ability to coordinate iron in the N-lobe.
This is non-glycosylated authentic recombinant His tagged apo-hTF mutant (Y95F/Y188F/Y426F/Y517F).
Over the past 40 years, the mM ε280 values of apo- and iron-bound hTF have been determined by a variety of techniques (Table 3). The mM ε280 values for apo-hTF show significant variation (83.8 to 93.0). In the earlier studies [40,41], this variation can be directly attributed to differences in the molecular weight since conversion of our mM ε280 values to A280 (1%) brings them within experimental error. Variability in the mass is mainly due to inconsistency in estimates of the contribution of glycosylation. Electrospray mass analysis of hTF samples from three commercial sources provided experimental values ranging from 79,559 to 79,619 [8]. Our recombinant non-glycosylated hTF has a mass of 75,143 and the His tagged version of this construct has a mass of 76,861. The highest mM ε280 value reported for apo-hTF [42] came from titration with iron, which, as described above, is experimentally challenging. For oTF (apo- and iron-bound) the values from the dry weight method and the Co titration correlate well with our determinations from this study. Overall, comparison of the mM ε280 values of apo-and iron-bound hTF and oTF clearly demonstrate the accuracy, sensitivity and reproducibility of our modified Edelhoch method.
Table 3. Millimolar absorption coefficients (ε) and molecular masses for hTF and oTF.
| Method | ε280 (Apo) | ε280 (Fe3+) | A(280,1%) Apo, Iron | MW hTF | Glycan | Reference |
|---|---|---|---|---|---|---|
| hTF | ||||||
| Fe titration | 93.0 | - | - | - | [42] | |
| Dry weight | 92.3 | 114.0 | 11.4, 14.1 | 81,000 | Yes | [40] |
| Not specified | 91.2 | - | - | - | [46] | |
| Not specified | 88.9 | - | 11.4, - | 78,000 | Yes | [44] |
| Dry weight | 87.2 | 112.0 | 10.9, 14.0 | 80,000 | Yes | [41] |
| Not specified | 87.2 | - | - | - | [48] | |
| Not specified | 86.6 | - | - | - | [45] | |
| Co titration | 85.2 | - | - | - | [36] | |
| Calculated | 85.1* | - | 11.1, - | 75,181 | No | [23] |
| Calculated | 84.9* | - | 11.0, - | 75,181 | No | [31] |
| Not specified | 83.8 | - | - | - | [43] | |
| Fe titration | - | 113.0 | - | - | [47] | |
| 84.0 | 103.9 | 11.1, 13.8 | 75,143 | No | This study | |
| oTF | ||||||
| Dry weight | 92.1 | 111.4 | 11.5, 14.0 | 79,882 | No | [49] |
| Co titration | 90.8 | 11.4 | 79,882 | No | [36] | |
| 87.9 | 109.3 | 11.6, 14.4 | 75,759 | No | This study |
These values were corrected because the number of cystine residues was given as 5 instead of the correct number which is 19.
Conclusions
In summary we report a protocol to experimentally determine accurate ε280 values of apo- and iron-bound hTF and oTF that is rapid and results in destruction of a minimal amount of sample. We verify that the method to calculate the ε280 from the amino acid composition [23] provides reliable estimates of ε280 values for all apo-samples tested. Importantly, our modification of the Edelhoch method allows a reliable estimate of the ε280 for the Fe3+ bound form of all mutants regardless of the strength of metal binding. This approach should be applicable to any metalloprotein in which metal binding makes a significant contribution to the A280.
Acknowledgements
We would like to thank Drs Stephen Everse and Tom Orfeo and Shaina Byrne for helpful comments and suggestions during the preparation of this manuscript.
