Crystallization of the cystine-knot protein Spätzle occurred following serendipitous limited degradation of the pro-Spätzle propeptide during the crystallization experiment.
Keywords: degradation, growth factors, Toll pathway
Abstract
The Spätzle protein is involved in both the definition of the dorsal–ventral axis during embryonic development and in the adult innate immune response. The disulfide-linked dimeric cystine-knot protein has been expressed as a proprotein in inclusion bodies in Escherichia coli and refolded in vitro by rapid dilution. Initial orthorhombic crystals that diffracted to 7 Å resolution were obtained after three months by the sitting-drop vapour-diffusion method. Optimization of the crystallization conditions resulted in orthorhombic crystals (space group P212121, with unit-cell parameters a = 53.0, b = 59.2, c = 62.5 Å) that diffracted to 2.8 Å resolution in-house. The small volume of the asymmetric unit indicated that it was not possible for the crystals to contain the complete pro-Spätzle dimer. Mass spectrometry, N-terminal sequencing and Western-blot analysis revealed that the crystals contained the C-terminal disulfide-linked cystine-knot dimer. Comparison of various crystallization experiments indicated that degradation of the N-terminal prodomain was dependent on the buffer conditions.
1. Introduction
The Spätzle protein is the precursor of a nerve-growth-factor-like ligand in Drosophila melanogaster (Stein et al., 1991 ▶). It plays a role in definition of the dorsal–ventral axis of the Drosophila embryo (Schneider et al., 1994 ▶) and acts in the initiation of immune responses to fungal and bacterial infection in adult flies (Hoffmann, 2003 ▶). An extracellular proteolytic cascade results in activation of Spätzle, which in turn leads to signalling by the transmembrane receptor Toll. In the developmental pathway, the trypsin-like serine protease Easter cleaves Spätzle to generate a 12 kDa C-terminal fragment. The recently discovered Spätzle-processing enzyme (SPE; Jang et al., 2006 ▶) activates Spätzle in the infectional pathway via the same cleavage, initiating an immune response that leads to drosomycin generation. Dimers of the C-terminal fragment have been found in vivo (DeLotto & DeLotto, 1998 ▶) as have dimers of the proprotein in vitro (Hoffmann et al., 2008 ▶; Weber et al., 2005 ▶, 2007 ▶). Binding of activated Spätzle to Toll leads to receptor dimerization and autophosphorylation of the cytosolic Toll/interleukin-1 receptor (TIR) domains (Weber et al., 2003 ▶, 2005 ▶). The receptor shares significant sequence similarity to the vertebrate interleukin-1 receptor (Schneider et al., 1994 ▶) as well as to mammalian Toll-like receptors (Jin et al., 2007 ▶).
The spacing of the cysteine residues and the sequence homology of the C-terminal 106 amino acids of Spätzle to coagulogen (Bergner et al., 1996 ▶) and mature human nerve growth factor (NGF; McDonald et al., 1991 ▶) suggest a cystine-knot motif: two disulfide bridges that form a ring through which a third disulfide bridge is threaded (Mizuguchi et al., 1998 ▶). Here, we describe the crystallization of the recombinantly expressed and in vitro folded Spätzle C-terminal domain derived from pro-Spätzle isoform Spz11.7.
2. Materials and methods
2.1. Protein purification
The expression of C-terminally His6-tagged pro-Spätzle isoform Spz11.7 as insoluble inclusion bodies with subsequent solubilization and refolding has been described elsewhere (Hoffmann et al., 2008 ▶). Briefly, inclusion-body protein dissolved in 6 M guanidinium hydrochloride was renatured by rapid dilution (Rudolph & Lilie, 1996 ▶) into 1 M l-arginine, 0.1 M Tris–HCl, 5 mM EDTA pH 8.5 with an oxidized/reduced glutathione redox-shuffling system in the ratio 1 to 5 mM. After concentration and dialysis in 20 mM sodium phosphate, 20 mM sodium chloride pH 7.4, dimers were separated from monomers and aggregates by size-exclusion chromatography (HiLoad 26/60 Superdex 75pg, GE Healthcare). Pooled dimer fractions were concentrated to 20 mg ml−1 according to the optical density OD280nm using Amicon filter devices (10 000 Da molecular-weight cutoff; Millipore) and used for further crystallization trials.
