Abstract
The formation of antibodies to interferon-beta (IFN-β), a protein-based disease-modifying agent for multiple sclerosis (MS), is a problem in clinical practice. These antibodies may neutralize the biological effects of the protein drug, potentially decreasing its therapeutic effects. By high-resolution HLA class I and II typing we identified two HLA class II alleles associated with the development of antibodies to IFN-β. In two independent continuous and binary-trait association studies, HLA-DRB1∗0401 and HLA-DRB1∗0408 (odds ratio: 5.15)—but not other HLA alleles—were strongly associated with the development of binding and neutralizing antibodies to IFN-β. The associated HLA-DRB1∗04 alleles differ from nonassociated HLA-DRB1∗04 alleles by a glycine-to-valine substitution in position 86 of the epitope-binding alpha-helix of the HLA class II molecule. The peptide-binding motif of HLA-DRB1∗0401 and ∗0408 might promote binding and presentation of an immunogenic peptide, which may eventually break T cell tolerance and facilitate antibody development to IFN-β. In summary, we identified genetic factors determining the immunogenicity of IFN-β, a protein-based disease-modifying agent for the treatment of MS.
Introduction
Multiple sclerosis (MS [MIM 126200]) is a chronic inflammatory disease of the central nervous system. Whereas the etiology of MS is likely to be heterogeneous, the influence of genetic factors in the development of MS has been confirmed by association and linkage studies.1–4 The crucial role of HLA alleles in the susceptibility to MS is well established. Whereas HLA-DRB1∗15 alleles are strongly associated with MS in Europeans, DRB1∗0301 and DRB1∗0401 are overrepresented in MS patients from southern Europe.5,6 Overrepresentation of DRB1∗04 alleles was reported for patients from Turkey and the Canary Islands. Although the genetic association of susceptibility to MS with loci outside the HLA complex has been confirmed in some cases, the search for susceptibility genes continues.4
Interferon-β is the most widely used immunomodulatory drug for MS therapy. It is approved for treatment of clinically isolated syndrome and relapsing-remitting MS. Furthermore, it has antiviral activity and remains an option in the therapy of diseases such as Hepatitis-C (HCV [MIM 609532]) or the severe acute respiratory syndrome (SARS).7–9 Like other protein-based disease-modifying agents, IFN-β exhibits immunogenicity.10 Even though the technical improvement of purification processes and the introduction of recombinant preparations have helped to reduce the immunogenicity of IFN-β, up to 50% of patients may develop antibodies to IFN-β (binding antibodies [BABs]), of which a significant proportion neutralize the activity of IFN-β (neutralizing antibodies [NABs]) in vitro and vivo.11 Currently, three distinct recombinant preparations of IFN-β are in use for the treatment of MS. Interferon β-1b (165 residues) is modified from naturally occurring IFN-β by deletion of the leading methionine and a single amino acid substitution (C17S).12 It is recombinantly expressed in Escherichia coli (IFN-β-1b; Betaferon or Betaseron). IFN-β-1a, recombinantly expressed in Chinese hamster ovary cells, is identical to natural human interferon beta (166 residues) (IFN-β-1a; Avonex, Rebif). Although preparation and administration influence the immunogenic properties and the rate of antibody development, little is known about host factors that determine immune responses to IFN-β.13
Antibody production and generation of B cell responses to proteins depend on the help of antigen-specific CD4+ T cells.14 The development of antigen-specific CD4+ T cell responses itself is strongly influenced by the individual repertoire of HLA class II molecules, which present peptide antigens for recognition to the T cell receptor.15,16 Given the central role of CD4+ T cells for the development of antigen-specific B cell responses, we performed high-resolution typing for HLA-A, -B, -C, -DRB1, and -DQB1 alleles in MS patients receiving long-term IFN-β therapy.
