Abstract
The size and organization of the brain neocortex has dramatically changed during primate evolution. This is probably due to the emergence of novel genes after duplication events, evolutionary changes in gene expression, and/or acceleration in protein evolution. Here, we describe a human Ret finger protein-like (hRFPL)1,2,3 gene cluster on chromosome 22, which is transactivated by the corticogenic transcription factor Pax6. High hRFPL1,2,3 transcript levels were detected at the onset of neurogenesis in differentiating human embryonic stem cells and in the developing human neocortex, whereas the unique murine RFPL gene is expressed in liver but not in neural tissue. Study of the evolutionary history of the RFPL gene family revealed that the RFPL1,2,3 gene ancestor emerged after the Euarchonta-Glires split. Subsequent duplication events led to the presence of multiple RFPL1,2,3 genes in Catarrhini (∼34 mya) resulting in an increase in gene copy number in the hominoid lineage. In Catarrhini, RFPL1,2,3 expression profile diverged toward the neocortex and cerebellum over the liver. Importantly, humans showed a striking increase in cortical RFPL1,2,3 expression in comparison to their cerebellum, and to chimpanzee and macaque neocortex. Acceleration in RFPL-protein evolution was also observed with signs of positive selection in the RFPL1,2,3 cluster and two neofunctionalization events (acquisition of a specific RFPL-Defining Motif in all RFPLs and of a N-terminal 29 amino-acid sequence in catarrhinian RFPL1,2,3). Thus, we propose that the recent emergence and multiplication of the RFPL1,2,3 genes contribute to changes in primate neocortex size and/or organization.
Introduction
The neocortex, which is the most recent brain structure, emerged with mammals approximately 200 million years ago (mya) and increased in size in many branches of mammalian evolution, mostly in the lineage of anthropoid primates.1 With the emergence of the Homo genus (∼2.5–3 mya), acceleration in cortical enlargement occurred, especially over the last two million years in which the total brain size went from 500–800 cm3 (Homo habilis) to 1200–1400 cm3 (Homo sapiens). Together with the size increase, transformations in the organization of the brain occurred as the two cerebral hemispheres developed specializations that led to the emergence of specific behavioral traits in human, such as the development of complex languages or tools, self-awareness, and cultures.2
At a developmental level, the generation of the cortical cells is triggered by modifications in cell-cycle kinetics of the neural progenitors leading to their differentiation.3 It has been reported that the cell-cycle parameters of the progenitors evolved in primates increasing the number of cortical cells produced;4 such an increase contributes to changes in neocortex development.3,5,6 However, the genetic basis of primate brain evolution remains obscure. Genome-wide approaches comparing human to other primates revealed that humans present an expansion in the copy number of genes involved in brain structure and/or function7 and an increase in gene expression in the neocortex.8,9 Furthermore, genes involved in brain development show higher rates of protein evolution in the primate lineage leading to human.10 The latter has been particularly documented for the cell-cycle regulating MCPH gene family (MIM 251200), which are the only genes known to date to determine brain size.11–13
The transcription factor Pax6 controls the specification of neural progenitors during corticogenesis.14,15 In mouse, PAX6-deficient cortical progenitors show during neurogenesis a shorter cell-cycle duration that impairs their differentiation,16–18 and in human, heterozygous PAX6 mutation leads to cognitive impairments by altering cortical regionalization.19 However, the molecular mechanisms involved in Pax6-mediated corticogenesis are largely unknown. Interestingly, they could differ between species. Although Pax6 is found in neural progenitors of the ventricular and subventricular zones common to rodents and primates, its expression has also been detected in a large population of primate-specific neural progenitors located in the outer subventricular zone.6,20 Furthermore, different Pax6 binding sites have been identified in rodents and human, suggesting that Pax6 transactivates different gene networks in those species.21
In an effort to identify genes involved in neocortex evolution we set out to (1) identify new Pax6 target genes involved in human neurogenesis and (2) assess the evolutionary behavior of those genes. We identified the human Ret-finger protein-like (hRFPL)1,2,3 genes as Pax6 targets and observed that they were highly expressed in human embryonic stem cell-derived neurogenesis and in human fetal neocortex. Evolutionary analyses and expression profiling of the RFPL1,2,3 genes indicated that they showed signs of positive selection, an increase in gene copy number in the hominoid lineage, and a striking increase in cortical expression in human in comparison to chimpanzee and macaque.
Material and Methods
Cell Culture
Human HeLa-TAT cell lines were obtained from the National Institutes of Health AIDS Research and Reference Reagent; HEK293T cells were kindly provided by Dr. P. Salmon (Department of Fundamental Neurosciences, University of Geneva, Switzerland). Cells were cultured at 37°C with 5% CO2 in DMEM medium supplemented with 10% fetal-bovine serum, penicillin and streptomycin (all from GIBCO-Invitrogen). SA-01 human embryonic stem cells (Cellartis AB) were maintained on a layer of mitotically inactivated STO feeder cells. They were cultured in DMEM/F12 Glutamax supplemented with 20% knockout serum replacement, 1 mM nonessential amino acids, penicillin and streptomycin, 0.55 mM β-mercaptoethanol (all from GIBCO-Invitrogen), and 10 ng/ml recombinant human FGF2 (Invitrogen). Cultures were fed daily and manually passed every 5–7 days.
Neuronal Differentiation of Human Embryonic Stem Cells
Induction of neural progenitors and neuronal differentiation of SA-01 human embryonic stem cells were performed according to Perrier et al.,22 with slight modifications. In brief, the cells were plated on mitotically inactivated murine bone-marrow-derived stromal feeder MS5 cells in serum replacement (KSR) medium containing DMEM/F12 supplemented with 15% knockout serum replacement, 1 mM nonessential amino acids, penicillin/streptomycin, and 0.55 mM β-mercaptoethanol (all from GIBCO-Invitrogen). After 12 days in these conditions, KSR medium was replaced by N2 medium (DMEM/F12 supplemented with 25 μg/ml insulin, 50 μg/ml transferrin, 100 μM putrescine, 30 nM selenium chloride, 20 nM progesterone [all from GIBCO-Invitrogen], penicillin and streptomycin). Medium was changed every 2–3 days, and growth factors were added. At day 24 of differentiation, rosette structures were mechanically collected and transferred to 15 μg/ml polyornithine- and 1 μg/ml laminin-coated culture dishes in N2 medium supplemented with growth factors. After 8–12 days, cells were exposed to Mg2+-free Hanks' Balanced Salt Solution for 3 hr at 37°C, spun at 500 × g for 5 min, resuspended in N2 medium, and plated onto polyornithine- and laminin-coated culture dishes (105 cells per cm2). After two other passages in the same conditions, we differentiated the cells by replating them onto polyornithine-and-laminin-coated culture dishes (25 to 50 × 103 cells per cm2).
Microarray Transcriptome Analysis
Two days after transduction of HeLa cells with GFP or PAX6, total RNA from each sample was isolated, labeled, and hybridized to Affymetrix oligonucleotide-array-containing probes to 38,500 human genes. Total RNA was isolated from HeLa cells with Qiashredder kit (QIAGEN) and purified with QIAGEN RNeasy kit in accordance with the “RNA cleanup” protocol. All RNAs were of high and comparable quality as determined with the 2100 Bioanalyzer (Agilent Technologies). Biotinylation of 10 μg RNA, hybridization to Affymetrix Human Genome U133 Plus 2.0 arrays, washing steps, and array scanning were carried out in accordance with Affymetrix protocols. Expression data were analyzed with GeneSpring GX 7 (Agilent Technologies). Arrays were scaled to the same average intensity with all probes on the array.