Abbreviations
- TF
transferrin
- hTF
human serum transferrin
- hTF N-lobe
recombinant N-lobe of human serum transferrin comprising residues 1–337
- oTF
chicken ovotransferrin
- TFR
transferrin receptor 1
- LMCT
ligand to metal charge transfer
- WT
wild-type
- GdHCl
guanidine hydrochloride
- EDTA
ethylenediaminetetraacetic acid
- MES
morpholinoethanesulfonic acid
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wally J, Halbrooks PJ, Vonrhein C, Rould MA, Everse SJ, Mason AB, Buchanan SK. The crystal structure of iron-free human serum transferrin provides insight into inter-lobe communication and receptor binding. J. Biol. Chem. 2006;281:24934–24944. doi: 10.1074/jbc.M604592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klausner RD, Ashwell G, van Renswoude J, Harford JB, Bridges KR. Binding of apotransferrin to K562 cells: explanation of the transferrin cycle. Proc. Natl. Acad. Sci. U. S. A. 1983;80:2263–2266. doi: 10.1073/pnas.80.8.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zuccola HJ. The crystal structure of monoferric human serum transferrin. Atlanta, GA: Georgia Institute of Technology; 1993. [Google Scholar]
- 4.Lambert LA, Perri H, Halbrooks PJ, Mason AB. Evolution of the transferring family: Conservation of residues associated with iron and anion binding. Comparative Biochemistry and Physiology [B] 2005;142:129–141. doi: 10.1016/j.cbpb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 5.He QY, Mason AB, Templeton DM. Molecular and Cellular Iron Transport. New York: Marcel Dekker, Inc; 2002. Molecular aspects of release of iron from transferrins; pp. 95–123. [Google Scholar]
- 6.Funk WD, MacGillivray RTA, Mason AB, Brown SA, Woodworth RC. Expression of the amino-terminal half-molecule of human serum transferrin in cultured cells and characterization of the recombinant protein. Biochemistry. 1990;29:1654–1660. doi: 10.1021/bi00458a043. [DOI] [PubMed] [Google Scholar]
- 7.Mason AB, Funk WD, MacGillivray RTA, Woodworth RC. Efficient production and isolation of recombinant amino-terminal half-molecule of human serum transferrin from baby hamster kidney cells. Protein Expr. Purif. 1991;2:214–220. doi: 10.1016/1046-5928(91)90074-s. [DOI] [PubMed] [Google Scholar]
- 8.Mason AB, Miller MK, Funk WD, Banfield DK, Savage KJ, Oliver RWA, Green BN, MacGillivray RTA, Woodworth RC. Expression of glycosylated and nonglycosylated human transferrin in mammalian cells. Characterization of the recombinant proteins with comparison to three commercially available transferrins. Biochemistry. 1993;32:5472–5479. doi: 10.1021/bi00071a025. [DOI] [PubMed] [Google Scholar]
- 9.Mason AB, Halbrooks PJ, Larouche JR, Briggs SK, Moffett ML, Ramsey JE, Connolly SA, Smith VC, MacGillivray RTA. Expression, purification, and characterization of authentic monoferric and apo-human serum transferrins. Protein Expr.Purif. 2004;36:318–326. doi: 10.1016/j.pep.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 10.Zak O, Aisen P, Crawley JB, Joannou CL, Patel KJ, Rafiq M, Evans RW. Iron release from recombinant N-lobe and mutants of human transferrin. Biochemistry. 1995;34:14428–14434. doi: 10.1021/bi00044a020. [DOI] [PubMed] [Google Scholar]
- 11.Halbrooks PJ, Mason AB, Adams TE, Briggs SK, Everse SJ. The oxalate effect on release of iron from human serum transferrin explained. J. Mol. Biol. 2004;339:217–226. doi: 10.1016/j.jmb.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 12.Halbrooks PJ, Giannetti AM, Klein JS, Bjorkman PJ, Larouche JR, Smith VC, MacGillivray RTA, Everse SJ, Mason AB. Composition of pH sensitive triad in C-lobe of human serum transferrin. Comparison to sequences of ovotransferrin and lactoferrin provides insight into functional differences in iron release. Biochemistry. 2005;44:15451–15460. doi: 10.1021/bi0518693. [DOI] [PubMed] [Google Scholar]