Material and Methods
Patients
MS patients on IFN-β therapy, predominantly of northern European heritage, were recruited in Germany by primary-care physicians and neurologists. The initial study group included 268 individuals with known anti-IFN-β1 status. The ratio of males to females in the first cohort was 0.53. A total of 34 patients received IFN-β1a intramuscular injection (i.m.) once weekly (Avonex), 118 patients IFN-β1b subcutaneous injection (s.c.) every second day (Betaferon), 26 patients 22 μg and 90 patients 44 μg IFN-β1a s.c. three times a week (Rebif). The validation group compassed 242 patients with a male-female ratio of 0.36. Avonex treatment was administered to 68 patients, and Rebif was given to 88 individuals and 86 patients received Betaferon. As expected, IFN-β1a i.m. (Avonex)-treated patients were underrepresented in the study as a result of the lower frequency of antibody development in this group compared to patients treated with the formulations for s.c. application. Mean age was 39.8 yr in the first and 39.0 yr in the second group (Table 1). Average treatment duration was 57 months in the first and 52 months in the second group. The duration of the drug exposure did not differ significantly among treatment groups. All patients gave their written informed consent. The study was approved by the ethics committees of the Universities of Düsseldorf and Munich.
Table 1.
Clinical Characteristics of the Initial Study and Validation Groups
|
Initial Study Group (n = 268) |
Validation Group (n = 242) |
|||||||
|---|---|---|---|---|---|---|---|---|
|
Anti-IFN-β-Positive Patients |
Anti-IFN-β-Positive Patients |
|||||||
| Parameter | All AB+ (n = 136) | NAB+ (n = 85) | BAB+ (n = 51) | Anti-IFN-β-Negative Patients (n = 132) | All AB+ (n = 77) | NAB+ (n = 59) | BAB+ (n = 18) | Anti-IFN-β-Negative Patients (n = 132) |
| Age in years [mean (range)] | 40 (17–69) | 40 (19–64) | 41 (17–69) | 39 (16–66) | 39 (21–75) | 39 (21–60) | 39 (21–75) | 39 (16–67) |
| Gender (male/female) | 48/88 | 27/58 | 21/30 | 45/87 | 23/54 | 19/40 | 4/14 | 40/124 |
| Therapy duration in months [mean (range)] | 55 (16–147) | 53 (16–147) | 57 (17–112) | 59 (12–156) | 47 (7–127) | 47 (7–111) | 46 (20–127) | 55 (7–146) |
| IFN-β1a (i.m., Avonex)-treated (n) | 6 | 5 | 1 | 28 | 4 | 2 | 2 | 64 |
| IFN-β1b (s.c., Betaferon)-treated (n) | 70 | 37 | 33 | 48 | 36 | 24 | 12 | 50 |
| IFN-β1a (s.c., Rebif 22 μg)-treated (n) | 10 | 6 | 4 | 16 | 9 | 9 | 0 | 15 |
| IFN-β1a (s.c., Rebif 44 μg)-treated (n) | 50 | 37 | 13 | 40 | 28 | 24 | 4 | 36 |
| cELISA, antibody reactivity (%) [median (range)] | 74 (26–119) | 91 (35–119) | 48 (26–97) | 3 (−9–23.5) | 83 (35–140) | 85 (57–140) | 76 (35–96) | 2 (−17–24) |
| In vivo MXA assays, induction (%) [median (range)] | 23 (−64–135) | −4 (−64–50) | 89 (41–135) | 92.6 (−12–93) | 23 (−45–128) | 17 (−45–57) | 79 (53–128) | 95 (70–202) |
Both groups comprised a total of 510 IFN-β-treated subjects. Mean treatment duration exceeds 45 months in both cohorts. Ranges are given in brackets. Whereas the mean age of both groups was between 39 and 41 yr, the sex ratio differed among both groups. Females were slightly overrepresented in the validation group. Note that the number of BABs-positive subjects is higher in the IFN-β1b (Betaferon)-treated group.
HLA Genotyping
For the first study cohort, HLA genotyping for HLA-A, -B, -C, DRB1, and DQB1 was performed by hybridization of sequence-specific oligonucleotide probes immobilized on microspheres with amplified genomic DNA samples followed by flow fluorescence-intensity analysis according to the instructions of the manufacturer (One Lambda, Canoga Park, CA, USA). In the validation study cohort, the HLA analysis was restricted to HLA-DRB1∗04 only. The presence or absence of HLA-DRB1∗04 was determined by sequence-specific primer polymerase chain reaction (PCR-SSP) using a nested approach as previously described.17 In both study cohorts, the identification of HLA-DRB1∗04 alleles was accomplished by group-specific sequencing of exon 2 of HLA-DRB1∗04-positive samples according to the manufacturer's instruction (Protrans, Ketschau, Germany).