Real-Time Quantitative PCR
Total RNA from human embryonic stem cells was isolated with RNeasy Mini kit according to the manufacturer's instructions (QIAGEN). We removed residual genomic DNA by incubating the RNA solution with 30 u RNase-free DNase for 15 min at room temperature with RNase-Free DNase Set (QIAGEN). Human fetal brain total RNAs were obtained from Biochain Institute. Total RNA (1 μg) was reverse transcribed with the Superscript II kit according to the manufacturer's instructions (Invitrogen). Real-time PCR reactions were performed with a Power SYBR Green PCR Master Mix (Applied Biosystems) and a Chromo 4TM Real-Time system (Bio-Rad). The amplification efficiency of each pair of primers was determined by comparison with a standard curve generated with serially diluted cDNA of fetal brain. Quantification was performed at a threshold detection line (CT value). The CT of each target genes was normalized against that of the housekeeping genes, cyclophilin or eEF1A1. The 2–ΔΔCT method was used to determine the relative level of expression of each gene.23 cDNAs from fetal or adult brain and adult testis were used as calibrators. The list of the primers used is given in Table S1, available online.
Immunocytochemistry
Three days after transduction, cells were washed in PBS and fixed with 2% paraformaldehyde (w/v) for 30 min at room temperature. After two washes in PBS/0.05% Tween-20 buffer, cells were permeabilized in PBS containing 0.5% (v/v) Triton X-100 for 30 min at room temperature and washed twice more in PBS/0.05% Tween-20 buffer. The fixed cells were then treated with PBS containing 1% fetal-calf serum (blocking buffer) for 30 min and incubated with Pax6 (1:100, Santa Cruz Biotechnology) and hRFPL1,2,3 (1:100, Abnova) primary antibodies for 1 hr at room temperature. After two washes in blocking buffer, cells were incubated with the secondary antibodies (Alexa Fluor 488 or 555 conjugates, 1:1000, Molecular Probes) for 1 hr at room temperature. The cells were finally washed twice in PBS/0.05% Tween-20 buffer, once in PBS buffer and once in distilled water before mounting with Fluorsave (Calbiochem). Visualization analysis was performed with an AxioSkop 2 Plus microscope equipped for epifluorescence and recorded with an AxioCam HR CCD camera and the AxioVision 4 software (Zeiss).
Chromatin-Immunoprecipitation Assay
We treated HeLa cells for 8 min at room temperature 48 hr after transduction with GFP or PAX6 by adding to the culture medium 5 mM HEPES (pH 7.9), containing 10 mM NaCl and 1.1% formaldehyde. Fixation was stopped by an addition of 180 mM glycine. After one wash with PBS, we lyzed the cells by pipetting them up and down in ice-cold 10 mM Tris-HCl (pH 8.0), 0.5% NP-40 supplemented with complete protease inhibitor (Roche). Nuclei were pelleted and lyzed in 10 mM Tris-HCl (pH 8.0), 500 mM NaCl, 1% Triton X-100, and 0.5% sodium deoxycholate supplemented with complete protease inhibitor. Crosslinked chromatin was sheared into 200–600 bp fragments by sonication, cleared by centrifugation at 15,000 × g for 15 min, and stored at −80°C. A total of 20 μg chromatin supernatants were diluted (1:10) in ChIP buffer (200 mM HEPES [pH 7.9], 2 M NaCl, and 20 mM EDTA) supplemented with 200 μg/ml salmon-sperm DNA and complete protease inhibitor. After preclearing the supernatants by rotating incubation for 30 min at 4°C with protein A–sepharose beads (Pierce), we stored half the chromatin at −20°C as the control chromatin input. The rest of the supernatant was then incubated overnight at 4°C with 20 μl of Pax6 antibody (Covance) and cleared by centrifugation at room temperature for 10 min at 8000 × g before a 2 hr rotating incubation at room temperature in 20 μl protein A-sepharose beads. The beads were washed as follows: twice in ChIP buffer; twice in ChIP buffer supplemented with 300 mM NaCl, 1% Triton X-100, and 0.1% sodium deoxycholate; twice in Tris-HCl (pH 8.0), 250 mM LiCl, 2 mM EDTA, and 0.5% sodium deoxycholate; once in 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 0.1% NP-40. We eluted immune complexes by incubating them for 10 min at 65°C in 111 mM Tris-HCl (pH 8.0) and 1.11% SDS. Crosslinks of the immunoprecipitated DNA and the chromatin input were reversed by incubation for 2 hr at 42°C after addition of 20 mg/ml Proteinase K (QIAGEN) and 100 mM NaCl and further incubation overnight at 67°C. After extraction with phenol-chloroform-isoamyl alcohol (25:24:1) and chloroform-isoamyl alcohol (24:1), DNA was precipitated with 100% ethanol in the presence of 20 μg glycogen (Fermentas) and 3 M sodium acetate. Chromatin pellets were resuspended in 100 μl TE buffer. The immunoprecipitated DNA and the input chromatin were analyzed by endpoint PCR (40 cycles) with Taq DNA Polymerase in a Q-solution-supplemented buffer in accordance with the manufacturer's instructions (QIAGEN) and promoter- and exon2-specific primers (Table S1). Design of the primers was performed according to the Pax6 putative binding sites identified in silico in the proximal region of the RFPL1,2,3 promoters with the MatInspector server.
Comparative RFPL1,2,3 Gene-Expression Array in Catarrhini
We used Affymetrix Human Exon 1.0 ST Arrays to measure gene-expression profiles in five to six humans, five to six chimpanzees, and two to four rhesus macaques from two age groups (adult and newborn) and in three tissues (neocortex, cerebellum and liver). All samples were sex and age matched to the best extent possible, and all had high RNA-quality preservation. To identify the expression differences between the species, we masked all probes that did not match the three genomes perfectly, leading to the use of three probes. Given the high homology of the three genes, we could not however discriminate them. To determine whether the signal intensity of a given probe was above the expected level of background noise, for each probe we compared its signal intensity to a distribution of signal intensities of the “antigenomic” probes with the same GC content provided by Affymetrix as an estimator of the unspecific background hybridization. The probe signal was classified as detected above the background if its intensity was greater than the 95% percentile of the background probes with the same GC content.24 To further remove the possible systematic experimental differences among the arrays, we performed a PM-GCBG correction and quantile normalization by using “affy” R package. We calculated gene-expression intensities as the means of all probe intensities mapped to the RFPL genes. The ratios of expression levels were calculated within the species and/or tissues.
Genetic Analyses
We created the alignment of RFPL gene family with ENCODE genome alignment (ENM 004)25 or by blasting human RFPL genes in the UCSC genome browser. To reveal the structure of the family, we first determined the internal structure of the catarrhinian RFPL1,2,3 subfamily. Given the loss of the first exon of some genes (Table S6), we created the sequence alignment including the genes' intron and the second exon, which are present in all RFPL1,2,3 sequences. The tree was rooted with a macaque RFPL gene closest to the RFPL1,2,3 cluster. We next created the coding sequence alignment including all RFPL1,2,3 genes found in the databases and the best BLAST hits of hRFPL4,5,6 genes in Catarrhini. Out of 26 RFPL genes, we selected a set of 21 most likely functional genes by using the following criteria: standard gene structure (two coding exons), start of CDS with a start codon, termination with a stop codon, and no frameshift. ML phylogenetic analysis revealed the robust monophyly of RFPL1,2,3 and RFPL4,5,6 primate-specific clades. The phylogenetic reconstructions were performed by constraining the topology of the clusters using the best BLAST hits. We then rooted the topology with the three RFPL genes found in Laurasiatheria and subsequently the bifurcation between the 21 likely functional catarrhinian genes and the two genes found in Glires.