- 13.Byrne SL, Leverence R, Klein JS, Giannetti AM, Smith VC, MacGillivray RTA, Kaltashov IA, Mason AB. Effect of glycosylation on the function of a soluble, recombinant form of the transferrin receptor. Biochemistry. 2006;45:6663–6673. doi: 10.1021/bi0600695. [DOI] [PubMed] [Google Scholar]
- 14.James NG, Berger CL, Byrne SL, Smith VC, MacGillivray RTA, Mason AB. Intrinsic Fluorescence Reports a Global Conformational Change in the N-Lobe of Human Serum Transferrin following Iron Release. Biochemistry. 2007;46:10603–10611. doi: 10.1021/bi602425c. [DOI] [PubMed] [Google Scholar]
- 15.Patch MG, Carrano CJ. The origin of the visible absorption in metal transferrins. Inorg.Chim.Acta. 1981;56:L71–L73. [Google Scholar]
- 16.Lehrer SS. Fluorescence and absorption studies of the binding of copper and iron to transferrin. J. Biol. Chem. 1969;244:3613–3617. [PubMed] [Google Scholar]
- 17.Egan TJ, Zak O, Aisen P. The anion requirement for iron release from transferring is preserved in the receptor-transferrin complex. Biochemistry. 1993;32:8162–8167. doi: 10.1021/bi00083a016. [DOI] [PubMed] [Google Scholar]
- 18.Muralidhara BK, Hirose M. Anion-mediated iron release from transferrins - The kinetic and mechanistic model for N-lobe of ovotransferrin. J. Biol. Chem. 2000;275:12463–12469. doi: 10.1074/jbc.275.17.12463. [DOI] [PubMed] [Google Scholar]
- 19.Jaenicke L. A rapid micromethod for the determination of nitrogen and phosphate in biological material. Anal. Biochem. 1974;61:623–627. doi: 10.1016/0003-2697(74)90429-1. [DOI] [PubMed] [Google Scholar]
- 20.Benson AM, Suruda AJ, Talalay P. Concentration-dependent association of delta5-3-ketosteroid isomerase of Pseudomonas testosteroni. J. Biol. Chem. 1975;250:276–280. [PubMed] [Google Scholar]
- 21.Nozaki Y. Determination of the concentration of protein by dry weight--a comparison with spectrophotometric methods. Arch. Biochem. Biophys. 1986;249:437–446. doi: 10.1016/0003-9861(86)90020-2. [DOI] [PubMed] [Google Scholar]
- 22.Hunter MJ. A Method for the Determination of Protein Partial Specific Volumes. 1966:3285–3292. [Google Scholar]
- 23.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wetlaufer DB. Ultraviolet spectra of proteins and amino acids. Adv. Protein Chem. 1962;17:303–390. [Google Scholar]
- 25.Edelhoch H. Spectroscopic Determination of Tryptophan and Tyrosine in Proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 26.Edelhoch H, Brand L, Wilchek M. Fluorescence studies with tryptophyl peptides. Biochemistry. 1967;6:547–559. doi: 10.1021/bi00854a024. [DOI] [PubMed] [Google Scholar]
- 27.Butler AP, Revzin A, von Hippel PH. Molecular parameters characterizing the interaction of Escherichia coli lac repressor with non-operator DNA and inducer. Biochemistry. 1977;16:4757–4768. doi: 10.1021/bi00641a001. [DOI] [PubMed] [Google Scholar]
- 28.Elwell ML, Schellman JA. Stability of phage T4 lysozymes. I. Native properties and thermal stability of wild type and two mutant lysozymes. Biochim. Biophys. Acta. 1977;494:367–383. doi: 10.1016/0005-2795(77)90166-0. [DOI] [PubMed] [Google Scholar]
- 29.Ford-Hutchinson AW, Perkins DJ. Chemical Modifications of the Tryptophan Groups of Transferrin. Eur. J. Biochem. 1972;25:415–419. doi: 10.1111/j.1432-1033.1972.tb01710.x. [DOI] [PubMed] [Google Scholar]
- 30.Gill SC, von Hippel PH. Calculation of Protein Extinction Coefficients from Amino Acid Sequence Data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 31.Mach H, Middaugh CR, Lewis RV. Statistical determination of the average values of the extinction coefficients of tryptophan and tyrosine in native proteins. Anal. Biochem. 1992;200:74–80. doi: 10.1016/0003-2697(92)90279-g. [DOI] [PubMed] [Google Scholar]
- 32.Gelb MH, Harris DC. Correlation of proton release and ultraviolet difference spectra associated with metal binding by transferrin. Arch. Biochem. Biophys. 1980;200:93–98. doi: 10.1016/0003-9861(80)90335-5. [DOI] [PubMed] [Google Scholar]