Assays for Binding and Neutralizing Antibodies
To detect antibodies against IFN-β, we performed a standardized capture ELISA as described.18 Defined positive and negative serum samples were included in each assay to obtain a standard curve for comparison of different assays. The standard curve ranged from 100% (highest positive control) to 0% (no antibodies). Serum positivity of all tested samples was normalized on the basis of these standard curves. Reactivity above 25% of the highest positive control was considered to be antibody positive (the cut-off value corresponded to the median of 50 control donors + five standard deviations). The biological in vivo activity of IFN-β was measured by MX1 (MxA) gene expression with TaqMan real-time PCR.19 Twelve hours after IFN-β injection, blood samples were drawn from subjects and mRNA was isolated according to the manufacturer's instructions. TaqMan reverse-transcription reagents (Applied Biosystems, Foster City, USA) were used to convert mRNA to cDNA. cDNA levels were detected with ABI's GeneAmp via the protocols described. The housekeeping gene GAPDH was used as internal control. Normalization was done according to the ΔΔCt-method. In each assay, the same positive and negative samples were included to ensure the quality of the assay. Antibodies were considered biologically active when the MX1 (MxA) induction was decreased by more than 50% compared to newly treated antibody-negative control donors.
In 203 patients from the first patient group, an MX1 RNA NAB assay was also performed to determine the in vitro neutralizing antibody activity. A human lung carcinoma cell line (A549, American Type Culture Collection, #CLL-185) was diluted in culture medium (2% BCS, 2 mM L-glutamine, 50 U/ml penicillin G, and 0.05 mg/ml streptomycin sulfate in alpha-MEM) at a concentration of 105 cells/ml, and each well of a microtiter plate was incubated overnight with 50 μl of this solution. Screening samples are diluted 1:10 and incubated with Betaferon 25 IU/ml for 45 min at 37°C and 5% CO2 on a microtiter plate. These samples are then transferred to the plate containing the A549 cells and incubated for 6.5 hr. For the standard curve, Betaferon with concentrations from 0 to10 IU was used. After that, the supernatant was decanted and cells were lysed with a lysis buffer.
A ΔCt value of a sample of less than 50% of the highest concentration on the standard curve was considered NAB negative. Otherwise, the sample was titrated. Titration samples were diluted serially beginning at 1:20 up to 1:10,240, and each dilution was incubated with Betaferon 25 IU/ml for 45 min at 37°C and 5% CO2 with an extra microtiter plate. These samples were then transferred to the plate containing the A549 cells and incubated for 6.5 hr. The supertnatant was decanted and cells were lysed with a lysis buffer followed by RNA extraction and transcription as described below. MX1 RNA was isolated semiautomatically with 6100 ABI PRISM Nucleic Prep Station. Lysates were filtered through a membrane to capture RNA. After several washing steps with washing buffers (by Applied Biosystems), RNA was eluted by addition of a purification elution solution into an extraction plate and stored at −80°C. After Master Mix was prepared (high-capacity cDNA Kit by Applied Biosystems) and applied to a new microtiter plate, the samples from the above RNA plate were thawed and transferred. RNA was transcribed into cDNA according to the manufacturer's instructions. Endogenous control (housekeeping gene) and target-detecting primers were mixed with the appropriate Master Mix (Applied Biosystems), and c-DNA was added. A predefined program (Applied Biosystems) for MX1 and house keeping gene (eukaryotic 18 s rRNA) detection was applied. A threshold is set in the exponential phase of rtPCR and the difference between the MX1 threshold cycles and the housekeeping gene cycles was determined (= ΔCt).