Estimations of KA/KS ratios (omegas) were made with CODEML program implemented in PAML package with model 1 (Figure S1). Assessment of selection acting on RFPL1,2,3 gene family in Catarrhini after the duplication events was performed with branch-site model A (model = 2, NSsites = 2, fix_omega = 0; model = 2 options allowing two omegas, one for the foreground clade and the other for the rest of the tree). The null model was also branch-site model A with omega for the foreground clade (Catarrhini RFPL1,2,3) fixed to 1 (model = 2, NSsites = 2, fix_omega = 1, mega = 1). We also applied the branch model for the same clade (model = 2, NSsites = 0) with the null model specified by model = 0 and NSsites = 0. We used a likelihood ratio test (LRT) to assess the significance of the difference between the two models. Positively selected sites were estimated with the same method as well as the M2 and M8 models, whereas M1 and M7 were used as the null models respectively.
Statistical Analysis
Results are displayed as mean ± standard error (SEM) because the criteria studied are all ratio level measurements (i.e., continuous variables). Analysis of the Pax6-elicited hRFPL1 and hRFPL2,3 expressions by qPCR was done with Student's unpaired t test for two group comparisons. Analyses of the hRFPL1, hRFPL2,3, and hRFPL4,5,6 expressions during neural differentiation of human embryonic stem cells were performed by a one-way analysis of variance; this was followed by post hoc multiple comparisons with Tukey test. Analysis of correlations was performed with the Pearson product moment correlation. Expression changes in catarrhinian RFPL1,2,3 genes were analyzed either by one-way ANOVA and then post hoc multiple comparisons with Tukey test (intraspecies divergence) or by Student's t test (interspecies divergence). For all tests, a p value that is less than 0.05 was taken as statistically significant.
Results
Pax6 Elicits the Expression of the Human RFPL1,2,3 Genes
Given the importance of cell-cycle regulation during corticogenesis, we performed a microarray-expression analysis in Pax6-overexpressing HeLa cells, a reference human cell system for cell-cycle study.
Pax6 overexpression resulted in marked changes in the transcriptome (Table S6) and particularly modified the expression pattern of gene clusters involved in the control of the cell cycle (Figure S2). The most striking change was observed with the 800-fold increase in the human Ret Finger Protein-Like 1 (hRFPL1) transcript level (Table S6), which was also obtained with real-time PCR (Figure 1A). We also observed a more moderate increase in hRFPL2 and hRFPL3 gene expressions (Table S6 and Figure 1A). Given the high homology between those two genes, we could not however discriminate their individual expression by real-time PCR. Putative Pax6 binding sites were predicted in silico on each hRFPL1,2,3 promoters; chromatin-immunoprecipitation assay showed that Pax6 interacts in vivo with all of these promoters (Figure 1B). Finally, we examined whether Pax6-induced transcriptional changes led to hRFPL1,2,3 protein expression. Using a pan-hRFPL1,2,3 antibody, we observed hRFPL immunofluorescence in Pax6-expressing cells (Figure 1C). Taken together, those data suggest that Pax6 could induce hRFPL1,2,3 protein expressions through direct promoter transactivation.
Figure 1.
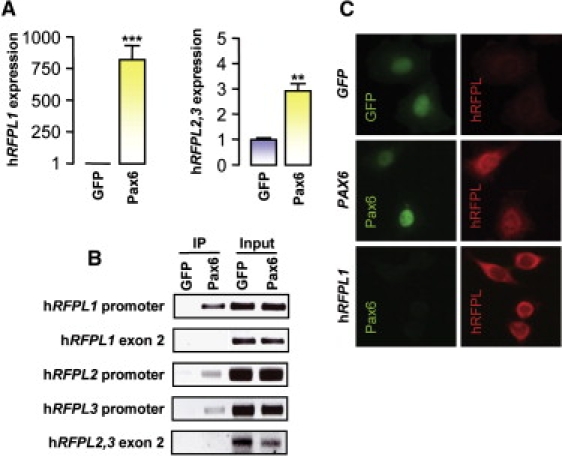
Pax6 Interacts with the hRFPL1,2,3 Gene Promoters and Induces Their Transcript and Protein Expressions
(A) hRFPL1 and hRFPL2,3 expressions were determined by real-time PCR after overexpression of GFP or Pax6 in HeLa cells. Transcript levels are indicated as the fold increase relative to the control level (GFP-transduced cells). hRFPL2 and hRFPL3 are detected by a common set of primers. Results are displayed as mean ± standard error (SEM). Statistical analyses were done with Student's t test, ∗∗p < 0.01 and ∗∗∗p < 0.001.
(B) In vivo binding of Pax6 to hRFPL1,2,3 promoters was assessed by chromatin-immunoprecipitation assay. After chromatin immunoprecipitation with a Pax6 antibody, endpoint PCRs were performed with primers specific for each hRFPL1,2,3 promoter or for exon 2 of each gene for visualization of nonspecific immunoprecipitation. “Input” represents dilutions of input chromatin used as PCR controls.
(C) Immunocytochemical detection of hRFPL1,2,3 proteins in GFP- and Pax6-expressing HeLa cells with a pan-hRFPL1,2,3 antibody. hRFPL1 overexpression was used as a control for antibody specificity.
hRFPL1,2,3 Genes Are Expressed during Neurogenesis In Vitro and In Vivo
We next examined hRFPL1,2,3 expression profiles during neurogenesis in vitro and in vivo. During neural differentiation of human embryonic stem cells, virtually no hRFPL1 and hRFPL,2,3 transcript levels were detected in NANOG-positive human embryonic stem cells, nor in SIX3- and SOX1-expressing neural precursors. Their expressions increased significantly at the onset of neurogenesis and were correlated to those of the GABAergic neuronal marker, GAD67 (Figure 2A).
Figure 2.
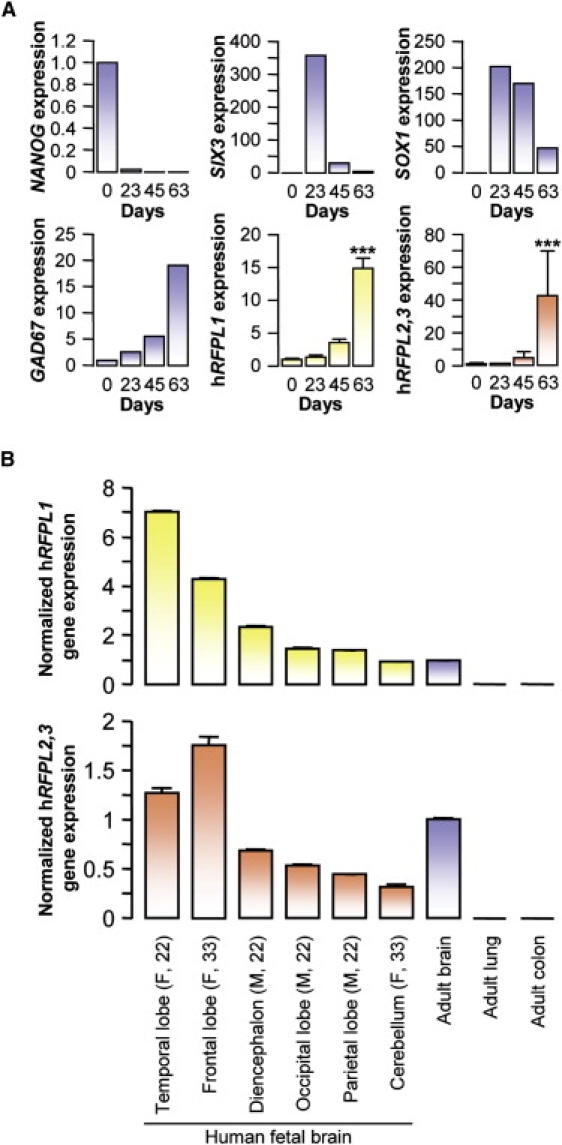
hRFPL1,2,3 Genes Are Expressed during Human Neurogenesis In Vitro and In Vivo
(A) hRFPL1 and hRFPL2,3 expressions were determined by real-time PCR during human embryonic-stem-cells-derived neurogenesis and normalized to the cDNA level in fetal brain. Results are displayed as mean ± standard error (SEM). ∗∗∗p < 0.001 versus day 0 with one-way ANOVA followed by post hoc Tukey's test. hRFPL1 and hRFPL2,3 expressions were correlated to those of the neuronal marker GAD67 (R2 = 0.99, p = 0.003 for hRFPL1 versus GAD67; R2 = 0.98, p = 0.012 for hRFPL2,3 versus GAD67 with Pearson product moment correlation).