2.2. Crystallization and optimization
Initial sitting-drop vapour-diffusion crystallization experiments were carried out with a variety of commercially available crystallization screens using a Cartesian Microsys NQ crystallization robot (Zinsser Analytik, Frankfurt, Germany). Crystallization in 96-well plates (CrystalQuick, Greiner) with 200 nl drops and 110 µl reservoir volumes was monitored using an OASIS LS3 imaging system (Veeco, Tucson, Arizona, USA). The first crystals appeared after three months in 50 mM Tris–HCl, 20% PEG 3350 pH 8.0 (Sigma–Aldrich, low ionic strength crystallization kit) at 293 K. Optimization using hanging-drop vapour diffusion (in EasyXtal plates, Qiagen) involved mixing the contents of an additive screen (Hampton Research) with the crystallization drops (1 µl protein solution + 1 µl reservoir solution) in a 1:10 ratio and equilibrating against 500 µl reservoir solution. Two additive conditions (0.1 M betaine monohydrate and 0.1 M phenol in 50 mM Tris–HCl, 20% PEG 3350 pH 8.0) yielded crystals that grew within three weeks.
2.3. Data collection
Crystals were mounted directly in a 100 K cryostream (XSTREAM2000; Rigaku/MSC, Japan) without any additional cryoprotection. Diffraction data were collected in-house using Cu Kα radiation with an R-AXIS IV++ imaging-plate system (Rigaku/MSC, Japan) mounted on a Rigaku MM-007 rotating-anode generator. Initial crystals from the 96-well plates diffracted anisotropically to 7 Å resolution. Larger crystals obtained using betaine monohydrate from the additive screen also diffracted anisotropically, but the data could be scaled and processed to 2.8 Å resolution using XDS (Kabsch, 1993 ▶).
2.4. Analysis of the crystallized fragment
2.4.1. Mass spectrometry
Samples from crystallization drops with similar conditions to those containing crystals were taken for MALDI-MS (matrix-assisted laser desorption/ionization mass spectrometry) analysis. Protein bands excised from nonreducing denaturing SDS gels were washed three times with water, twice with 10 mM ammonium bicarbonate and finally with 10 mM ammonium bicarbonate in 50% acetonitrile. The gel slices were dried under a gentle stream of nitrogen, re-swollen in 20 µl 10 mM ammonium bicarbonate pH 8.0 and digested with trypsin (Promega, USA) overnight at 310 K. 0.5 µl of a saturated solution of α-cyano-4-hydroxycinnamic acid in acetone was deposited onto the anchor target and 1 µl of the digest was added to the matrix layer. After evaporation of the solvent, the spot was recrystallized with 1 µl ethanol:acetone:0.1% trifluoroacetic acid (6:3:1). The peptide mass-fingerprint spectra were recorded on an Ultraflex-II TOF/TOF mass spectrometer (Bruker Daltonic, Germany) equipped with MALDI source, nitrogen laser, LIFT cell for fragment-ion post-acceleration and gridless ion reflector. The software Flex Control 2.4, Flex Analysis 2.4 and Biotools 3.0 were used to operate the instrument and analyse the data. A peptide-calibration mixture (Bruker Daltonics, Germany) was used for external calibration.
2.4.2. N-terminal sequencing
Three large intergrown crystals from a 2 µl drop with 2 µg protein were fished out from the drop and washed in crystallization buffer before being dissolved in SDS loading buffer. Protein bands from the unstained SDS gel were blotted on a methanol-activated polyvinylidene fluoride membrane (PALL; Gelman Laboratory, USA) in 50 mM sodium borate, 20% methanol pH 9.0 buffer at 60 mA for 2 h. The membrane was stained in 0.006% Coomassie G250, 10% acetic acid and 30% methanol solution and air dried. The single protein band was excised and sent for N-terminal sequencing (Alphalyse Europe, Denmark).
2.4.3. Western blot
Reduced and nonreduced samples from the crystallization drop, as well as the starting material as a positive control, were loaded onto a denaturing SDS gel. Protein bands from the unstained SDS gel were blotted onto a nitrocellulose transfer membrane (BioRad Laboratories) at 60 mA for 2 h. The membrane was incubated in 1× PBS (10 mM sodium phosphate, 150 mM sodium chloride pH 7.2), 4%(w/v) milk powder, 0.2%(v/v) Tween-20 for 1.5 h before overnight application of the first antibody, mouse anti-His (Roche). After three washing steps with 1× PBS, 4% milk powder, 0.2% Tween-20, the second antibody, peroxidase-coupled goat anti-mouse (Chemicon International, USA), was applied for 4 h. The blot was treated with chemiluminescent reagents (Amersham Biosciences) after three washing steps in the absence of milk powder. His-tagged proteins were visualized after development and fixing (Kodak, Sigma–Aldrich) of the detection film (Roche).
