Abstract
DNA polymerases attach to the DNA sliding clamp through a common overlapping binding site. We identify a small-molecule compound that binds the protein-binding site in the Escherichia coli β-clamp and differentially affects the activity of DNA polymerases II, III, and IV. To understand the molecular basis of this discrimination, the cocrystal structure of the chemical inhibitor is solved in complex with β and is compared with the structures of Pol II, Pol III, and Pol IV peptides bound to β. The analysis reveals that the small molecule localizes in a region of the clamp to which the DNA polymerases attach in different ways. The results suggest that the small molecule may be useful in the future to probe polymerase function with β, and that the β-clamp may represent an antibiotic target.
Keywords: antibiotic target, rational drug design, fluorescence anisotropy, crystallography
The replication machinery of all cells utilizes a ring-shaped, sliding-clamp protein that encircles DNA and slides along the duplex, thus acting as a mobile tether to hold the chromosomal replicase to DNA for high processivity (1–3). Sliding clamps from the three domains of life are remarkably similar in architecture (4–6). They consist of six domains of similar chain fold. The bacterial β-clamp is a homodimer, and each protomer consists of three domains, whereas eukaryotic and archaeal proliferating cell nuclear antigen (PCNA) clamps are homotrimers formed from protomers containing two domains each.
Initially, β and PCNA were identified as processivity factors for chromosomal replicases, but sliding clamps are now known to function with diverse DNA polymerases, repair factors, and cell cycle-control proteins (reviewed in ref. 1). Proteins typically bind PCNA through a conserved sequence, referred to as a PCNA interaction peptide (PIP) (7). The detailed interaction of a PIP sequence with PCNA was originally observed for human PCNA bound to a C-terminal peptide of the p21CIP1/WAF1 cyclin kinase inhibitor (8). Proteins that bind to the bacterial β-clamp contain a five- or six-residue consensus sequence, QL[S/D]LF and QLxLx[L/F] (9).
The peptide-binding pocket of sliding clamps is located between two domains on each protomer (8, 10). The binding pocket of the bacterial β-clamp is located between domains II and III, as demonstrated by structures of β bound to the δ-clamp loading subunit (11) and the β-Pol IV complex (10, 12). The protein-binding pocket of β consists of two subsites (10). Subsite 1 is 8 Å × 10 Å and 8.5 Å deep, whereas subsite 2 is 14 × 7.5 Å and 4.5 Å deep. Clamp-binding proteins can have additional points of contact with the clamp, as exemplified by Escherichia coli Pol IV, which also interacts with the edge of the β-ring (12).
E. coli harbors five DNA polymerases. Pols II, III, IV, and V are greatly stimulated by interaction with the β-clamp (12). Pol III is the chromosomal replicase, whereas Pols II, IV, and V are induced upon DNA damage and function in repair and chromosome maintenance (13). Pol IV and Pol V are Y-family, error-prone DNA polymerases that lack 3′–5′ proofreading exonuclease activity and are thought to advance replication forks over template lesions that block the Pol III replicase. Pol V is detectable only after DNA damage and is the main DNA polymerase responsible for mutagenic lesion bypass. Interestingly, whereas Pol II and Pol IV are induced 7- to 10-fold upon DNA damage, they are also present in undamaged cells (50 and 250 copies per cell, respectively) and may play roles during normal cell growth as well as during the DNA damage response. The roles of Pol II and Pol IV are relatively obscure.
The fact that the β-clamp is an essential protein and uses the same peptide-binding pocket for all of the DNA polymerases makes it difficult to apply classic genetic approaches to study how diverse polymerases function with β. Thus, a chemical may be used in the future to probe and better define the function of Pol II and Pol IV with β and their interplay with Pol III. To further this endeavor, the current report identifies a small-molecule compound that binds to the peptide-binding pocket of the β-clamp and selectively inhibits Pol III, compared with Pol II and Pol IV. To determine the molecular basis by which the compound selectively alters the function of β with these different DNA polymerases, we solve the structures of β bound to the compound as well as the related peptides of Pol II and Pol III with β, and compare them with the Pol IV-β structure. The analysis indicates how the chemical compound may discriminate among these different DNA polymerase-β-clamp interactions. Interestingly, the compound inhibits the bacterial Pol III replicase without disrupting the eukaryotic replicase. Hence, the β-clamp may represent a target for antibiotic compounds.
Results
Identification of a Small-Molecule Compound That Binds the Peptide-Binding Pocket of the β-Clamp.