(B) hRFPL1 and hRFPL2,3 expressions were assessed in different structures of the developing brain and in adult neocortex by real-time PCR normalized to the level of expression observed in adult brain. The gender of the embryo (M, male; F, female) and its age in weeks are indicated in the parentheses. Results are displayed as mean ± standard error (SEM).
hRFPL1 and hRFPL2,3 transcripts were also detected in the developing human fetal brain. hRFPL1 transcripts were highest in the temporal lobe, and hRFPL2,3 transcripts were highest in the frontal lobe. All transcripts were also detected in adult brain (Figure 2B).
Interestingly, a murine RFPL gene (mRFPL) was previously identified, but its expression had not been detected in the mouse adult brain.26–28 Thus, we further compared the expression profile of mRFPL with that of hRFPL1,2,3 by PCR and did not detect any mRFPL transcript during mouse embryonic stem cell-derived neurogenesis, during in vitro maturation of primary cortical neurons, or in fetal, newborn, and adult brains (Figure S3).
Identification of the Likely Functional RFPL Genes
Given the absence of mRFPL brain expression, we investigated at what point during evolution the RFPL genes acquired brain expression by tracing their evolutionary history. We first searched for likely functional RFPL genes by using the ENCODE TBA genomic alignments (ENM 004)25 and BLAT alignment in the UCSC genome browser. They were found only in boreoeutherian mammals, suggesting that the RFPL gene ancestor emerged ∼100 mya (Table S2). Whereas one gene was identified in Laurasiatheria (dog, cat, and horse) and Glires (mouse and rat), multiple RFPL genes were found in Catarrhini (Great Apes and Old World monkeys), including six in human. Maximum-likelihood phylogenetic analysis showed that the set of likely functional genes in Catarrhini formed two isolated clusters: The first one grouped the genes orthologous to the human hRFPL1,2,3 genes, whereas the second cluster grouped the genes orthologous to the human hRFPL4,5,6 genes (Figure 3A).
Figure 3.
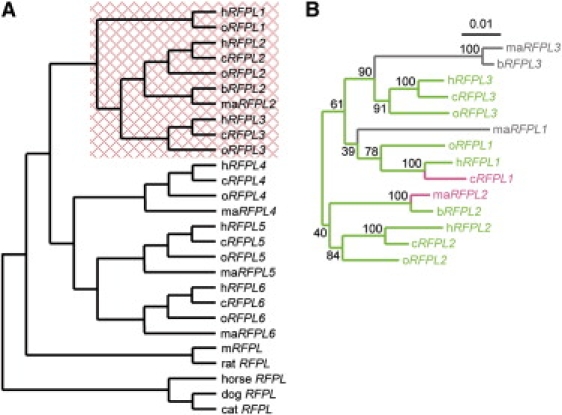
Topology of the RFPL Gene Family
(A) Phylogenetic tree based on RFPL coding sequences encoding the likely functional proteins in Laurasiatheria, Glires, and Catarrhini. The RFPL1,2,3 clade shows a significant (p < 0.0001) positive selection or constraint relaxation that is indicated by the red box.
(B) ML phylogenetic tree of the catarrhinian RFPL1,2,3 genes. The topology and branch lengths are based on concatenated alignments of the intron and the second exon. Branch lengths are scaled to the number of substitutions per site. Bootstrap support values are indicated at each node. Likely functional genes are shown in green, truncated genes are shown in pink, and nonfunctional genes are shown in gray. The following abbreviations are used in both panels: h, human; c, chimpanzee; o, orangutan; b, baboon; ma, macaque; and m, mouse.
The RFPL1,2,3 Genes Are Restricted to Catarrhini
In addition to those likely functional genes, we identified in the marmoset genome a single degenerated RFPL1,2,3-like sequence and nonfunctional RFPL4, RFPL5, and RFPL6 genes (Table S2), indicating that the duplication of the RFPL gene ancestor to generate the RFPL1,2,3 and RFPL4,5,6 gene progenitors originated at least ∼57 mya, but after the Euarchonta-Glires split. Further, whereas the intrachromosomal duplication of the RFPL4,5,6 gene progenitor was already present in marmoset, the lack of multiple RFPL1,2,3-like sequences in New World monkey suggested that the duplication of the RFPL1,2,3 gene ancestor happened only in the Catarrhini lineage (∼34 mya). This timing was supported by the calculation of pairwise Ks values among human, chimpanzee, and orangutan paralogs; such Ks values were at least two times lower than the average Ks divergence between human and marmoset genomes29 (Table S3).
Not All RFPL1,2,3 Genes Are Functional in Catarrhini
Analysis of the topology of the RFPL1,2,3 clade with the sequences of the intron and the second exon of each gene showed that this subfamily forms three distinct clusters corresponding to the RFPL1, RFPL2, and RFPL3 genes (Figure 3B).
However, only human and orangutan, among the genomes available, kept the complete open reading frame in the three RFPL1,2,3 genes. In contrast, some RFPL-predicted polypeptides showed truncations (macaque RFPL2 coding sequence has a frameshift leading to a stop codon; chimpanzee RFPL1 5′ region including exon 1 was inverted, which might lead to the synthesis of a truncated protein only partially homologous to the other RFPLs). Macaque RFPL1 and RFPL3 and baboon RFPL3 also lack an ORF after the loss of the first exon (Table S2).
According to those results, Figure 4 illustrates the presumptive timing of emergence and conservation of the RFPL genes over the course of evolution.
Figure 4.
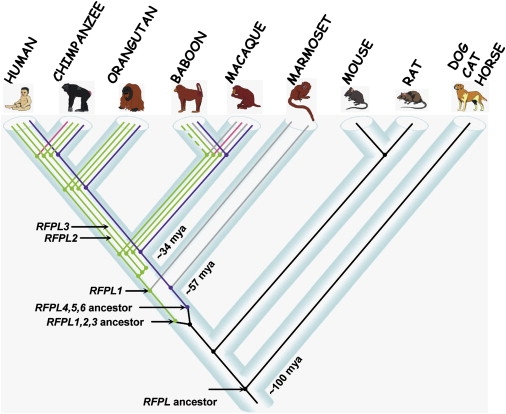
Timing of the Emergence of the RFPL Genes during Evolution
The RFPL gene ancestor is shown in black. Each likely functional RFPL1, RFPL2, and RFPL3 gene is indicated in green, truncated genes are indicated in pink, and genes with no ORF are indicated in gray. Putative baboon RFPL1 gene is indicated in a dashed line. The RFPL4,5,6 gene cluster is represented by a single purple line. The estimated time of the gene-ancestor emergence and subsequent duplication events are indicated in millions of years (mya).