3. Results and discussion
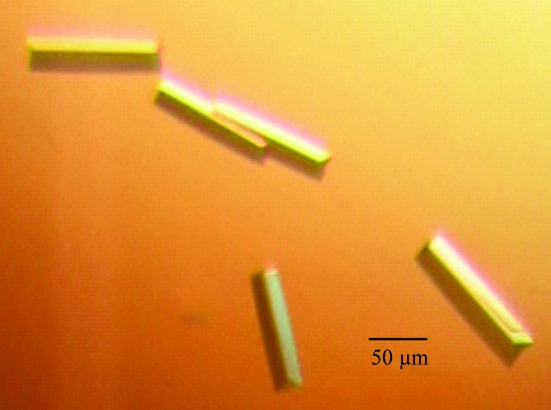
Initial crystals of the Spätzle isoform Spz11.7 appeared after three months and diffracted in-house to ∼7 Å resolution. These could be reproduced and improved using the crystallization additive betaine monohydrate (Fig. 1 ▶). The optimized crystals, which grew within three weeks, diffracted to 2.8 Å resolution. This large jump in resolution may be attributable to the presence of the additive, the larger volume of the crystals, their accelerated growth or a combination of these factors. The crystals belonged to the orthorhombic space group P212121, with unit-cell parameters a = 53.0, b = 59.2, c = 62.5 Å (data-collection statistics are summarized in Table 1 ▶). As the unit-cell parameters and space group precluded the presence of a dimer of pro-Spätzle Spz11.7 (molecular weight 57 kDa) in the crystals, their contents were subjected to further analysis.
Figure 1.
Crystals of Spz11.7 from 50 mM Tris–HCl, 20% PEG 3350 pH 8.0 with 10 mM betaine monohydrate as an additive.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| Space group | P212121 |
| Unit-cell parameters | |
| a (Å) | 53.0 |
| b (Å) | 59.2 |
| c (Å) | 62.5 |
| Wavelength (Å) | 1.5418 |
| Mosaicity (°) | 0.5 |
| Resolution range (Å) | 30.0–2.8 (2.9–2.8) |
| Total no. of reflections | 84797 (5203) |
| Unique reflections | 8348 (500) |
| Redundancy | 16.3 (16.7) |
| Completeness (%) | 99.8 (100.0) |
| I/σ(I) | 24.6 (4.5) |
| Rmerge† (%) | 9.8 (75.9) |
| Wilson B factor (Å2) | 54.7 |
| Molecules per ASU | 2 |
| Matthews coefficient (Å3 Da−1) | 2.23 |
| Solvent content (%) | 45 |
R
merge = 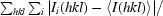
 .
.
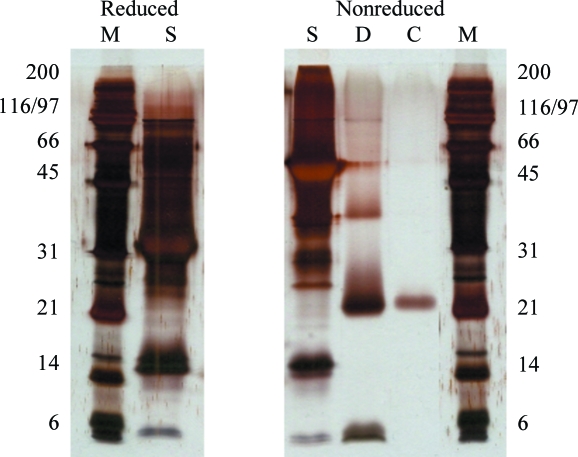
The starting material and dissolved crystals as well as selected negative crystallization experiments were analysed using denaturing SDS–PAGE. While the starting material yielded a major band corresponding to a molecular weight of 54 kDa (nonreducing conditions; 27 kDa under reducing conditions), analysis of the dissolved crystals revealed a fragment of ∼25 kDa (nonreduced samples, Fig. 2 ▶; ∼12 kDa under reducing conditions, data not shown). The crystals therefore contain a partially degraded disulfide-linked dimer of Spätzle. In order to identify this fragment, samples were subjected to mass spectrometry, N-terminal sequencing and Western blot analysis.