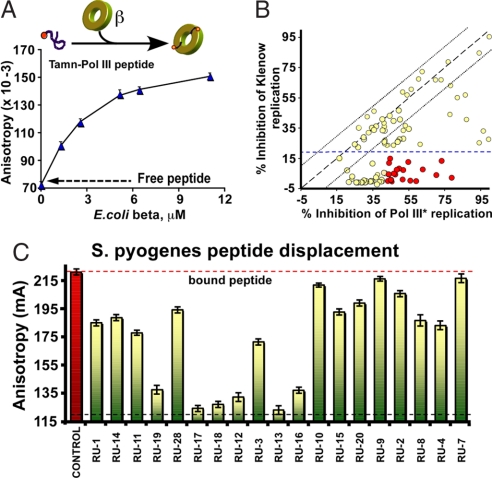
To identify small-molecule compounds that bind the peptide-binding pocket of β, we developed a fluorescence anisotropy assay that is easily adapted to a high-throughput approach. The assay uses a TAMN-labeled 20-mer peptide derived from the Pol III C terminus. Titration of β into the TAMN-peptide yields an apparent Kd of 2.7 ± 0.4 μM (Fig. 1A). Compounds that disrupt this interaction should displace the TAMN-peptide, resulting in a decrease in anisotropy. The peptide displacement assay was used to screen the Rockefeller University chemical library consisting of ≈30,600 polar organic compounds. An example result from one 386-well plate is shown in supporting information (SI) Fig. S1. The screen gave baseline dispersion values that grouped within 5%, with a Z-score of 0.901 ± 0.032 (14). Using a threshold value of 30% displacement, the screen yielded 91 different compounds. The screen was repeated, and chemicals that reproducibly displayed 30% displacement of Pol III peptide from β were screened further in Pol III β-dependent DNA synthesis assays on a singly primed M13mp18 ssDNA substrate (Fig. 1B). Compounds were also tested for nonspecific inhibition in β-independent assays by using Pol I Klenow fragment (Fig. 1B). Inhibition of Pol I Klenow fragment is indicative of nonspecific inhibition, for example, by binding DNA or protein denaturation, because Pol I, in contrast to the other Pols, is only stimulated 2-fold by the β-clamp.
Fig. 1.
High-throughput screen for chemical inhibitors that displace a Pol III peptide from the β-clamp. (A) Titration of E. coli β into TAMN-labeled Pol III C-terminal 20-mer peptide is monitored by fluorescence anisotropy. (B) Inhibition of DNA replication by compounds identified in the peptide-displacement assay. The plot shows the percentage of inhibition of E. coli DNA Pol I Klenow versus β-dependent synthesis by Pol III* in the presence of 20 μM compound. (C) Chemicals (i.e., at 50 μM) that displace E. coli Pol III α-peptide from E. coli β were tested for ability to displace S. pyogenes Pol C peptide from S. pyogenes β.
Compounds that inhibit Pol III substantially greater than Pol I (red circles in Fig. 1B) were tested in a TAMN-peptide displacement assay by using Streptococcus pyogenes β and a TAMN-20-mer peptide derived from the C terminus of S. pyogenes Pol C replicase (Kd = 1.5 μM; data not shown). Residues lining the peptide-binding pocket of E. coli β are conserved across diverse bacterial species, and accordingly, a subset of the compounds that disrupt the Gram-negative E. coli Pol III-β interaction also score positive in the Gram-positive S. pyogenes peptide displacement assay (Fig. 1C).
A Small-Molecule Compound Differentiates Between Function of Pols II, III, and IV with β.
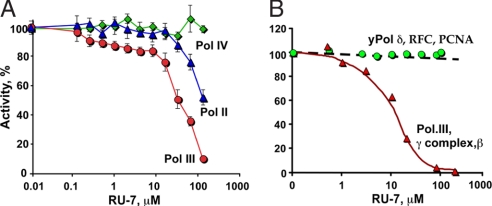
With a long-term goal of understanding the roles of Pol II and Pol IV with β, and their interplay with Pol III, we wished to identify a compound that differentiates between the actions of these DNA polymerases with β. Pols II, III, and IV display β-dependent activity with a 7.2-Kb M13mp18 ssDNA primed with a single oligonucleotide, and this assay revealed a compound, RU7, that differentially inhibits these DNA polymerases (Fig. 2A). At high concentrations of RU7, only Pol IV remains functional with β; the RU7 compound is at least a 50-fold less effective inhibitor of Pol IV than Pol III. At intermediate concentrations of RU7, the function of Pol III with β is inhibited to a greater extent than Pol II.
Fig. 2.
A small-molecule compound selectively inhibits Pol III. (A) The RU7 compound is titrated into β-dependent replication reactions by using Pol II (blue triangles), Pol III (red circles), or Pol IV (green diamonds). (B) The effect of RU7 on replication assays by using either the E. coli replication system (i.e., Pol III core, γ complex. and β clamp; red triangles) or the eukaryotic system (i.e., yeast Pol δ, RFC, and PCNA; green circles).
Despite the similar structures of bacterial β and eukaryotic PCNA, the amino acid sequence of these clamps is highly divergent, and sequence comparison algorithms do not detect similarity between them (4, 5). Hence, a small-molecule compound that binds β and inhibits Pol III would not be expected to bind PCNA and inhibit the eukaryotic replicase. In Fig. 2B, the RU7 compound is titrated into replication reactions by using the E. coli Pol III replicase or the PCNA-dependent yeast Pol δ replicase. The results show that RU7 inhibits the E. coli system with a Ki of ≈10 μM but does not inhibit the PCNA-dependent Saccharomyces cereviase Pol δ. The assays in Fig. 2A contain an additional 50 mM NaCl, compared with the assays of Fig. 2B, which results in slightly different Ki values.