Computational Biology and Statistics
In order to attenuate the problem of deliberately setting up thresholds for ELISA and MX1 values to declare cases and controls, a binary-trait case-control association study was supported by continuous trait analysis for single HLA alleles and haplotypes. This bilateral approach was used for the initial study group as well as for the validation group. For identification of genetic variation of anti-IFN-β formation, the continuous normalized antibody reactivities were directly compared among carriers of different alleles. In the binary-trait case-control study, patients were considered antibody positive (cases) if the anti-IFN-β levels exceeded the 25% cutoff value. Otherwise, patients were considered antibody negative (controls). Additionally, antibody-positive patients were considered NABs positive if their MX1 mRNAs were reduced below 50% of MX1 levels of newly treated patients. The remaining antibody-positive patients who had not significantly reduced MX1 mRNA levels were considered BABs positive. For the testing of differences of location parameters for MX1 gene expression and anti-IFN-β levels, the two-sided nonparametric Wilcoxon test was applied. p values were adjusted for multiple testing with the false-discovery rate (FDR). For identification of haplotype-specific associations with continuous (MX1 and anti-IFN-β levels) and binomial trait types (NABS, BABs, and control subgroups), the genetic analysis package (gap) of the R Project for statistical computing was applied with score tests for association as described by 20,21. Haplotype-specific scores have been calculated with a score test for ambiguous haplotypes. The score statistics are given by
where denotes the measured trait, a mean trait value, and a measure of variation. denotes expectation value over the posterior distribution of the null hypothesis. The score statistics provide a measure for the covariation of a haplotype and the examined trait.21 However, this analysis has explorative character, because the results are based on the maximization of probabilities rather than a known linkage phase.
Multiple sequence alignments were generated with the CLUSTALW application. The sequences of HLA alleles were obtained from the HLA repository at the European Bioinformatics Institute (EBI). Linkage disequilibrium (LD) tests for HLA alleles as well as multilocus analysis for LD were done with the PyPop package.
Results
HLA Class II but Not HLA Class I Alleles Are Associated with Antibody Occurrence
Two hundred and sixty-eight MS patients on long-term IFN-β therapy were included in the first group of the study. One hundred and thirty-six patients of the study group had developed antibodies to IFN-β (determined by cELISA), of whom 85 developed neutralizing antibodies as determined by the in vivo induction of the IFN-β response gene MX1. One hundred and thirty-two patients were antibody negative (Table 1). In the second group, 76 patients were antibody positive, including 59 with neutralizing in vivo activity. The remaining 164 individuals were antibody negative. For analysis of possible confounding effects, treatment-independent allele distribution was tested. The null hypothesis of independency holds in the study (chi-square = 55.34, p value = 0.65) as well as in the validation group (chi-square = 8.333, p value = 0.97).
All patients from the initial study group were typed for HLA class I and II alleles. Although we expected an association of HLA class II genes with antibody development, we also typed class I genes as internal negative control. As expected, only small differences between antibody-negative and -positive patients were found for HLA class I alleles. Median anti-IFN-β levels were lower in HLA-C∗16 carriers (p value = 0.007, FDR = 0.095) and higher in HLA-C∗2 and HLA-B∗57 (p value = 0.03, FDR ≤ 0.1) carriers, although the associations did not reach significance after adjustment for multiple testing.
By contrast, significant differences were observed for the HLA class II alleles. The largest difference of median anti-IFN-β levels was observed between DRB1∗04-positive (n = 58) and DRB1∗04-negative subjects (n = 210) (p = 0.006). DRB1∗04-positive patients showed significantly higher anti-IFN-β levels with a median normalized antibody titer of 55.1% compared to DRB1∗04-negative patients (median = 17.1%) (Table 2). Significantly reduced median anti-IFN-β reactivities were also found for DRB1∗1101 (p value = 0.013) and DRB1∗1104 (p value = 0.02). However, null hypothesis was kept after the FDR procedure. Likewise, elevated anti-IFN-β reactivities in DRB1∗1601-positive patients (p value = 0.02) were not significant after correction.
Table 2.