Only the RFPL1,2,3 Gene Cluster Acquired Brain Expression during Evolution
We then examined by real-time PCR whether the RFPL4,5,6 gene cluster also acquired neural expression during evolution. Similarly to the absence of mRFPL expression in mouse neural tissue, we did not detect any hRFPL4,5,6 transcripts during human embryonic stem cell-derived neurogenesis or in human fetal and adult brains (Figure S4). Thus, RFPL expression in neural tissue seemed to be restricted to the catarrhinian-specific RFPL1,2,3 gene cluster.
Cortical RFPL1,2,3 Gene Expression Increases in Human in Comparison to Chimpanzee and Macaque
Several studies reported changes in gene expression in the brain of human and other nonhuman primates, and these findings seem to account for most phenotypic changes between those species.8,9 We therefore investigated whether RFPL1,2,3 genes had different cerebral expression levels among humans, chimpanzees, and macaques and determined RFPL1,2,3 gene-expression profiles in newborn and adult neocortex, cerebellum, and liver by using data from microarray transcriptome studies.
First, this approach showed that RFPL1,2,3 expressions in newborn and adult brains were also detected in chimpanzees and macaques, indicating that those genes can also exert a role in brain development and function in other Catarrhini. Second, intraspecies comparisons of RFPL1,2,3 expressions in different tissues pointed out a change directed toward brain expression of the RFPL1,2,3 genes through evolution. This was particularly marked in humans in which RFPL1,2,3 transcript levels significantly increased in newborn and adult neocortex and cerebellum in comparison to the liver, used as a nonneural, RFPL1,2,3-expressing control tissue (Figure 5A). Furthermore, we observed in humans that RFPL1,2,3 transcript levels in the neocortex were higher than in the cerebellum, both in newborn and adult tissues (Figure 5A). However, this divergence was not observed in chimpanzees and macaques, suggesting that the adaptation of RFPL1,2,3 toward cortical expression is specific to human (Figure 5A). This was confirmed by interspecies comparisons that showed that cortical RFPL1,2,3 transcript levels were significantly higher in humans than in chimpanzees or macaques, whereas only marginal interspecies differences were observed in the cerebellum (Figure 5B), and none was seen in the liver (data not shown).
Figure 5.
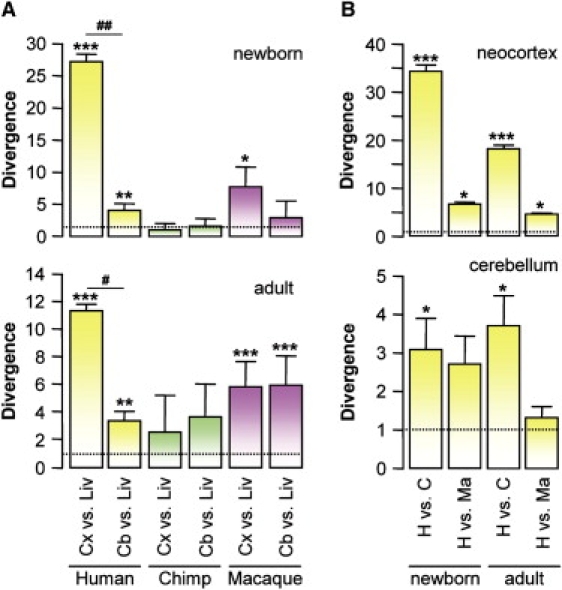
Expression Divergence between Human, Chimpanzee and Macaque RFPL1,2,3 Genes in Brain
RFPL1,2,3 gene expressions were assessed in newborn and adult neocortex, cerebellum, and liver by microarray transcriptome studies in humans, chimpanzees, and macaques. Results are displayed as mean ± standard error (SEM). The following abbreviations are used: Cx, neocortex; Cb, cerebellum; Liv, liver; H, human; C, chimpanzee; and Ma, macaque. (A) shows intraspecies divergence in RFPL1,2,3 tissue expression. Cortical and cerebellar RFPL1,2,3 transcript levels were normalized to that of the liver. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 for neocortex or cerebellum versus liver; #p < 0.05 and ##p < 0.01 for neocortex versus cerebellum with one-way ANOVA followed by post hoc Tukey test. (B) shows interspecies divergence in cortical and cerebellar RFPL1,2,3 expression levels. hRFPL1,2,3 expressions were normalized to those in chimpanzee or macaque. ∗p < 0.05 and ∗∗∗p < 0.001 with Student's t test.
Positive Selection Acted on the RFPL1,2,3 Genes
We next examined the evolutionary forces that acted on the RFPL1,2,3 coding sequences to determine the RFPL1,2,3 protein evolution. First, we analyzed the structure of the RFPL proteins in silico to examine the acquisition of neofunctionalization (Figure 6). All RFPL proteins shared a RING domain and a B30.2 domain composed of the PRY and SPRY motifs. In addition, we identified a protein motif (Pfam B-20538) bridging those RING and B30.2 domains. Using BLAST search, we found that this domain was present in all RFPLs but was exclusively restricted to those proteins. We referred this first sign of neofunctionalization to as RFPL-defining motif (RDM; Figure 6). We found a second neofunctionalization event with the appearance of an upstream translational initiation site only in the catarrhinian RFPL1,2,3 genes (except for chimpanzee and orangutan RFPL3). This would lead to a predicted synthesis of 29 additional amino acids on N-terminus (Figure 6). No putative protein domain could be defined from the different analyses we performed. Nonetheless, secondary structure predictions indicated that this sequence contained a hydrophobic region potentially forming an alpha helix loop that we termed RFPL1,2,3-specifying helix (RSH). No other protein that contained the complete 29 amino acid sequence or RSH could be identified.
Figure 6.
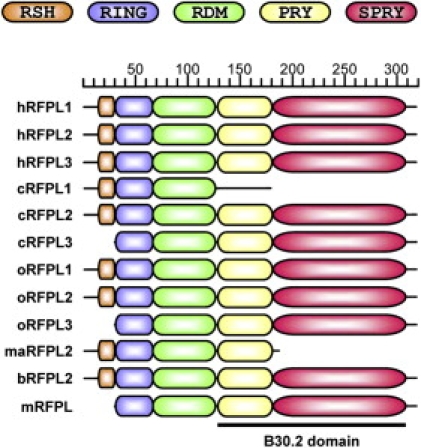
RFPL-Protein-Domain Predictions Reveal the Acquisition of RDM and RSH as Neofunctionalization Events
Identification of RFPL-specific RDM (RFPL-defining motif) and RSH (RFPL1,2,3-specifying helix) were obtained after the alignment of the likely functional catarrhinian RFPL1,2,3 and murine mRFPL proteins and in silico predictions of secondary structures and protein domains. The following abbreviations are used: h, human; c, chimpanzee; o, orangutan; b, baboon; ma, macaque; and m: mouse.
Second, we investigated whether the RFPL1,2,3 genes evolved under positive selection by using the previously described topology (Figure 3A). We tested the a priori hypothesis that positive selection occurred after the duplications that led to the generation of the brain-expressed RFPL1,2,3 cluster because selective pressure is often observed after duplication events to retain the functionality of the daughter genes.30 The ratio of nonsynonymous (KA) over synonymous (KS) substitutions per site in RFPL genes was 0.53 on average, indicating that the whole family was under strong purifying selection. However, we observed a significant increase of nonsynonymous substitutions in the RFPL1,2,3 cluster (KA/KS = 0.73), with both branch-site and branch models, suggesting an accelerated protein evolution driven by positive selection or a relaxation of constraints (Figure 3A).
Positive selection was highly variable across sites in RFPL1,2,3 genes. Estimates of KA/KS under models that allow for positive selection indicated that a fraction of sites (6.6% to 9.8%) evolved under positive selection (Table S4). The amino acid changes that occurred at sites inferred to have been under positive selection in RFPL1,2,3 proteins in Catarrhini were mainly located in the B30.2 domain (Table S5).