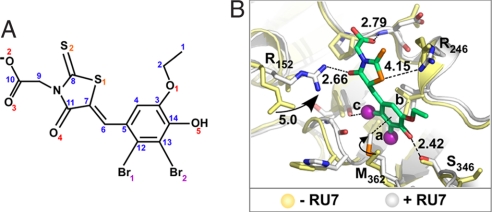
The RU7 compound is shown in Fig. 3A. RU7 presumably binds to the peptide-binding pocket of β because it was identified by using a peptide-displacement assay. However, the peptide-displacement assay is indirect and could be explained in other ways. For example, RU7 could bind the Pol III peptide or could bind another position on β and displace the TAMN-peptide by an allosteric mechanism. To obtain direct evidence that the RU7 compound binds to the peptide-binding pocket in β, we solved the structure of a RU7-β complex.
Fig. 3.
Structure of a small-molecule inhibitor bound to the β-clamp. (A) The RU7 compound. (B) Distances between RU7 and the β-clamp. Side-chain movements upon binding RU7 are indicated. Yellow and white are residues of β in the absence or presence of RU7, respectively. Distances between RU7 and protein residues are marked in black; the distances 3.73, 3.41, and 3.03 Å are marked a, b, and c, respectively.
Cocrystal Structure of β Bound to a Chemical Inhibitor.
The RU7-β cocrystal structure was solved by molecular replacement by using the apoenzyme structure (4). The region containing the compound was particularly well ordered (Fig. S2). The structure, refined to 1.64 Å resolution, reveals that RU7 occupies subsite 1 of the peptide-binding pocket on both subunits (Fig. 3B). Subsite 1 is the deeper of the two subsites and is shaped by residues R152, L155, L177, V247, V347, and V360. The chemical compound interacts with V247, P242, R152, R246, M362, and T172 (Fig. 3B). With the exception of the R152 side chain that swings 5 Å to form a H-bond with the carbonyl oxygen 4 of RU7 (2.7 Å), the peptide-binding pocket residues do not undergo a significant conformational change upon complex formation (Fig. 3B).
To understand how RU7 derives specificity for Pol III over Pol II and Pol IV, we solved structures of β with peptides of Pol II and Pol III and compared them with the previously determined Pol IV-β structure and the structure of RU7 bound to β.
Cocrystal Structure of β Bound to a Pol III Peptide.
Studies have identified two sequences within the Pol III α subunit that bind to β; one is located at the extreme C terminus of α (15), and the other is an internal site 220 residues upstream of the α C terminus (16). The two sequences differ to some extent, but both fall within the conserved consensus sequence defined for β-binding peptides (9). Two 9-mer peptides, corresponding to the internal and C-terminal clamp-binding sequences of Pol III α, were studied for β-binding affinity by using isothermal titration calorimetry (ITC) (Fig. S3). The analysis shows that the C-terminal peptide of Pol III α binds β >10-fold tighter than the internal β-binding sequence. We also examined β-binding to C-terminal Pol III peptide sequences of different lengths (9-mer, 20-mer, and 30-mer) to determine whether longer peptides bind β tighter than the 9-mer (Fig. S3). Overall, the analysis indicates that the major contribution of binding enthalpy is contained in the 9-mer β-clamp interactions (Fig. S3). We also used phage display to determine whether there exists a peptide that binds to β tighter than Pol III and other enzymes. The predominant β-binding peptide sequence selected by phage display belonged to the six-residue β-binding motif (Fig. S4). ITC analysis of this sequence, LQLELDF, shows that it binds to β with similar affinity as the Pol III 9-mer peptide (Kd ≈ 1.8 μM; data not shown).
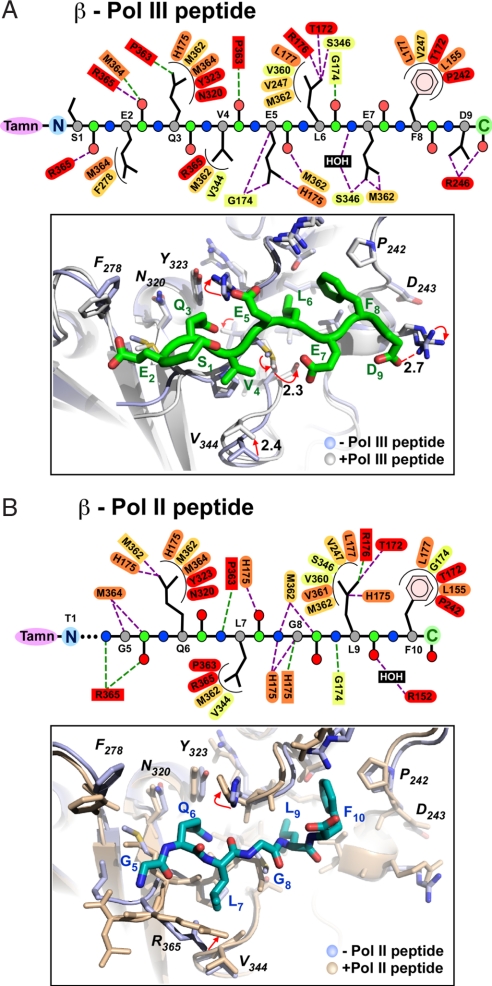
We tried to form cocrystals of β with both the C-terminal and the internal Pol III β-binding peptides, but only cocrystals using the Pol III α C-terminal peptide were obtained (Fig. S2). The structure of the Pol III C-terminal 9-mer (TAMN-S1E2Q3V4E5L6E7F8D9) peptide bound to β was solved by molecular replacement (Fig. 4A and Table S1). Superposition of the Cα atoms of the two 9-mer peptides occupying the two identical binding sites of the β-dimer yields a weighted rmsd value of only 0.26 Å. Hence, the Pol III 9-mer peptide binds each site in nearly the same manner. Although all 9 aa can be modeled into the electron density, only the C-terminal six residues fill the peptide-binding pocket. TAMN is also clearly visible in the electron density (Fig. S2). Residues L6, E7, and F8 of the Pol III 9-mer lie within subsite 1 and establish interactions with several side chains of the clamp; residues L6 and F8 penetrate deep into this site (Fig. 4A). Residues Q3, V4, and E5 are positioned in subsite 2. The side chains of the first two residues of the 9-mer (S1 and E2) do not appear to form significant interactions with β, although the main-chain carbonyl of E2 aligns structurally with R10 of the Pol IV peptide and forms a clear H-bond (2.7 Å) with the amide NH of R365.