Anti-IFN-β Levels and Low-Resolution HLA Class II Typing in the Study Group
|
Allele Carriersa |
Anti-IFN Reactivity (%)b |
Statistics |
|||
|---|---|---|---|---|---|
| Specificity | n | Median | Range | pc | FDRd |
| DR 1 | 31 | 14.6 | −0.7–100.5 | 0.2 | 0.43 |
| DR 3 | 48 | 17.9 | −3.2–110.8 | 0.34 | 0.55 |
| DR 4 | 58 | 55.9 | −3.2–119.3 | 0.004 | 0.025 |
| DR 7 | 50 | 18.2 | −9.2–119.3 | 0.41 | 0.59 |
| DR 8 | 16 | 37 | 0.2–103.5 | 0.87 | 0.87 |
| DR 10 | 5 | 6.7 | 1.8–43.4 | 0.14 | 0.43 |
| DR 11 | 40 | 6.3 | −4.5–112.1 | 0.003 | 0.02 |
| DR 12 | 7 | 46.6 | −4.7–78.0 | 0.69 | 0.75 |
| DR 13 | 53 | 17.5 | −9.22–110.8 | 0.2 | 0.43 |
| DR 14 | 4 | 41.8 | 9.92–91.2 | 0.55 | 0.65 |
| DR 15 | 155 | 28.3 | −4.6–112.1 | 0.26 | 0.48 |
| DR 16 | 21 | 90.8 | 0.0–103.0 | 0.006 | 0.03 |
| DQ 2 | 90 | 17.7 | −9.2–110.8 | 0.1 | 0.5 |
| DQ 3 | 121 | 33.7 | −4.7–119.3 | 0.99 | 0.99 |
| DQ 4 | 15 | 54.4 | 0.3–103.5 | 0.46 | 0.74 |
| DQ 5 | 60 | 18.3 | −9.2–102.9 | 0.59 | 0.74 |
| DQ 6 | 180 | 28.3 | −4.6–112.1 | 0.28 | 0.7 |
The anti-IFN-β levels are compared among different HLA class II alleles. Median anti-IFN-β levels are significantly elevated in patients carrying the DR4 and DR16 alleles (in bold). Subjects carrying the DR11 serotype show a significant decrease of anti-IFN-β, indicating a protective effect of DR11 alleles (in bold). Significant results for HLA-DQ serotypes were not observed.
For all alleles listed, note that heterozygous allele carriers occur twice, and homozygotic allele carriers occur only once. Only alleles that occurred more than twice are shown.
Anti-IFN reactivity was determined as described in the Material and Methods. Values above 25% identified patients with anti-IFN antibodies.
p value was determined by comparison of the anti-IFN level of allele carriers to that of all nonallele carriers.
p values have been adjusted to multiple testing by using the false-discovery rate method (FDR). Adjusted p values below 0.05 were considered to be statistically significant (in bold).
HLA-DRB1∗04 Alleles Confer a Higher Risk of Developing Antibodies during IFN-β Therapy
A majority of DRB1∗04-positive patients included in the initial study group carried the DRB1∗0401 allele (n = 42), whereas only 12 subjects were DRB1∗0404-positive. Only four patients carried DRB1∗0408 (Table 3). Differences of location parameters for DRB1∗0401 (FDR = 0.012) and DRB1∗0408 (FDR = 0.048) were significant after correction for multiple testing. On the other hand, average cELISA levels for DRB1∗0404 were lower compared to DRB1∗04-negative patients. After FDR correction, null hypothesis was kept for DRB1∗0404 despite a significant single test (p value = 0.031, FDR = 0.16).
Table 3.
