XometC, a cystathionine γ-lyase-like protein from X. oryzae pv. oryzae and an antibacterial drug-target protein against bacterial blight, was cloned, purified and crystallized. Preliminary X-ray crystallographic analysis of XometC crystals was carried out.
Keywords: bacterial blight, cystathionine γ-lyase, reverse transsulfuration pathway, Xanthomonas oryzae pv. oryzae
Abstract
Xanthomonas oryzae pv. oryzae (Xoo) causes bacterial blight of rice (Oryza sativa L.), one of the most devastating diseases of rice in most rice-growing countries. XometC, a cystathionine γ-lyase (CGL) like protein that is an antibacterial drug-target protein against Xoo, was cloned, expressed, purified and crystallized. CGL catalyzes the second step in the reverse-transsulfuration pathway, which is essential for the metabolic interconversion of the sulfur-containing amino acids cysteine and methionine. Crystals of two different shapes, plate-shaped and pyramid-shaped, diffracted to 2.9 and 3.2 Å resolution and belonged to the primitive orthogonal space group P212121 and the tetragonal space group P41 (or P43), with unit-cell parameters a = 73.0, b = 144.9, c = 152.3 Å and a = b = 78.2, c = 300.7 Å, respectively. For the P212121 crystals, three or four monomers exist in the asymmetric unit with a corresponding V M of 3.02 or 2.26 Å3 Da−1 and a solvent content of 59.3 or 45.7%. For the P41 (or P43) crystals, four or five monomers exist in the asymmetric unit with a corresponding V M of 2.59 or 2.09 Å3 Da−1 and a solvent content of 52.5 or 40.6%.
1. Introduction
Rice (Oryza sativa L.) is an important staple for human consumption especially in East, South and Southeast Asia. Bacterial blight (BB) caused by Xanthomonas oryzae pv. oryzae (Xoo) is the most destructive bacterial disease of rice (Ezuka & Kaku, 2000 ▶). In 2006, BB resulted in a loss worth 100 million dollars in South Korea. However, no effective antibacterial drugs against Xoo have been developed. Recently, the whole genome sequence of Xoo has been determined (Lee et al., 2005 ▶). Based on this information, we selected approximately 100 essential enzymes of Xoo (Payne et al., 2004 ▶, 2007 ▶) as drug-target proteins and started cloning the corresponding genes systematically. In this study, we present the cloning, purification and crystallization of XometC (renamed from MetB), a cystathionine γ-lyase (CGL) like protein from Xoo.
Recently, mutational analysis showed that CysB, XometC, XomtA (renamed from SmtA) and WxoABCD, which are involved in lipopolysaccharide (LPS) biosynthesis cluster I in Xoo, were necessary for pathogenicity towards rice as well as for the production of LPS O-antigen and exopolysaccharide (Wang et al., 2008 ▶). CGL (EC 4.4.1.1), a member of the pyridoxal 5′-phosphate (PLP) γ-family, catalyzes the second step in the reverse-transsulfuration pathway, which is essential for the metabolic interconversion of the sulfur-containing amino acids cysteine and methionine (Steegborn et al., 1999 ▶). It converts cystathionine to cysteine, α-oxobutyrate and ammonia via an α,γ-elimination reaction (Dobric et al., 2000 ▶). PLP-dependent enzymes catalyze a vast array of reactions such as transamination, racemization, decarboxylation and side-chain elimination and replacement in amino-acid metabolism (Clausen et al., 1996 ▶). Based on amino-acid similarity, PLP-dependent enzymes can be divided into three different families (α, β and γ; Alexander et al., 1994 ▶). XometC shows 54% sequence identitity to human cystathione γ-lyase, 48% to yeast cystathione γ-lyase, 46% to Pseudomonas putida methionine γ-lyase, 45% to Escherichia coli cystathione γ-synthase and 44% to Arabidopsis thaliana cystathione β-lyase.
This study describes the cloning, expression, purification, crystallization and preliminary X-ray crystallographic analysis of XometC. Three-dimensional structural studies will elucidate the molecular basis for the enzymatic reaction mechanism of XometC, a CGL-like protein, and will be useful for the design of a potential antibacterial drug against Xoo.