Fig. 4.
Structures of Pol II and Pol III peptides bound to the β clamp. (A Upper) Main-chain atoms of the Pol III peptide are shown as circles (C, green; Cα, gray; N, blue; and O, red). β-Clamp residues (ovals, side-chain interactions; rectangles, main chain) are colored according to conservation as described in the legend to Fig. 5. Hydrophilic and hydrophobic interactions are shown as straight purple and curved black lines, respectively. (Lower) Pol III 9-mer peptide (green) bound to β. Peptide-free β (light blue) is superimposed onto peptide-bound β (white). (B Upper) Pol II peptide atoms and β side chains are color coded as in A. (Lower) Pol II 10-mer peptide (blue) bound to β. Peptide-free β (light blue) is superimposed onto the peptide-bound β (wheat).
Pol II Peptide-β Structure.
The Pol II peptide binds to β with a Kd value of 1.7 μM (ITC; data not shown). We determined that the structure of a Pol II C-terminal peptide (TAMN-T1L2M3T4G5Q6L7G8L9F10) bound to β by similar methods as for the Pol III peptide-β complex. The Pol II peptide-β complex crystallized in space group P1 (Fig. 4B, Fig. S2, and Table S1). Interestingly, only one of the two peptide-binding sites in β contains the Pol II peptide. A similar half-site occupancy was observed in the study of a Pol IV peptide bound to β (10) due to a crystal contact that occludes peptide from binding to the second site. This may also be the case for the Pol II peptide-β cocrystal. The Pol II β-binding sequence consists of five residues and thus lacks 1 aa compared with the six-residue β-binding sequence in the Pol III 9-mer peptide that is inserted between the last two C-terminal hydrophobic residues of the clamp-binding motif. Within subsite 2, the Pol II peptide forms similar contacts to β, as observed for the Pol III peptide, but there are substantial differences in the way the C-terminal sequences of the Pol II and Pol III peptides bind to subsite 1. These differences are likely due to the extra residue in the six-residue Pol III motif compared with the five-residue Pol II β-binding motif. The differences in subsite 1 are also due to L9, which superimposes with L7 of the Pol III 9-mer. L9 appears to be more efficiently buried than its Pol III peptide homolog (L7) and establishes two additional hydrophobic contacts with the His175 and Val361 side chains of β.
Comparison of Pol II, Pol III, Pol IV, and RU7 in the Peptide-Binding Site of β.
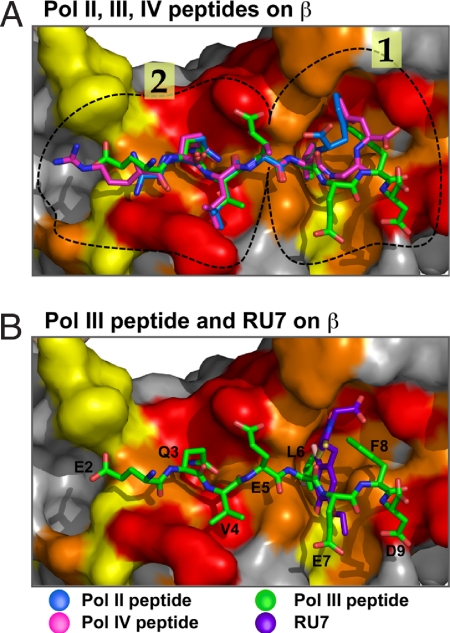
Superposition of the Pol II and Pol III peptides bound to β is shown in Fig. 5A. The Pol II and Pol III peptide residues in subsite 2 are held in nearly the same conformation (all atoms align within an rms of 0.125 Å). Subsite 1 is the deepest pocket and would be expected to form the tightest interactions with bound ligands. However, superposition of Pol II and Pol III peptides in subsite 1 reveals a remarkable difference in the position of the last few residues of the two peptides (see Fig. 5A). It is interesting to note that the side chains in subsite 1 that bind RU7 (R246, V247, S346, M362, T172, R152, and backbone amide of P242) also bind the Pol III peptide, with the exception of R152. In contrast, the Pol II peptide makes contact with six of these seven residues. The Pol II peptide makes unique contacts to residues His175 and Val361 in subsite 1, which do not appear to contact RU7 or the Pol III 9-mer. Superposition of the Pol IV peptide, which was solved in an earlier study (10), with the Pol II and Pol III peptides show that the Pol IV peptide binds to subsite 2, similar to Pols II and III, but the Pol IV peptide binds to subsite 1 of β in yet a different way than either Pol II or Pol III (Fig. 5B). Even though the Pol IV peptide is of the six-residue class of the β-binding motif, it does not appear to contact some of the residues that bind RU7 and the Pol III peptide. For example, E7 of the Pol III 9-mer (fifth residue of the six-residue consensus) is a glycine in the Pol IV peptide. Hence, the two interactions with β, formed by the Pol III E7 residue, are lacking in the Pol IV peptide. One of the Pol III 9-mer E7 interactions is with the side chain of M362, and the second is a water-mediated H-bond with the amide nitrogen of S346. Both of these residues contact RU7 and thus likely contribute to the greater inhibition by RU7 of Pol III compared with Pol IV. Furthermore, Pol IV is known to form additional contacts to β outside of the peptide-binding site (12). These additional contacts of Pol IV to β may contribute to the relative resistance of Pol IV to inhibition by the RU7 compound.