Discussion
Understanding the genetic basis of human brain evolution remains a major challenge in neuroscience. Here we identified the human RFPL1,2,3 genes (MIM 605968, 605969, and 605970) as Pax6 targets and showed that they are highly expressed at the onset of neurogenesis during neural differentiation of human embryonic stem cells. Expression of hRFPL1,2,3 was also detected in vivo in human developing brain, and the highest levels were found in the temporal and frontal cortical lobes, which have been previously described to be developmentally influenced by Pax6.19 Given the similarity in the structure of all RFPLs and the interaction of mRFPL with cyclin B1,27 hRFPL1,2,3 could therefore affect cell-cycle parameters and exert a role in Pax6-dependent generation of cortical cells. Further, the present genetic analysis shows that the hRFPL1,2,3 gene evolution exhibits the same features as those observed at the genome-wide level in the hominoid lineage, namely an expansion in gene copy number, an increase in cortical expression, and an acceleration in protein evolution.
First, we observed that the RFPL gene family emerged recently in evolutionary terms and expanded during evolution after duplication events. However, only the RFPL1,2,3 gene cluster acquired brain expression during this period. The RFPL1,2,3 progenitor gene emerged after the Euarchonta-Glires split. Subsequently, intrachromosomal segmental duplications occurred in Catarrhini to generate the RFPL1,2,3 gene cluster ∼34 mya. Yet, the maintenance of the functionality of the three RFPL1,2,3 genes was not ensured in Old World monkeys because of degeneration of some gene duplicates. Thus, an increase in RFPL1,2,3 gene copy number occurred in the hominoid lineage; this is in accordance with the increase in gene copy number previously observed at the genome-wide level in the hominoid lineage with interspecies cDNA or BAC array-based comparative genomic hybridization.7,31,32 Interestingly, those changes were particularly pronounced in genes with a role in brain development and function.7 The regions enriched in copy-number variations have been found to be not randomly distributed in the genome but are preferentially present in regions thought to be genomically and evolutionary dynamic. This is particularly the case for the 22q11-q13 region,7,31 which corresponds to the chromosomal location of the hRFPL1,2,3 gene cluster (22q12.2-q12.3).
Second, changes in transcript levels of the neocortex have been reported in several comparative studies between human and other primates.8,33–35 Those changes were predominantly directed toward an increase in mRNA levels in human,9 suggesting that constraints imposed by gene-dosage requirements are a major mechanism for the extensive modifications of brain development and physiology during primate evolution. Fitting with this concept, we observed an increase in brain RFPL1,2,3 transcript level in humans, both in newborn and adult tissues, in comparison to chimpanzees and macaques. Such a divergence was dramatic in the neocortex. However, the fact that the number of likely functional RFPL1,2,3 genes in Catarrhini (human = orangutan ≥ chimpanzee > macaque) is not correlated with their neocortex expression level (human > macaque > chimpanzee) or with neocortex evolution in size (human > chimpanzee ≈ orangutan > macaque) suggests that the increase in RFPL1,2,3 transcript level can not be fully explained by the increase in gene copy number in the hominoid lineage. Hence, mutations in the regulatory sequences of those genes during evolution could have led to distinct transcriptional regulation of RFPL1,2,3 expression in the developing neocortex of those different species. Remarkably, human is the only catarrhinian species among the three studied to have a RFPL1,2,3 expression significantly higher in the neocortex than in the cerebellum, suggesting that the RFPL1,2,3 gene expression was selectively directed toward the neocortex in the human lineage. In addition, the influence of posttranscriptional regulatory mechanisms could also be essential. Indeed, noncoding mRNAs of hRFPL1S (MIM 605972) and hRFPL3S (MIM 605971) antisense genes covering large portions of the sense hRFPL1 and hRFPL3 genes have been detected.36 In addition, the study of the human chromosome 22 revealed the presence of two hRFPL pseudogenes (hRFPLψ1 and hRFPLψ2).36 It has been shown that pseudogenes can still play important functional roles, notably by controlling, as antisense sequences, the stability of the messenger RNA of their homologous coding sequences.37 Therefore, the hRFPL pseudogene copies may not be necessarily functionally silent; such a finding could also be important for the evolutionary differences in RFPL1,2,3 expression profiles between Catarrhini.
Third, we found an acceleration in the evolution of the RFPL proteins. Neofunctionalization events, considered as signs of positive selection, are usually acquired to retain the functionality of the duplicate genes during evolution by providing them a new function.30 In this line, the protein domains found in RFPLs appeared at different times of evolution and as such corroborates our estimated period of emergence of the RFPL gene ancestor. Whereas SPRY appeared in eukaryotes, PRY is limited to vertebrates in which it associated with SPRY to form the B30.2 domain. The acquisition of the RING-type zinc-finger domain in B30.2-containing proteins appeared later and led to the generation of the TRIM protein family that is restricted to primates, rodents, and Xenopus.38 Here, we reported the emergence of RFPL-specific neofunctionalization events. One of these events was the acquisition of the RFPL-defining motif possibly after the gene-duplication event that led to the emergence of the RFPL gene ancestor. A second sign of neofunctionalization consisted of the appearance of an upstream transcriptional initiation site in catarrhinian RFPL1,2,3 genes. Moreover, analysis of positive selection indicated that the RFPL1,2,3 cluster showed an accelerated evolution. This was particularly notable after the duplication events that led to the emergence of the RFPL1 (KA/KS = 1.1) and RFPL2 (KA/KS = 1.0) genes. The observed positive selection is quantitatively moderate, and one could argue that it only reflects a relaxation of constraints. However, the different events we underlined above (gene duplication and retention, acquisition of brain expression, and emergence of new domains) are in favor of positive selection. We observed that the positive selection mainly affected the B30.2 domain, and this is accordance with the selective pressure observed in this domain in the human lineage of other TRIM proteins in order to provide specificity to their protein-protein interactions.39–42 Indeed, the amino acids lining the binding surface of the B30.2 domain are highly variable, and it was suggested that B30.2 is a protein-interacting module recognizing specific partners rather than a consensus sequence motif.39,41 Moreover, binding studies with mRFPL indicated that its protein partners were binding to the B30.2 domain.27 Therefore, the positive selection on the B30.2 domain in the RFPL1,2,3 cluster may have provided the acquisition of new partners and/or more selectivity toward those identified with mRFPL.
Only a few specific genes with an impact on brain evolution and/or development have been identified so far; these are the MCPH genes (determining brain size), FoxP2 (involved in speech production [MIM 605317]), GLUD2 (a brain-specific glutamate dehydrogenase important for glutamate detoxification after neuron firing [MIM 300144]) and HAR1 (coexpressed with reelin in Cajal-Retzius cells in human developing brain [MIM 610556]).43–46 The late emergence of the RFPL1,2,3 gene cluster in Catarrhini is similar only to that of GLUD2, which appeared between ∼18 and 23 mya after the hominoid-Old World monkey separation.45 However, all those genes have in common a period of accelerated protein evolution that coincides together, as well as with a period of increased structural and functional complexity at the cerebral level.11–13,45–47 The increase in RFPL1,2,3 gene expression in the human neocortex is also in accordance with the importance of gene-dosage constraints reported with FoxP2 in human.44 Thus, RFPL1,2,3 duplications to generate a brain-expressed cluster could be an important feature for human brain development and evolution. This could be corroborated by the recent finding that a 1:22 chromosomal translocation in the q12.1-q12.3 region, where the hRFPL1,2,3 gene cluster is located, has been observed in patients suffering from the Costello syndrome,48 a disease showing mental retardation and multiple brain atrophies49 (MIM 218040).