2. Methods and results
2.1. Cloning
The XometC gene was amplified from its BAC plasmid via polymerase chain reaction (PCR) using a forward primer (XometC-F: 5′-CAGAATCTCATATGTCCAACCGCACCAC-3′) containing an NdeI restriction site (bold) at the start codon of the ORF and a reverse primer (XometC-R: 5′-CTGCTCGAGTCAATTTTGATTCACCAAC-3′) containing an XhoI restriction site after the stop codon. The PCR amplicon was double-digested with NdeI and XhoI, ligated into pET15b expression vector (Novagen) containing a 6×His tag upstream of a thrombin cleavage site and the multiple cloning site and transformed into E. coli BL21 (DE3) pLysS, yielding a recombinant clone, pET-XometC.
2.2. Overexpression and purification
E. coli BL21 (DE3) pLysS cells containing pET-XometC were grown at 310 K to an OD600 of 0.6 in Luria–Bertani medium supplemented with 50 µg ml−1 ampicillin and 37 µg ml−1 chloramphenicol. Protein expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM. The cells were cultured at 310 K for an additional 4 h and harvested by a 20 min centrifugation at 6000 rev min−1 (Vision VS24-SMTi V5006A rotor) at 277 K. The resulting cell pellet was then resuspended in ice-cold lysis buffer (25 mM Tris–HCl pH 7.5, 300 mM NaCl, 15 mM imidazole and 3 mM β-mercaptoethanol) and homogenized using a sonicator (Sonomasher). The crude cell extract was centrifuged for 30 min at 15 000 rev min−1 (Vision VS24-SMTi V508A rotor) at 277 K to remove cell debris. The supernatant was applied onto Ni–NTA His-Bind resin (Novagen) and affinity purification was performed according to the manufacturer’s protocol at 277 K. The column was washed using lysis buffer including 40 mM imidazole. The 6×His-tagged XometC protein was then eluted using lysis buffer containing 250 mM imidazole and dialyzed against thrombin cleavage buffer (25 mM Tris–HCl pH 7.5, 15 mM NaCl and 3 mM β-mercaptoethanol). The resulting protein was treated with 0.5 U thrombin (Sigma) per milligram of XometC for 3 h at 298 K in order to remove the N-terminal 6×His