Fig. 5.
Superposition of Pol II, III, and IV peptides and RU7 compound bound to β. (A) Superposition of the Pol III peptide (green), Pol II peptide (blue), and Pol IV peptide (purple; PDB ID code 1OK7) (10). (B) Superposition of Pol III peptide (green) and the RU7 compound (orange). The surface of the β-clamp is colored white, and the protein-binding pocket of β is colored according to sequence conservation of an alignment of 42 bacterial subunits; the color scale proceeds from red (90% conservation) to yellow (50% conservation). Circled regions in the peptide-binding pocket indicate subsites 1 and 2. Figures were prepared by using Pymol (27).
We cannot exclude the possibility that the N-terminal TAMN moiety on the Pol II and Pol III peptides used in the current study may affect their conformation upon binding to β (the structure of Pol IV peptide-β complex did not contain an N-terminal TAMN moiety). However, the structural differences in the three peptides localize to the last few C-terminal residues and the N-terminal residues are in essentially the same conformation for all three peptides, indicating that the N-terminal TAMN may not underlie the structural differences in the C-terminal residues of the polymerase peptides observed here. The TAMN moiety of the Pol III peptide is visible in the electron density and forms a crystal contact with an adjacent β-clamp. However, the TAMN moiety of the Pol II peptide is not visible in the electron density, indicating that it may be disordered and thus may not interact with β.
Discussion
A Chemical Distinguishes Polymerase Function with β.
The structures of polymerase peptides bound to β reveal unexpected features in the way different DNA polymerases engage the peptide-binding pocket in the β-clamp. Specifically, the clamp-binding peptides of Pol II, Pol III, and Pol IV contact β in different ways in subsite 1, the deepest part of the protein-binding pocket. The RU7 small-molecule compound binds to subsite 1 of the clamp and distinguishes the activity of these different polymerases with β. The RU7 compound inhibits β-dependent synthesis by Pol III at least 5- and 50-fold more efficiently than it inhibits β-dependent synthesis by Pol II and Pol IV, respectively. We propose that the underlying basis by which RU7 differentially inhibits Pols II, III, and IV lies in the different ways these DNA polymerases bind to subsite 1 of the clamp. In addition, the Pol IV-β structure shows that Pol IV has two major points of contact with β; Pol IV binds the peptide-binding pocket and also interacts with the side of the β-ring, which may contribute to the resistance of Pol IV to RU7 (12).
Both β and Pol III α subunit are essential proteins, and therefore the interaction of α with β cannot be eliminated by mutagenesis without killing the cell. Thus, a chemical that specifically disrupts this interaction may enable future chemical genetic approaches to probe the roles of Pol II and Pol IV with β. For example, both Pol II and Pol IV are present in normal cells, but their role in normal growth is obscure. At low concentrations, RU7 will first inhibit Pol III-β, leaving only Pol II and/or Pol IV to function with β. Chemical genetic studies could perhaps address whether Pol IV or Pol II play a role in chromosome replication during normal growth. Moreover, Pol II and Pol IV are induced to high levels in the SOS response to DNA damage, and a chemical probe may be useful to address the role of these enzymes during times of cellular stress. Chemical genetic approaches would require further studies beyond those reported here. For example, the chemical must enter the cell and not be pumped back out.
The β-Clamp As a Target for Antibacterial Compounds.
Cell growth can be stopped by using chemical compounds that inhibit a major information pathway such as transcription, translation, and replication. These pathways use numerous essential enzymes, each of which is a potential target for an antibiotic chemical inhibitor. Indeed, there are several well known, small-molecule inhibitors of the transcription and translation machinery. To mention a few, rifampicin targets RNA polymerase, and tetracyclin and erythromycin target the ribosome. Precedent for replication inhibition in antiviral and antibacterial therapies is based in chain-terminating nucleoside analogs such as AZT (polymerase inhibitors), topoisomerase inhibitors, and active-site competitive inhibitors of RNA/DNA polymerases.