Anti-IFN-β Levels for Alleles of the HLA-DRB1 Locus in the Study and Validation Groups
|
Allele Carriersa,b |
Anti-IFN Reactivity (%)c |
Statistics |
|||
|---|---|---|---|---|---|
| DRB1 | n | Median | Range | pd | FDRe |
| Inital Study Group | |||||
| 0101 | 27 | 10.8 | −0.83–100.5 | 0.12 | 0.45 |
| 0301 | 48 | 17.8 | −3.16–110.8 | 0.34 | 0.66 |
| 0401 | 42 | 66.6 | −3.18–119.3 | 4e-04 | 0.012 |
| 0404 | 12 | 6.6 | −1.75–96.54 | 0.031 | 0.16 |
| 0408 | 4 | 98.5 | 90.5–109.8 | 0.003 | 0.048 |
| 0701 | 50 | 18.2 | −9.2–119.3 | 0.41 | 0.66 |
| 0801 | 15 | 39.4 | 0.2–109.5 | 0.74 | 0.8 |
| 1001 | 5 | 6.7 | 1.79–43.4 | 0.14 | 0.47 |
| 1101 | 32 | 27.0 | −4.6–112.1 | 0.013 | 0.13 |
| 1104 | 8 | 3.1 | −0.6–48.62 | 0.02 | 0.14 |
| 1201 | 7 | 46.6 | −4.8–78.0 | 0.69 | 0.8 |
| 1301 | 25 | 38.1 | −2.1–102.9 | 0.74 | 0.8 |
| 1302 | 18 | 15.8 | −9.2–110.8 | 0.35 | 0.66 |
| 1303 | 12 | 11.9 | −1.61–81.7 | 0.24 | 0.6 |
| 1501 | 153 | 30.5 | −4.6–112.1 | 0.43 | 0.66 |
| 1502 | 3 | 87.3 | 3.4–103.2 | 0.31 | 0.66 |
| 1601 | 17 | 90.8 | 0.1–102.9 | 0.02 | 0.13 |
| 1602 | 3 | 77.6 | 3.9–95.29 | 0.44 | 0.66 |
| Validation Group | |||||
| DR4− | 183 | 5.5 | −17.3–140.3 | 0.12 | 0.21 |
| DR4+ | 59 | 10.1 | −8.3–123.1 | 0.12 | 0.21 |
| 0401 | 31 | 71.8 | −5.4–120.4 | 0.005 | 0.03 |
| 0402 | 6 | 0.3 | −7.0–23.17 | 0.01 | 0.21 |
| 0403 | 6 | 3.4 | −8.3–89.6 | 0.37 | 0.43 |
| 0404 | 9 | 1.9 | −5.9–84.1 | 0.22 | 0.31 |
| 0407 | 2 | 47.8 | 2.8–92.8 | 0.51 | 0.51 |
| 0408 | 5 | 91.6 | −3.7–123.1 | 0.04 | 0.15 |
A significant elevation of IFN-β reactivity is seen for DRB1∗0401 and DRB1∗0408 (in bold) after adjustment for multiple testing. Only alleles that occurred more than twice are shown. In contrast to the findings in the low-resolution analysis, a significant increase of anti-IFN-β levels in DRB1∗1601-positive subjects vanishes after the FDR procedure. Likewise, the potential protective effect of DRB1∗0404 does not pass the significance criterion after adjustment for multiple testing. In the validation group, a significant elevation of IFN-β reactivity is seen in single tests for DRB∗0401 (in bold) as well as for DRB1∗0408. Because of an insufficient number of cases, however, significance vanishes after correction for multiple tests for DRB1∗0408. Because of the small number of cases, a potential protective effect of DRB1∗0402 did not pass the FDR significance criterion. The results in the validation group support our previous findings.
For all alleles listed, note that heterozygous allele carriers occur twice, and homozygotic allele carriers occur only once. Only alleles that occurred more than twice are shown.
Heterozygous allele carriers (∗0401+ and ∗0408+) occur twice.
Anti-IFN reactivity was determined as described in the Material and Methods. Values above 25% identified patients with anti-IFN antibodies.
p value was determined by comparison of the anti-IFN level of allele carriers to that of all nonallele carriers.
p values have been adjusted to multiple testing by using the false-discovery rate method (FDR). Adjusted p values below 0.05 were considered to be statistically significant (in bold).
To further investigate the role of DR4 alleles for antibody development, we determined the activity of IFN-β antibodies by another assay, which was used in a number of clinical studies. The in vitro MX1 test was performed on 203 randomly selected samples. As expected, a significant increase (p value = 0.003) of neutralizing in vitro activity was found in sera of HLA-DRB1∗0401- and ∗0408-positive subjects (median activity = 8462 NU) compared to sera from HLA-DRB1∗0401- and ∗0408-negative patients (median activity = 0) (data not shown).