tag. After cleavage, three additional residues (Gly-Ser-His) from the pET15b vector remained at the N-terminus of XometC. The cleavage product was further purified using an UNO Q6 column (Biorad). The homogeneity of the purified protein was analyzed via SDS–PAGE (Fig. 1 ▶). SeMet XometC was expressed using the modified methionine-pathway inhibition protocol (Khandekar et al., 2000 ▶; Van Duyne et al., 1993 ▶) and purified as described for the native protein. For crystallization, the protein solution was concentrated using Centri-Prep (Millipore) to a final concentration of 8 mg ml−1 in a buffer consisting of 25 mM Tris–HCl pH 7.5, 15 mM NaCl and 3 mM β-mercaptoethanol.
Figure 1.
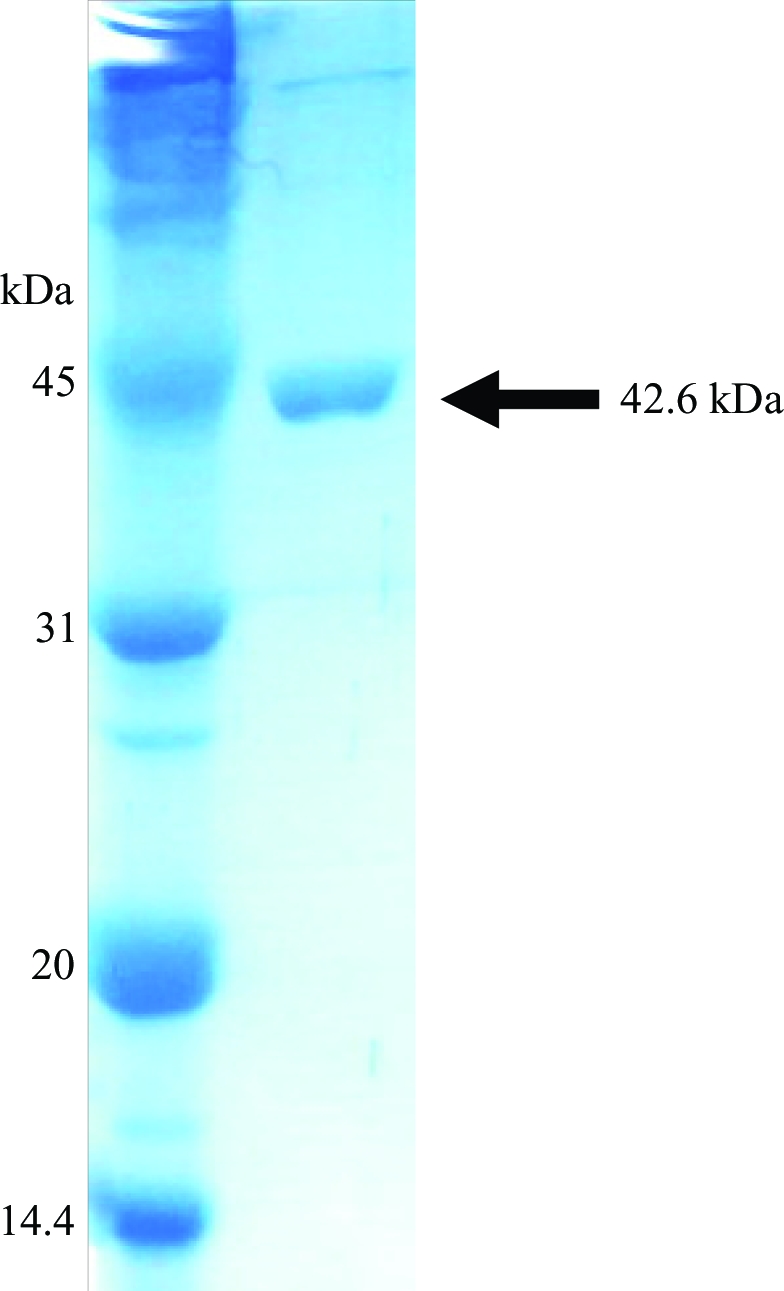
Purified XometC is shown on a 12% SDS–PAGE gel.
2.3. Crystallization and X-ray diffraction data collection
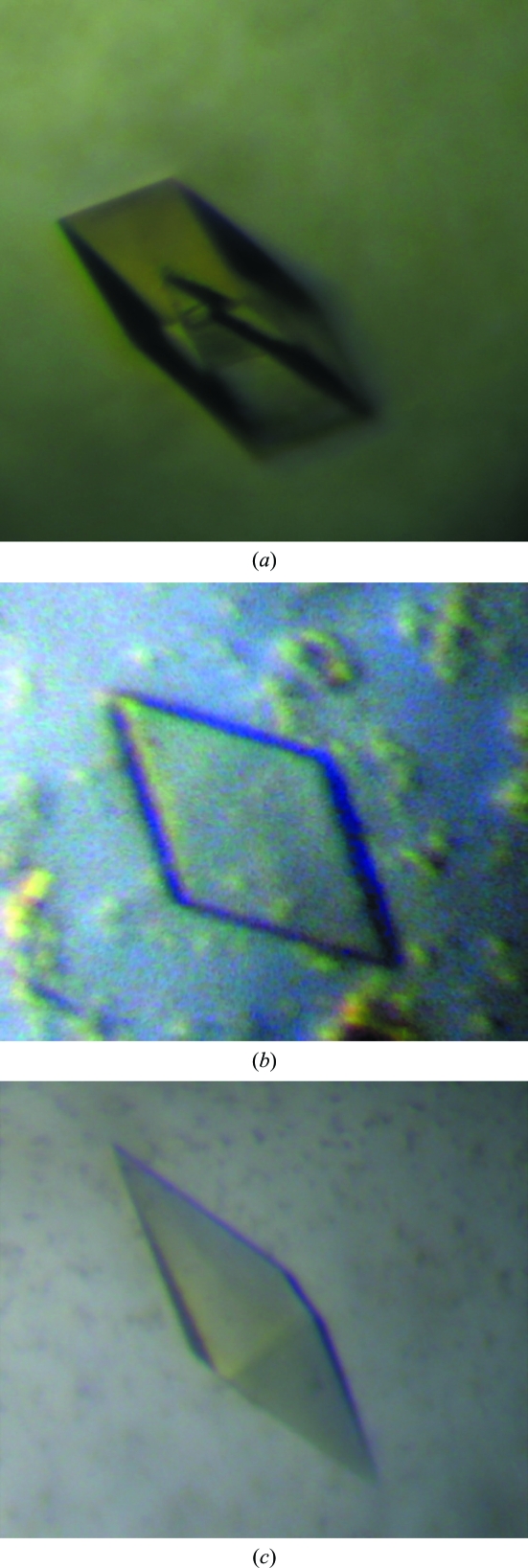
Crystallization was initiated at 283 K using the Hydra II e-drop automated pipetting system (Matrix) on a 96-well Intelliplate (Art Robbins) and the Crystal Screen HT, Index HT and Salt Rx HT screening kits (Hampton Research). Initially, hollow crystals were observed and reproduced in hanging drops consisting of 0.9 µl protein solution mixed with 0.9 µl reservoir solution containing 25% PEG 3350, 0.2 M lithium sulfate monohydrate and 0.1 M bis-Tris pH 5.5 (Fig. 2 ▶). Each hanging drop was then positioned over 1 ml reservoir solution. The well was sealed with a cover slip using vacuum grease. Additive screening was applied to improve the quality of crystals. For additive screening, drops were made up of 0.9 µl protein solution plus 0.9 µl reservoir solution plus 0.2 µl additive screen (Hampton Research Additive Screen HR2-428). Improved crystals of two different shapes, plates and pyramids, were obtained using sodium thiocyanate solution as additive (Fig. 2 ▶). The fully grown crystals were flash-cooled at 100 K in liquid nitrogen with 20%(v/v) glycerol as a cryoprotectant. X-ray diffraction data were collected from frozen crystals of both