The β sliding clamp would appear to be an attractive pharmaceutical target because it is essential for cell viability yet shares no sequence homology with the eukaryotic PCNA clamp. Consistent with the extensive sequence difference between PCNA and β, RU7 does not inhibit PCNA-dependent synthesis by yeast Pol δ. Thus, one may ask why antibacterial compounds that target the β-clamp have not been found long ago. A large factor in this stems from the fact that <1% of terrestrial microorganisms have been isolated and cultured for study of antimicrobial compounds (17). Another factor is that the bacterial replicase has historically been difficult to study because the intracellular level is exceedingly low, and some components of the replicase machinery separate during purification. Only recently has the bacterial replication apparatus been expressed to high levels and reconstituted for chemical screens, such as those reported here. In contrast, RNA polymerase and ribosomes are plentiful in cells, making these tightly associated machineries relatively easy to purify for chemical screens and biochemical analysis.
The β-clamp functions in a unique manner that could enhance its suitability as an antibacterial target. For instance, Pol III and the clamp loader bind to β by using the same peptide-binding pocket, and they function with β in a very dynamic way (1, 18, 19). After the clamp loader assembles β onto DNA, the clamp is left on DNA and must wait for Pol III to associate with it. This handoff from the clamp loader to Pol III provides a window of opportunity for a chemical compound to target the unoccupied peptide-binding pockets of the clamp. Moreover, during lagging strand synthesis, Pol III repeatedly dissociates and reassociates thousands of times, from one β-clamp to another. This may provide additional opportunity for a chemical inhibitor to bind β. The fact that many different proteins bind β could also make development of resistant cells difficult. For example, if cells mutate the peptide-binding pocket of β to circumvent the compound, the mutated clamp may no longer bind one or more essential proteins.
Comparison of β-clamp sequences shows that residues that bind the Pol III 9-mer peptide are rather well conserved (see Fig. S5), as noted (10). Moreover, the structure of the S. pyogenes β-clamp has been solved (PDB ID code 2AVT) (20), and superposition of residues that form the protein-binding pocket of the Gram-negative E. coli β-clamp and the β-clamp of the Gram-positive S. pyogenes bacterium, results in only minor differences between corresponding α-carbon atoms (i.e., up to 0.36 Å) (Fig. S5B). The structure of peptide-β complexes and sequence alignment of bacterial β-clamps suggests a consensus sequence for the peptide-binding pocket of bacterial clamps (Fig. S5C).
The E. coli Pol III C-terminal peptide inhibits β-dependent DNA synthesis by the Gram-positive Pol C replicases of S. pyogenes and Staphylococcus aureus (21). In fact, the C-terminal regions of the Pol C subunits of these Gram-positive replicases also inhibit β-dependent synthesis by E. coli Pol III core, and E. coli β even functions with the Pol C replicase of these Gram-positive organisms (22). These observations support functional conservation, in addition to sequence conservation of this important polymerase/β-clamp connection and underscore the possibility that a chemical compound may inhibit protein–protein interactions from diverse organisms. Indeed, we demonstrate here that some compounds that bind the Gram-negative E. coli β-clamp also bind to Gram-positive S. pyogenes β.
Structure of RU7 As a Starting Point for Rational Drug Design.
The RU7 compound and its high-resolution structure with β provide a starting point for rational drug design. RU7 contains nine H-bond acceptors, six of which contact side chains in β. Most side chains of β that bind RU7 also form H-bonds to the Pol III 9-mer peptide. Although β uses only 6 side chains to bind RU7, 11 side chains are used in this same vicinity to bind the Pol III 9-mer peptide. Therefore, it seems possible that additional atoms could be strategically placed on RU7 to enhance its affinity for β. It is also interesting to note that the carboxyl moiety of RU7 is directed toward subsite 2 of the β-binding pocket. Subsite 2 contains several conserved residues that form important contacts with the Pol III 9-mer peptide. Thus additional interactive atoms could be engineered into RU7 at the carboxyl moiety that may fit into subsite 2 and enhance its potency.
Interestingly, some compounds are specific for the Gram-negative clamp, indicating that the β-clamp target offers possibilities to develop either “species-limited” or “broad-spectrum” compounds (Fig. 1C). Thus, the high-resolution structure analysis of RU7 underscores the possibility that specific pharmaceuticals may be developed for β-clamps from different organisms.
Methods
Materials.
HPLC-purified peptides were from Bio-Synthesis Inc.: Pol III C-terminal 9-mer (double-underlined), 20-mer (underlined), 30-mer (italic) peptides (GATWRVSPSDRLLNDLRGLIGSEQVELEFD), Pol III internal β-binding motif (IGQADMFGV), Pol II C-terminal 10-mer (TAMN-T1L2M3T4G5Q6L7G8L9F10), S. pyogenes PolC C-terminal 20-mer (TAMN-MGILGNMPEDNQLSLFDDFF), and S. aureus PolC 20-mer (TAMN-DELGSLPNLPDKAQLSIFDM). M13mp18 ssDNA was primed with a DNA 60-mer. Genes encoding Pol II and Pol IV were cloned into pET11 (Novagen), followed by transformation, induction, and purification from BL21(DE3) cells. Pol III core and Pol III* were constituted from recombinant subunits.