For validation of our initial findings, we used a second study cohort and analyzed the most significant results for DRB1∗04. HLA genotyping of the validation group supported the previous findings. Fifty-nine of 242 individuals were DRB1∗04 positive. DRB1∗0401 carriers showed significantly higher levels of anti-IFN-β antibodies. Median cELISA positivity was 71.8% (p value = 0.005, FDR = 0.03) (Table 3). Five subjects carried the DRB1∗0408 allele, with a median level of 91.6%. Whereas the elevation of ELISA reactivity was significant in a single test, null hypothesis was kept after correction for multiple testing (p value = 0.04, FDR = 0.15).
In a combined analysis of anti-IFN-β levels for DR4-positive and DR4-negative carriers of validation and study groups, a highly significant elevation of anti-IFN-β levels was found in carriers of DR4 alleles (median reactivity = 55.1%, p value ≤ 0.01) (Figure 1). Fifty-four of 74 HLA-DRB1∗0401 carriers showed antibodies during IFN-β treatment. In DRB1∗0401- and ∗0408-negative subjects, 153 of 429 showed seroconversion, resulting in an odds ratio of 4.9 for HLA-DRB1∗0401-positive carriers. The odds ratio for DRB1∗0408 was 14.4, with four of five allele carriers being antibody positive. A highly significant increase of ELISA reactivities was observed in a comparison of all HLA-DRB1∗0401- and 0408-positive carriers versus HLA-DRB1∗0401- and ∗0408-negative carriers (Figure 2) (p value ≤ 0.01). Furthermore, the difference of anti-IFN-β levels among DRB1∗0401 (median reactivity = 70.6%) and DRB1∗0408 (median reactivity = 96.4%) is significant (p value = 0.006). Because of the small number of homozygous DRB1∗04 carriers, a comparison with DRB1∗04-negative or heterozygous carriers was not possible.
Figure 1.
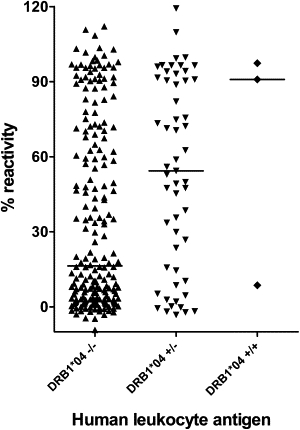
Comparison of Anti-IFN-β Reactivities for HLA-DR Serological Equivalents in the Study Group
Median ELISA optical density (OD) of heterozygous DR4 carriers was significantly elevated compared to DR4-negative individuals (p value ≤ 0.01).
Figure 2.
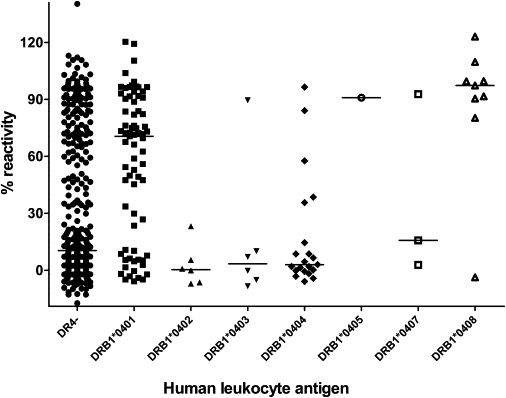
Comparison of Anti-IFN-β Reactivities for HLA-DRB1 Alleles in the Study and Validation Groups
A significant increase of ELISA reactivities was measured for HLA-DRB1∗0401 and HLA-DRB1∗0408 alleles (p value ≤ 0.01). DRB1∗0402, DRB1∗0403, and DRB1∗0404 seem to have a protective effect.