The impact of the RFPL1,2,3 genes on neocortex development remains to be determined. Given their presumptive role on cell-cycle regulation,27 one could establish parallels with other cell-cycle-progression mediators, such as p27 Kip1 or the MCPH genes, and hypothesize that RFPL1,2,3 genes may similarly control the balance between proliferative and neurogenic divisions and thereby the number of cortical cells generated. Hence, the RFPL1,2,3 genes could alter the patterning of specific neocortical areas like the cyclin E-regulating p27Kip1, whose differential expression in areas 17 and 18 of the neocortex affects neuron production that contributes to distinct areal cytoarchitectonics.50 Alternatively, the RFPL1,2,3 genes could affect brain morphology and size determination, like the MCPH1-6 genes. Indeed, those genes control the type of cell division (proliferative versus neurogenic) by regulating mitotic-spindle orientation or centrosome assembly,3,51 and their mutations were shown to be responsible for primary microcephaly, a disorder characterized by a reduction in neocortex size due to a decrease in neuron production.51
In conclusion, our observations suggest that the recent emergence and multiplication of the RFPL1,2,3 genes may be important for brain development and function in Catarrhini. Their role could be even more essential in humans because this species maintained the complete open reading frames in all three genes and shows a striking increase in cortical RFPL1,2,3 expressions in comparison to chimpanzees and macaques. We therefore propose that the RFPL1,2,3 genes may contribute to changes in cortical organization leading to the acquisition of human-specific behavioral traits.
Supplemental Data
Supplemental Data include four figures and six tables and can be found with this article online at http://www.ajhg.org/.
Supplemental Data
Microarray expression analysis was performed in GFP- and PAX6-expressing HeLa cells with the Affymetrix Human Genome U133 Plus 2.0 arrays. The results were analyzed with the GeneSpring GX 7 software.
Web Resources
The URLs for data presented herein are as follows:
“affy” R package, http://www.bioconductor.org/packages/release/Software.html
InterProScan server, http://www.ebi.ac.uk/Tools/InterProScan/
MatInspector server, http://www.genomatix.de
MyHits server, http://myhits.isb-sib.ch/cgi-bin/index
NCBI GenBank Database, http://www.ncbi.nlm.nih.gov
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/sites/entrez?db=omim
Pfam server, http://pfam.sanger.ac.uk/
University of California Santa Cruz Genome Browser and Database, http://www.genome.ucsc.edu/
Acknowledgments
The authors thank Philippe Rousseau for his advice with the chromatin-immunoprecipitation assay, the genomic core facility of the University of Geneva for their assistance with the microarray transcriptome analysis, and Dr. Karen Bedard for helpful discussions with the manuscript. J.B. is supported by the Auvergne Regional Council and the Geneva Department of Public Education. S.E.A. is supported by grants from the Swiss National Science Foundation, the European Union, and the National Institutes of Health. K.H.K. is supported by the Swiss National Science Foundation.
References
- 1.Kaas J.H. From mice to men: The evolution of the large, complex human brain. J. Biosci. 2005;30:155–165. doi: 10.1007/BF02703695. [DOI] [PubMed] [Google Scholar]
- 2.Kaas J.H. Evolution of the neocortex. Curr. Biol. 2006;16:R910–R914. doi: 10.1016/j.cub.2006.09.057. [DOI] [PubMed] [Google Scholar]
- 3.Gotz M., Huttner W.B. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 4.Kornack D.R., Rakic P. Changes in cell-cycle kinetics during the development and evolution of primate neocortex. Proc. Natl. Acad. Sci. USA. 1998;95:1242–1246. doi: 10.1073/pnas.95.3.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kornack D.R. Neurogenesis and the evolution of cortical diversity: Mode, tempo, and partitioning during development and persistence in adulthood. Brain Behav. Evol. 2000;55:336–344. doi: 10.1159/000006668. [DOI] [PubMed] [Google Scholar]
- 6.Dehay C., Kennedy H. Cell-cycle control and cortical development. Nat. Rev. Neurosci. 2007;8:438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- 7.Fortna A., Kim Y., MacLaren E., Marshall K., Hahn G., Meltesen L., Brenton M., Hink R., Burgers S., Hernandez-Boussard T. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2:E207. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caceres M., Lachuer J., Zapala M.A., Redmond J.C., Kudo L., Geschwind D.H., Lockhart D.J., Preuss T.M., Barlow C. Elevated gene expression levels distinguish human from non-human primate brains. Proc. Natl. Acad. Sci. USA. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Preuss T.M., Caceres M., Oldham M.C., Geschwind D.H. Human brain evolution: Insights from microarrays. Nat. Rev. Genet. 2004;5:850–860. doi: 10.1038/nrg1469. [DOI] [PubMed] [Google Scholar]
- 10.Dorus S., Vallender E.J., Evans P.D., Anderson J.R., Gilbert S.L., Mahowald M., Wyckoff G.J., Malcom C.M., Lahn B.T. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 11.Evans P.D., Anderson J.R., Vallender E.J., Choi S.S., Lahn B.T. Reconstructing the evolutionary history of microcephalin, a gene controlling human brain size. Hum. Mol. Genet. 2004;13:1139–1145. doi: 10.1093/hmg/ddh126. [DOI] [PubMed] [Google Scholar]
- 12.Evans P.D., Anderson J.R., Vallender E.J., Gilbert S.L., Malcom C.M., Dorus S., Lahn B.T. Adaptive evolution of ASPM, a major determinant of cerebral cortical size in humans. Hum. Mol. Genet. 2004;13:489–494. doi: 10.1093/hmg/ddh055. [DOI] [PubMed] [Google Scholar]
- 13.Evans P.D., Vallender E.J., Lahn B.T. Molecular evolution of the brain size regulator genes CDK5RAP2 and CENPJ. Gene. 2006;375:75–79. doi: 10.1016/j.gene.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 14.Heins N., Malatesta P., Cecconi F., Nakafuku M., Tucker K.L., Hack M.A., Chapouton P., Barde Y.A., Gotz M. Glial cells generate neurons: The role of the transcription factor Pax6. Nat. Neurosci. 2002;5:308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- 15.Quinn J.C., Molinek M., Martynoga B.S., Zaki P.A., Faedo A., Bulfone A., Hevner R.F., West J.D., Price D.J. Pax6 controls cerebral cortical cell number by regulating exit from the cell cycle and specifies cortical cell identity by a cell autonomous mechanism. Dev. Biol. 2007;302:50–65. doi: 10.1016/j.ydbio.2006.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gotz M., Stoykova A., Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron. 1998;21:1031–1044. doi: 10.1016/s0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 17.Guillemot F. Cellular and molecular control of neurogenesis in the mammalian telencephalon. Curr. Opin. Cell Biol. 2005;17:639–647. doi: 10.1016/j.ceb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Warren N., Caric D., Pratt T., Clausen J.A., Asavaritikrai P., Mason J.O., Hill R.E., Price D.J. The transcription factor, Pax6, is required for cell proliferation and differentiation in the developing cerebral cortex. Cereb. Cortex. 1999;9:627–635. doi: 10.1093/cercor/9.6.627. [DOI] [PubMed] [Google Scholar]
- 19.Ellison-Wright Z., Heyman I., Frampton I., Rubia K., Chitnis X., Ellison-Wright I., Williams S.C., Suckling J., Simmons A., Bullmore E. Heterozygous PAX6 mutation, adult brain structure and fronto-striato-thalamic function in a human family. Eur. J. Neurosci. 2004;19:1505–1512. doi: 10.1111/j.1460-9568.2004.03236.x. [DOI] [PubMed] [Google Scholar]