Structures.
β-Pol III 9-mer.
β (290 μM) and Pol III C-terminal 9-mer (290 μM) were mixed in 10 mM Tris (pH 7.4), 0.5 mM EDTA, and 50 mM NaCl, 5% glycerol and allowed to crystallize upon equilibrating 1.0 μl of protein-peptide solution with 1.0 μl of precipitant buffer (27.5% PEG 400, 100 mM Mes (pH 6.2), 100 mM CaCl2, ans 1% DMSO) in a hanging drop at 22°C. Trapezoidal crystals (0.3 × 0.5 × 0.6 mm3) diffracted to 1.9 Å resolution at 100 K by using the X4a beamline at the National Synchrotron Light Source, Brookhaven, NY. E. coli β crystallized in space group P3 (2) with two dimers in the asymmetric unit and a solvent content of 53.4%. Data reduction was achieved by using HKL2000 (23). Subsequently, the model was rebuilt and refined at 2.0 Å by using the program Crystallography and NMR System (CNS) (24) and ONO (25). The structure was solved by molecular replacement by using the monoclinic structure (PDB ID code 2POL) (4). Residual electron density indicated peptides in the protein-binding pockets of both protomers. All refinements were performed against ∣F0∣ > 0σ data, by using the CNS program suite (24).
β-Pol II 10-mer.
A TAMN-labeled,10-mer peptide encompassing the C-terminal residues of E.coli Pol II (TAMN-T1L2M3T4G5Q6L7G8L9F10) was cocrystallized with β and analyzed as described above (1.74 Å resolution at 100 K). β-Pol II 10-mer crystallized in space group P1 with one dimer in the asymmetric unit and a solvent content of 56.4%.
β-RU7.
1 μl of β (300 μM) and RU7 (600 μM) in 20 mM Tris·HCl 7.4, 0.5 mM EDTA, 10% glycerol, and 5% DMSO was mixed with 1.0 μl of precipitant buffer [25% PEG 400, 100 mM Mes (pH 6.1), 100 mM CaCl2, and 3% DMSO] in a hanging drop at 22°C. Crystals (0.3 × 0.4 × 0.5 mm3) formed in space group P1 and diffracted to 1.52 Å resolution at 100 K (see Table S1). Structure determination was by molecular replacement and refined to 1.64 Å as described above.
Fluorescence Anisotropy.
Increasing amounts of E. coli β or S. pyogenes β were titrated into reactions containing 1 μM TAMN-20-mer C-terminal peptide derived from E. coli Pol III α, S. pyogenes Pol C, or S. aureus Pol C in 20 mM Tris·HCl (pH 7.5) and 0.5 mM EDTA at 25°C. Anisotropy was measured in a PTI spectrofluorometer (535-nm excitation; 575-nm emission).
High-Throughput Screening.
Reactions (15 μl) contained E. coli β (1 μM) and TAMN-labeled Pol III C-terminal 20-mer peptide (5 μM) and compound (50 μM) in 20 mM Tris·HCl (pH 7.5), 1 mM DTT, and 0.5 mM EDTA in 384-well plates. The Rockefeller University chemical library contained 30,600 compounds at the time of this work. Control wells included TAMN-labeled peptide with no β and β with different concentrations of unlabeled peptide competitor. Assay plates were gently agitated, centrifuged at 1,500 × g, and incubated 10–15 min at 22°C. Fluorescence anisotropy was measured by using a plate reader (excitation 535 nm; emission 575 nm). Results were stable for 16 h at 22°C. Reactions were performed in duplicate, and compounds were analyzed for inhibition of Pol I Klenow and for β-dependent synthesis by Pol III* as described below.
DNA Replication Assays.
Compounds (0.1 μl of 5 mM stocks) were added to 25-μl reactions containing 20 mM Tris·Cl (pH 7.5), 0.5 mM EDTA, 40 μg/ml BSA, 5 mM DTT, 0.5 mM ATP, 8 mM MgCl2, 60 μM concentrations of each of the four dNTPs, 30 fmol of singly primed M13mp18 ssDNA, 425 pmol of SSB, and 124 fmol of β. DNA synthesis was initiated upon adding 80 fmol of Pol III*. Plates were incubated at 22°C for 10 min on a shaker. The same protocol was followed for Pol I Klenow reactions, except 2 units of Pol I Klenow was added to initiate replication, and reactions did not contain β. Reactions were quenched with 25 μl of 1% SDS/40 mM EDTA containing Quant-iT PicoGreen (1/150 dilution) (Invitrogen) as described (26). Fluorescence intensity was measured by using a plate reader (excitation 480 nm; emission 520 nm).
Assays comparing RU7 in yeast and E. coli replication systems were performed as above except that reactions contained 20 μM α-32P-dTTP, and DNA synthesis was monitored by radioactive incorporation. Yeast Pol δ replicase was assayed by using 60 ng of PCNA, 115 ng of replication factor C (RFC), and 20 ng of Pol δ. Assays comparing RU7 with Pols II, III, and IV were also performed by using α-32P-dTTP as above except that β (100 fmol) was loaded onto singly primed M13mp18 DNA by γ-complex (20 fmol) in a 5-min preincubation before adding DNA polymerase (600 fmol per reaction). After 1 min at 37°C, 1 μl of RU7 was added, and replication was initiated by adding dATP and α-32P-dTTP.