shapes with 1° oscillations using a Bruker Proteum 300 CCD detector on beamline 6C1 of the Pohang Light Source (PLS), South Korea and an ADSC Quantum 270 CCD detector on beamline 17A of Photon Factory, High Energy Accelerator Research Organization (KEK), Japan. X-ray diffraction data were collected to 2.9 and 3.3 Å resolution for the native and SeMet crystals, respectively. Each data set was integrated and scaled using DENZO and SCALEPACK, respectively (Otwinowski & Minor, 1997 ▶).
Figure 2.
(a) Initial hollow crystal. (b) Plate-shaped crystal of dimensions 0.3 × 0.3 × 0.02 mm. (c) Pyramid-shaped crystal of dimensions 0.4 × 0.1 × 0.1 mm
3. Results and discussion
The plate-shaped and pyramid-shaped crystals belonged to space groups P212121 and P41 (or P43), with unit-cell parameters a = 73.0, b = 144.9, c = 152.3 Å and a = b = 78.22, c = 300.7 Å, respectively. The space group was derived by autoindexing and data-collection statistics are provided in Table 1 ▶. According to the Matthews coefficient calculation (Matthews, 1968 ▶), the P212121 crystal contains three or four molecules in the asymmetric unit, with a solvent content of 59.3% or 45.7%, and the P41 (or P43) crystal contains four or five molecules in the asymmetric unit, with a solvent content of 52.5% or 40.6%. When the SOLVE/RESOLVE programs were applied, SeMet sites were not identified in the MAD data (data not shown). However, molecular replacement (MR) using Phaser from the CCP4 program package (McCoy et al., 2007 ▶) with cystathionine γ-lyase (PDB code 2nmp) as a search model showed four monomers in the asymmetric unit for the native data from pyramid-shaped crystals. The MR solution provided 2F o − F c and F o − F c electron-density maps that were informative for an improved model. The structural details will be described in a separate paper. Our structural data for XometC will provide an insight into its enzymatic mechanism and be useful for developing a potential antibacterial drug against Xoo.
Table 1. Data-collection statistics.