Figure 2.
Silver-stained SDS–PAGE of Spz11.7 used for crystallization (lane S, starting material), a crystallization drop (lane D) and a dissolved crystal (lane C); Lane M, molecular-weight markers (kDa).
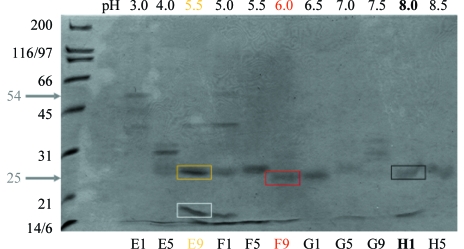
Protein bands excised from nonreducing denaturing SDS gels (Fig. 3 ▶) were subjected to MALDI-MS. The fingerprints of three samples of the same size as the crystallized fragment showed peptide masses corresponding to amino acids 139–143, 148–162, 193–204, 205–217 and 228–237 (summarized in Fig. 4 ▶). These peptides are all located within the C-terminal disulfide-knot domain; the missing peptides of the C-terminal domain are either those containing cysteines expected to be disulfide-linked or smaller than 500 Da. A low-molecular-weight degradation product that did not appear in the gel sample of the crystallized fragment (Fig. 3 ▶) was also measured as a negative control. The fingerprint corresponded to residues 10–25, 26–39, 40–54, 55–66, 71–77, 78–84, 110–123 and 148–162 (Fig. 4 ▶), which with the exception of peptide 148–162 are all located in the N-terminal propeptide of Spz11.7.
Figure 3.
Nonreducing SDS–PAGE of Spz11.7 crystallization screening experiments using 4% PEG 3350 as precipitant and 0.05 M buffer. The pH values of individual crystallization drops are indicated and were achieved using the following buffers: sodium citrate (E1–F5), MES (F9), bis-tris–HCl (G1), imidazole–HCl (G5), sodium HEPES (G9) and Tris–HCl (H1–H5). Note the varying degradation bands observed. Lanes E9, F9 and H1 exhibit comparable bands (∼25 kDa) to those present in the crystals (see Fig. 2 ▶). Marked gel slices from E9, F9 and H1 were further analysed using MALDI-MS (see Fig. 4 ▶).
Figure 4.
Recombinant Spz11.7 sequence, indicating the N-terminal propeptide as well as the mature cystine knot (bold). The peptides detected by MALDI-MS are underlined in black (from sample H1), red (sample F9), gold (sample E9, high-molecular-weight band) or grey (negative control, E9 low-molecular-weight band, see text for details). The dashed line indicates a peptide with ambiguous assignment (peptide mass 55Y–R66, 1437.807 Da; 205S–K217, 1437.686 Da).
To identify the N-terminus of the crystallized fragment, which according to the MALDI-MS analysis could be anywhere between amino acids 123 and 139, N-terminal sequencing of washed and dissolved crystals was performed. Six cycles of Edman degradation yielded the sequence V/A-X-X-Y/S-D-E/Q with low signal strength and glycine contamination, which corresponds perfectly and uniquely to Spz11.7 residues 124V-G-G-S-D-E129. Thus, the N-terminus (which also corresponds to the N-terminus of mature Spätzle after activation cleavage) is in agreement with the MALDI-MS data, where the last non-included peptide is 110–123 and the first included peptide is 139–143. As the His6 tag peptide 228–237 only occurred in one of three MALDI-MS samples, the presence of an intact C-terminus was examined by Western blotting against the C-terminal His6 tag. As signals were obtained from both reduced and nonreduced samples of the starting material (positive control) but not from the crystallization drop sample (data not shown), the C-terminal His6 tag of Spz11.7 does not appear to be present in the crystals.