Accession Code.
Coordinates are deposited in the Protein Data Bank under PDV ID codes 3D1E (β-Pol II peptide), 3D1F (β-Pol II peptide), and 3DIG (β-RU7).
Supplementary Material
Acknowledgments.
We thank Chuck Karan for help at the Rockefeller University Screening facility and the staff at beamlines X4a and X25 of the National Synchrotron Light Source, Brookhaven, NY. This work was supported by National Institutes of Health Grants GM38839 (to M.O.D.), GM70841 (to X.-P.K.), and GM45547 (to J.K.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3D1E, 3D1F, and 3DIG).
This article contains supporting information online at www.pnas.org/cgi/content/full/0804754105/DCSupplemental.
References
- 1.Johnson A, O'Donnell M. Cellular DNA replicases: Components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 2.Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu Rev Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 3.McHenry CS. Chromosomal replicases as asymmetric dimers: Studies of subunit arrangement and functional consequences. Mol Microbiol. 2003;49:1157–1165. doi: 10.1046/j.1365-2958.2003.03645.x. [DOI] [PubMed] [Google Scholar]
- 4.Kong XP, Onrust R, O'Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: A sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 5.Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 6.Matsumiya S, Ishino Y, Morikawa K. Crystal structure of an archaeal DNA sliding clamp: Proliferating cell nuclear antigen from Pyrococcus furiosus. Protein Sci. 2001;10:17–23. doi: 10.1110/ps.36401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warbrick E. PCNA binding through a conserved motif. BioEssays. 1998;20:195–199. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 8.Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 9.Wijffels G, et al. Inhibition of protein interactions with the beta 2 sliding clamp of Escherichia coli DNA polymerase III by peptides from beta 2-binding proteins. Biochemistry. 2004;43:5661–5671. doi: 10.1021/bi036229j. [DOI] [PubMed] [Google Scholar]
- 10.Burnouf DY, et al. Structural and biochemical analysis of sliding clamp/ligand interactions suggest a competition between replicative and translesion DNA polymerases. J Mol Biol. 2004;335:1187–1197. doi: 10.1016/j.jmb.2003.11.049. [DOI] [PubMed] [Google Scholar]
- 11.Jeruzalmi D, et al. Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell. 2001;106:417–428. [PubMed] [Google Scholar]
- 12.Bunting KA, Roe SM, Pearl LH. Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the beta-clamp. EMBO J. 2003;22:5883–5892. doi: 10.1093/emboj/cdg568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tippin B, Pham P, Goodman MF. Error-prone replication for better or worse. Trends Microbiol. 2004;12:288–295. doi: 10.1016/j.tim.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 15.Lopez de Saro FJ, Georgescu RE, O'Donnell M. A peptide switch regulates DNA polymerase processivity. Proc Natl Acad Sci USA. 2003;100:14689–14694. doi: 10.1073/pnas.2435454100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dohrmann PR, McHenry CS. A bipartite polymerase-processivity factor interaction: Only the internal beta binding site of the alpha subunit is required for processive replication by the DNA polymerase III holoenzyme. J Mol Biol. 2005;350:228–239. doi: 10.1016/j.jmb.2005.04.065. [DOI] [PubMed] [Google Scholar]
- 17.Rappe MS, Giovannoni SJ. The uncultured microbial majority. Annu Rev Microbiol. 2003;57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 18.Naktinis V, Turner J, O'Donnell M. A molecular switch in a replication machine defined by an internal competition for protein rings. Cell. 1996;84:137–145. doi: 10.1016/s0092-8674(00)81000-4. [DOI] [PubMed] [Google Scholar]
- 19.Fujii S, Fuchs RP. Defining the position of the switches between replicative and bypass DNA polymerases. EMBO J. 2004;23:4342–4352. doi: 10.1038/sj.emboj.7600438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Argiriadi MA, Goedken ER, Bruck I, O'Donnell M, Kuriyan J. Crystal structure of a DNA polymerase sliding clamp from a Gram-positive bacterium. BMC Struct Biol. 2006;6:2. doi: 10.1186/1472-6807-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruck I, Georgescu RE, O'Donnell M. Conserved interactions in the Staphylococcus aureus DNA PolC chromosome replication machine. J Biochem. 2005;280:18152–18162. doi: 10.1074/jbc.M413595200. [DOI] [PubMed] [Google Scholar]
- 22.Klemperer N, Zhang D, Skangalis M, O'Donnell M. Cross-utilization of the beta sliding clamp by replicative polymerases of evolutionary divergent organisms. J Biochem. 2000;275:26136–26143. doi: 10.1074/jbc.M002566200. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. In: Macromolecular Crystallography, Part A. Carter CWJ, Sweet RM, editors. New York: Academic; 1997. pp. 307–326. [Google Scholar]
- 24.Brunger AT, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 26.Seville M, West AB, Cull MG, McHenry CS. Fluorometric assay for DNA polymerases and reverse transcriptase. BioTechniques. 1996;21:664–672. doi: 10.2144/96214st04. [DOI] [PubMed] [Google Scholar]
- 27.DeLano WL. The PyMOL User's Manual. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.