Values in parentheses are for the outer shell.
| SeMet, plate-shaped | |||||
|---|---|---|---|---|---|
| Native, plate-shaped | Native, pyramid-shaped | Peak | Edge | Remote | |
| X-ray source | PF-BL17A | PLS-6C1 | |||
| Unit-cell parameters (Å) | a = 73.0, b = 144.9, c = 152.3 | a = b = 78.2, 78.2, c = 300.7 | a = 72.8, b = 144.9, c = 153.0 | ||
| Wavelength (Å) | 1.00000 | 1.00000 | 0.97955 | 0.97967 | 0.97182 |
| Space group | P212121 | P41 (or P43) | P212121 | ||
| Resolution (Å) | 50.0–3.2 (3.3–3.2) | 50.0–2.9 (3.0–2.9) | 50.0–3.3 (3.4–3.3) | 50.0–3.3 (3.4–3.3) | 50.0–3.2 (3.3–3.2) |
| No. of observations (unique) | 220119 | 401799 | 107574 | 105900 | 107776 |
| No. of unique observations | 26614 | 39539 | 24964 | 24006 | 28134 |
| Completeness (%) | 100.0 (99.9) | 100.0 (100.0) | 96.3 (85.6) | 98.3 (93.4) | 97.5 (88.2) |
| Rmerge† (%) | 15.2 (41.4) | 7.8 (40.2) | 16.7 (47.7) | 17.1 (41.4) | 12.2 (45.4) |
| I/σ(I) | 18.5 (5.7) | 30.5 (4.9) | 7.8 (2.3) | 8.6 (2.8) | 9.7 (2.3) |
R
merge = 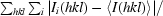
 , where I(hkl) is the intensity of reflection hkl,
, where I(hkl) is the intensity of reflection hkl,  is the sum over all reflections and
is the sum over all reflections and 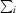 is the sum over i measurements of reflection hkl.
is the sum over i measurements of reflection hkl.
Acknowledgments
We are grateful to Dr S. S. Cha and Dr K. J. Kim for their assistance at beamline 6C1 of Pohang Light Source (PLS), Republic of Korea, to staff members at beamline 17A of Photon Factory (KEK), Japan and to Sun-Gou Ji, Yonsei University, Republic of Korea for his technical assistance. This work was supported by a grant (Code No. 20070501034003) from BioGreen 21 Program, Rural Development Administration, Republic of Korea.
References
- Alexander, F. W., Sandmeier, E., Mehta, P. K. & Christen, P. (1994). Eur. J. Biochem.219, 953–960. [DOI] [PubMed]
- Clausen, T., Huber, R., Laber, B., Pohlenz, H. D. & Messerschmidt, A. (1996). J. Mol. Biol.262, 202–224. [DOI] [PubMed]
- Dobric, N., Limsowtin, G. K., Hillier, A. J., Dudman, N. P. & Davidson, B. E. (2000). FEMS Microbiol. Lett.182, 249–254. [DOI] [PubMed]
- Ezuka, A. & Kaku, H. (2000). Bull. Natl Inst. Agrobiol. Resour. (Jpn), 15, 53–54.
- Khandekar, S. S., Konstantinidis, A. K., Silverman, C., Janson, C. A., McNulty, D. E., Nwagwu, S., Van Aller, G. S., Doyle, M. L., Kane, J. F., Qiu, X. & Lonsdale, J. (2000). Biochem. Biophys. Res. Commun.270, 100–107. [DOI] [PubMed]
- Lee, B. M. et al. (2005). Nucleic Acids Res.33, 577–586. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.32, 491–497. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.277, 307–326. [DOI] [PubMed]
- Payne, D. J., Gwynn, M. N., Holmes, D. J. & Pompliano, D. L. (2007). Nature Rev. Drug Discov.6, 29–40. [DOI] [PubMed]
- Payne, D. J., Gwynn, M. N., Holmes, D. J. & Rosenberg, M. (2004). Methods Mol. Biol.266, 231–259. [DOI] [PubMed]
- Steegborn, C., Clausen, T., Sondermann, P., Jacob, U., Worbs, M., Marinkovic, S., Huber, R. & Wahl, M. C. (1999). J. Biol. Chem.274, 12675–12684. [DOI] [PubMed]
- Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. (1993). J. Mol. Biol.229, 105–124. [DOI] [PubMed]
- Wang, J. C., So, B. H., Kim, J. H., Park, Y. J., Lee, B. M. & Kang, H. W. (2008). Plant Pathol., doi:10.1111/j.1365-3059.2008.01884.x.