Thus, the crystals contained the C-terminal cystine-knot-containing fragment Val124–Lys227. It is known that the propeptide of Spätzle isoforms is highly susceptible to trypsin cleavage (Weber et al., 2003 ▶, 2005 ▶). The N- and C-termini of the crystallized fragment are consistent with a trypsin-like activity cleaving Spz11.7 after Arg123 and Lys227, although we have no clear evidence for such contamination. Based on the unit-cell parameters and molecular weight of mature Spätzle, the crystals are likely to contain a dimer in the asymmetric unit (Matthews coefficient V M = 2.23 Å3 Da−1, corresponding to a solvent content of 45%; Matthews, 1968 ▶). A preliminary molecular-replacement search carried out using Phaser (McCoy et al., 2005 ▶) with diverse NGF monomer and dimer structures as search models [PDB codes 1btg (Holland et al., 1994 ▶), 1sg1 (He & Garcia, 2004 ▶), 1bet (McDonald et al., 1991 ▶) and 1sgf (Bax et al., 1997 ▶)] has so far failed to yield a satisfactory solution. A search for heavy-atom derivatives is under way, as are attempts to obtain crystals diffracting to higher resolution using synchrotron radiation.
Acknowledgments
This work was supported by the SFB610 ‘Protein-Zustände mit zellbiologischer und medizinischer Relevanz’ and the Graduiertenkolleg 1026 ‘Conformational transitions in macromolecular interactions’ of the DFG.
References
- Bax, B., Blundell, T. L., Murray-Rust, J. & McDonald, N. Q. (1997). Structure, 5, 1275–1285. [DOI] [PubMed]
- Bergner, A., Oganessyan, V., Muta, T., Iwanaga, S., Typke, D., Huber, R. & Bode, W. (1996). EMBO J.15, 6789–6797. [PMC free article] [PubMed]
- DeLotto, Y. & DeLotto, R. (1998). Mech. Dev.72, 141–148. [DOI] [PubMed]
- He, X. L. & Garcia, K. C. (2004). Science, 304, 870–875. [DOI] [PubMed]
- Hoffmann, A., Funkner, A., Neumann, P., Walther, M., Weininger, U., Schierhorn, A., Balbach, J., Reuter, G. & Stubbs, M. T. (2008). Submitted. [DOI] [PubMed]
- Hoffmann, J. A. (2003). Nature (London), 426, 33–38. [DOI] [PubMed]
- Holland, D. R., Cousens, L. S., Meng, W. & Matthews, B. W. (1994). J. Mol. Biol., 239, 385–400. [DOI] [PubMed]
- Jang, I. H., Chosa, N., Kim, S.-H., Nam, H. J., Lemaitre, B., Ochiai, M., Kambris, Z., Brun, S., Hashimoto, C., Ashida, M., Brey, P. T. & Lee, W. J. (2006). Dev. Cell, 10, 45–55. [DOI] [PubMed]
- Jin, M. S., Kim, S. E., Heo, J. Y., Lee, M. E., Kim, H. M., Paik, S. G., Lee, H. & Lee, J. O. (2007). Cell, 130, 1071–1082. [DOI] [PubMed]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800.
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed]
- McDonald, N. Q., Lapatto, R., Murray-Rust, J., Gunning, J., Wlodawer, A. & Blundell, T. L. (1991). Nature (London), 354, 411–414. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Mizuguchi, K., Parker, J. S., Blundell, T. L. & Gay, N. (1998). Trends Biochem. Sci.23, 239–242. [DOI] [PubMed]
- Rudolph, R. & Lilie, H. (1996). FASEB J.10, 49–56. [PubMed]
- Schneider, D. S., Jin, Y. S., Morisato, D. & Anderson, K. V. (1994). Development, 120, 1243–1250. [DOI] [PubMed]
- Stein, D., Roth, S., Vogelsang, E. & Nusslein-Volhard, C. (1991). Cell, 65, 725–735. [DOI] [PubMed]
- Weber, A. N. R., Gangloff, M., Moncrieffe, M. C., Hyvert, Y., Imler, J. L. & Gay, N. J. (2007). J. Biol. Chem.282, 13522–13531. [DOI] [PubMed]
- Weber, A. N. R., Moncrieffe, M. C., Gangloff, M., Imler, J. L. & Gay, N. J. (2005). J. Biol. Chem.280, 22793–22799. [DOI] [PubMed]
- Weber, A. N. R., Tauszig-Delamasure, S., Hoffmann, J. A., Lelievre, E., Gascan, H., Ray, K. P., Morse, M. A., Imler, J. L. & Gay, N. J. (2003). Nature Immunol.4, 794–800. [DOI] [PubMed]