- 33.Kretchmar SA, Raymond KN. Effects of Ionic Strength on Iron Removal from the Monoferric Transferrins. Inorganic Chemistry. 1988;27:1436–1441. [Google Scholar]
- 34.Harris WR. Equilibration constants for the complexation of metal ions by serum transferrin. Adv. Exp. Med. Biol. 1989;249:67–93. doi: 10.1007/978-1-4684-9111-1_6. [DOI] [PubMed] [Google Scholar]
- 35.Li YJ, Harris WR, Maxwell A, MacGillivray RTA, Brown T. Kinetic studies on the removal of iron and aluminum from recombinant and site-directed mutant N-lobe half transferrins. Biochemistry. 1998;37:14157–14166. doi: 10.1021/bi9810454. [DOI] [PubMed] [Google Scholar]
- 36.He QY, Mason AB, Woodworth RC. Spectrophotometric titration with cobalt (III) for the determination of accurate absorption coefficients of transferrins. Biochem. J. 1996;318:145–148. doi: 10.1042/bj3180145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mason AB, Woodworth RC, Oliver RWA, Green BN, Lin LN, Brandts JF, Savage KJ, Tam BM, MacGillivray RTA. Association of the two lobes of ovotransferrin is a prerequisite for receptor recognition. Studies with recombinant ovotransferrins. Biochem. J. 1996;319:361–368. doi: 10.1042/bj3190361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mason AB, He QY, Adams TE, Gumerov DR, Kaltashov IA, Nguyen V, MacGillivray RTA. Expression, Purification, and Characterization of Recombinant Nonglycosylated Human Serum Transferrin Containing a C-Terminal Hexahistidine Tag. Protein Expr. Purif. 2001;23:142–150. doi: 10.1006/prep.2001.1480. [DOI] [PubMed] [Google Scholar]
- 39.He QY, Mason AB, Lyons BA, Tam BM, Nguyen V, MacGillivray RTA, Woodworth RC. Spectral and metal-binding properties of three single-point tryptophan mutants of the human transferrin N-lobe. Biochem. J. 2001;354:423–429. doi: 10.1042/0264-6021:3540423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aisen P, Aasa R, Malmstrom BG, Vanngard T. Bicarbonate and the Binding of Iron to Transferrin. J. Biol. Chem. 1967;242:2484–2490. [PubMed] [Google Scholar]
- 41.Harris DC, Gray GA, Aisen P. 13-C nuclear magnetic resonance study of the spatial relation of the metal- and anion-binding sites of human transferrin. J. Biol. Chem. 1974;249:5261–5264. [PubMed] [Google Scholar]
- 42.Harris WR, Pecoraro VL. Thermodynamic binding constants for gallium transferrin. Biochemistry. 1983;22:292–299. doi: 10.1021/bi00271a010. [DOI] [PubMed] [Google Scholar]
- 43.Zweier JL, Wooten JB, Cohen JS. Studies of anion binding by transferrin using carbon-13 nuclear magnetic resonance spectroscopy. Biochemistry. 1981;20:3505–3510. doi: 10.1021/bi00515a031. [DOI] [PubMed] [Google Scholar]
- 44.Folajtar DA, Chasteen ND. Measurement of nonsynergistic anion binding to transferrin by EPR difference spectroscopy. J. Am. Chem. Soc. 1982;104:5775–5780. [Google Scholar]
- 45.Zak O, Aisen P. Nonrandom distribution of iron in circulating human transferrin. Blood. 1986;68:157–161. [PubMed] [Google Scholar]
- 46.Battistuzzi G, Calzolai L, Messori L, Sola M. Metal-induced conformational heterogeneity of transferrins: A spectroscopic study of indium(III) and other metal(III)-substituted transferrins. Biochem. Biophys. Res. Commun. 1995;206:161–170. doi: 10.1006/bbrc.1995.1023. [DOI] [PubMed] [Google Scholar]
- 47.Hamilton DH, Turcot I, Stintzi A, Raymond KN. Large cooperativity in the removal of iron from transferrin at physiological temperature and chloride ion concentration. J.Biol.Inorg.Chem. 2004;9:936–944. doi: 10.1007/s00775-004-0592-6. [DOI] [PubMed] [Google Scholar]
- 48.Abergel RJ, Raymond KN. Terephthalamide-containing ligands: fast removal of iron from transferrin. J Biol Inorg Chem. 2007 doi: 10.1007/s00775-007-0314-y. [DOI] [PubMed] [Google Scholar]
- 49.Brown-Mason A, Woodworth RC. Physiological levels of binding and iron donation by complementary half-molecules of ovotransferrin to transferrin receptors on chick reticulocytes. J. Biol. Chem. 1984;259:1866–1873. [PubMed] [Google Scholar]