- 20.Bayatti N., Moss J.A., Sun L., Ambrose P., Ward J.F., Lindsay S., Clowry G.J. A molecular neuroanatomical study of the developing human neocortex from 8 to 17 postconceptional weeks revealing the early differentiation of the subplate and subventricular zone. Cereb. Cortex. 2007;18:1536–1548. doi: 10.1093/cercor/bhm184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y.H., Zheng J.B., Gu X., Saunders G.F., Yung W.K. Novel PAX6 binding sites in the human genome and the role of repetitive elements in the evolution of gene regulation. Genome Res. 2002;12:1716–1722. doi: 10.1101/gr.188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perrier A.L., Tabar V., Barberi T., Rubio M.E., Bruses J., Topf N., Harrison N.L., Studer L. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2004;101:12543–12548. doi: 10.1073/pnas.0404700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Clark T.A., Schweitzer A.C., Chen T.X., Staples M.K., Lu G., Wang H., Williams A., Blume J.E. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Margulies E.H., Cooper G.M., Asimenos G., Thomas D.J., Dewey C.N., Siepel A., Birney E., Keefe D., Schwartz A.S., Hou M. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17:760–774. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajkovic A., Lee J.H., Yan C., Matzuk M.M. The ret finger protein-like 4 gene, Rfpl4, encodes a putative E3 ubiquitin-protein ligase expressed in adult germ cells. Mech. Dev. 2002;112:173–177. doi: 10.1016/s0925-4773(01)00635-9. [DOI] [PubMed] [Google Scholar]
- 27.Suzumori N., Burns K.H., Yan W., Matzuk M.M. RFPL4 interacts with oocyte proteins of the ubiquitin-proteasome degradation pathway. Proc. Natl. Acad. Sci. USA. 2003;100:550–555. doi: 10.1073/pnas.0234474100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A.F., Boguski M.S., Brockway K.S., Byrnes E.J. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 29.Nikolaev S.I., Montoya-Burgos J.I., Popadin K., Parand L., Margulies E.H., Antonarakis S.E. Life-history traits drive the evolutionary rates of mammalian coding and noncoding genomic elements. Proc. Natl. Acad. Sci. USA. 2007;104:20443–20448. doi: 10.1073/pnas.0705658104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lynch M., Conery J.S. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 31.Goidts V., Armengol L., Schempp W., Conroy J., Nowak N., Muller S., Cooper D.N., Estivill X., Enard W., Szamalek J.M. Identification of large-scale human-specific copy number differences by inter-species array comparative genomic hybridization. Hum. Genet. 2006;119:185–198. doi: 10.1007/s00439-005-0130-9. [DOI] [PubMed] [Google Scholar]
- 32.Wilson G.M., Flibotte S., Missirlis P.I., Marra M.A., Jones S., Thornton K., Clark A.G., Holt R.A. Identification by full-coverage array CGH of human DNA copy number increases relative to chimpanzee and gorilla. Genome Res. 2006;16:173–181. doi: 10.1101/gr.4456006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enard W., Khaitovich P., Klose J., Zollner S., Heissig F., Giavalisco P., Nieselt-Struwe K., Muchmore E., Varki A., Ravid R. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- 34.Khaitovich P., Muetzel B., She X., Lachmann M., Hellmann I., Dietzsch J., Steigele S., Do H.H., Weiss G., Enard W. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khaitovich P., Enard W., Lachmann M., Paabo S. Evolution of primate gene expression. Nat. Rev. Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- 36.Seroussi E., Kedra D., Pan H.Q., Peyrard M., Schwartz C., Scambler P., Donnai D., Roe B.A., Dumanski J.P. Duplications on human chromosome 22 reveal a novel Ret Finger Protein-like gene family with sense and endogenous antisense transcripts. Genome Res. 1999;9:803–814. doi: 10.1101/gr.9.9.803. [DOI] [PubMed] [Google Scholar]
- 37.Hirotsune S., Yoshida N., Chen A., Garrett L., Sugiyama F., Takahashi S., Yagami K., Wynshaw-Boris A., Yoshiki A. An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature. 2003;423:91–96. doi: 10.1038/nature01535. [DOI] [PubMed] [Google Scholar]
- 38.Rhodes D.A., de Bono B., Trowsdale J. Relationship between SPRY and B30.2 protein domains. Evolution of a component of immune defence? Immunology. 2005;116:411–417. doi: 10.1111/j.1365-2567.2005.02248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chae J.J., Wood G., Masters S.L., Richard K., Park G., Smith B.J., Kastner D.L. The B30.2 domain of pyrin, the familial Mediterranean fever protein, interacts directly with caspase-1 to modulate IL-1beta production. Proc. Natl. Acad. Sci. USA. 2006;103:9982–9987. doi: 10.1073/pnas.0602081103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newman R.M., Hall L., Connole M., Chen G.L., Sato S., Yuste E., Diehl W., Hunter E., Kaur A., Miller G.M. Balancing selection and the evolution of functional polymorphism in Old World monkey TRIM5alpha. Proc. Natl. Acad. Sci. USA. 2006;103:19134–19139. doi: 10.1073/pnas.0605838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woo J.S., Imm J.H., Min C.K., Kim K.J., Cha S.S., Oh B.H. Structural and functional insights into the B30.2/SPRY domain. EMBO J. 2006;25:1353–1363. doi: 10.1038/sj.emboj.7600994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sawyer S.L., Wu L.I., Emerman M., Malik H.S. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang B.L. Molecular genetic determinants of human brain size. Biochem. Biophys. Res. Commun. 2006;345:911–916. doi: 10.1016/j.bbrc.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 44.Vargha-Khadem F., Gadian D.G., Copp A., Mishkin M. FOXP2 and the neuroanatomy of speech and language. Nat. Rev. Neurosci. 2005;6:131–138. doi: 10.1038/nrn1605. [DOI] [PubMed] [Google Scholar]
- 45.Burki F., Kaessmann H. Birth and adaptive evolution of a hominoid gene that supports high neurotransmitter flux. Nat. Genet. 2004;36:1061–1063. doi: 10.1038/ng1431. [DOI] [PubMed] [Google Scholar]
- 46.Pollard K.S., Salama S.R., Lambert N., Lambot M.A., Coppens S., Pedersen J.S., Katzman S., King B., Onodera C., Siepel A. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- 47.Krause J., Lalueza-Fox C., Orlando L., Enard W., Green R.E., Burbano H.A., Hublin J.J., Hanni C., Fortea J., de la Rasilla M. The derived FOXP2 variant of modern humans was shared with Neandertals. Curr. Biol. 2007;17:1908–1912. doi: 10.1016/j.cub.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 48.Bertelli M., Cecchin S., Fabbri A., Lapucci C., Gasparini P. Exclusion of chromosome region 22q12.1-q12.3 as a second locus for Costello syndrome. Panminerva Med. 2006;48:145–146. [PubMed] [Google Scholar]
- 49.Roberts A., Allanson J., Jadico S.K., Kavamura M.I., Noonan J., Opitz J.M., Young T., Neri G. The cardiofaciocutaneous syndrome. J. Med. Genet. 2006;43:833–842. doi: 10.1136/jmg.2006.042796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lukaszewicz A., Savatier P., Cortay V., Giroud P., Huissoud C., Berland M., Kennedy H., Dehay C. G1 phase regulation, area-specific cell cycle control, and cytoarchitectonics in the primate cortex. Neuron. 2005;47:353–364. doi: 10.1016/j.neuron.2005.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woods C.G., Bond J., Enard W. Autosomal recessive primary microcephaly (MCPH): A review of clinical, molecular, and evolutionary findings. Am. J. Hum. Genet. 2005;76:717–728. doi: 10.1086/429930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Microarray expression analysis was performed in GFP- and PAX6-expressing HeLa cells with the Affymetrix Human Genome U133 Plus 2.0 arrays. The results were analyzed with the GeneSpring GX 7 software.