The haplotype analysis was performed with score statistics for ambiguous haplotypes. DRB∗11 alleles might have a protective influence: DRB1-DQB1 haplotype analysis shows a significant reduction of anti-IFN-β levels in DRB1∗1101-DQB1∗0301 and DRB∗1104-DQB1∗0301 carriers. Elevated MX1 mRNA levels in carriers of these haplotypes support this observation. A significant increase of anti-IFN-β reactivity could be observed for DRB1∗0401-DQB1∗0301 and DRB1∗0401-DQB1∗0302 haplotypes. The reduction of MX1 mRNA levels, however, was not significant (p value ≥ 0.5). Reduced MX1 levels as well as increased antibody reactivities were significantly changed in patients carrying the DRB1∗0408-DQB1∗0301 haplotype. Susceptibility to seroconversion was increased for DRB1∗1601-DQB1∗0502 carriers. The DRB1∗1601-DQB1∗0502-positive carriers also showed significantly decreased MX1 levels (Table 4).
Table 4.
Haplotype Analysis for the HLA Class II Locus
|
Haplotype |
MxA mRNA Levels (% Positivity) |
Anti-IFN Reactivity (%) |
||||
|---|---|---|---|---|---|---|
| DRB1 | DQB1 | Freq | Score | p | Score | p |
| 0101 | 0501 | 0.05 | 1.3 | 0.2 | −1.93 | 0.05 |
| 0301 | 0201 | 0.10 | 0.2 | 0.8 | −0.68 | 0.5 |
| 0401 | 0301 | 0.02 | −0.7 | 0.5 | 2.05 | 0.04 |
| 0401 | 0302 | 0.06 | −1.4 | 0.2 | 3.35 | 8e-04 |
| 0404 | 0302 | 0.02 | 0.9 | 0.4 | −2.13 | 0.03 |
| 0408 | 0301 | 0.01 | −2.6 | 0.01 | 2.76 | 0.006 |
| 0701 | 0201 | 0.09 | 0.2 | 0.8 | −0.7 | 0.5 |
| 0701 | 0303 | 0.01 | −0.2 | 0.8 | 0.63 | 0.5 |
| 0801 | 0402 | 0.03 | 0.8 | 0.4 | 0.25 | 0.8 |
| 1001 | 0501 | 0.01 | 1.1 | 0.3 | −1.61 | 0.1 |
| 1101 | 0301 | 0.06 | 2.8 | 0.004 | −2.16 | 0.03 |
| 1104 | 0301 | 0.02 | 2.1 | 0.04 | −2.23 | 0.03 |
| 1201 | 0301 | 0.01 | 0.1 | 0.9 | 0.49 | 0.6 |
| 1301 | 0603 | 0.04 | −0.7 | 0.5 | 0.15 | 0.9 |
| 1302 | 0604 | 0.02 | −0.6 | 0.5 | −0.44 | 0.7 |
| 1302 | 0609 | 0.01 | −2.1 | 0.03 | 1.68 | 0.09 |
| 1303 | 0301 | 0.02 | 1.4 | 0.2 | −1.58 | 0.1 |
| 1501 | 0602 | 0.30 | −0.5 | 0.7 | 0.54 | 0.6 |
| 1501 | 0603 | 0.02 | −0.5 | 0.6 | 0.13 | 0.9 |
| 1501 | 06XX | 0.01 | 0.4 | 0.7 | −1.14 | 0.3 |
| 1502 | 0601 | 0.01 | −0.9 | 0.4 | 1.16 | 0.2 |
| 1601 | 0502 | 0.03 | −2.7 | 0.006 | 2.77 | 0.006 |
| 1602 | 0502 | 0.01 | −1.1 | 0.3 | 0.9 | 0.4 |
A significant increase of anti-IFN-β activities can be observed for haplotypes containing HLA-DRB1∗0401, DRB1∗0408 and DRB1∗1601 alleles, HLA-DRB1∗11 and DRB1∗0404 alleles have a protective effect on seroconversion (in bold). Additionally, for DRB1∗1601 and DRB1∗0408 a significant reduction of MX1 levels is seen. Interestingly, even though not significant in single allele tests, the haplotype DRB1∗1601-DQB1∗0502 shows a significant reduction of MX1 levels and elevation of anti-IFN-β levels. Scores reflect the strength and the direction of the effects. The % reactivity of each sample is related to the highest measured OD of positive controls (MS patients with high titers of anti-IFN-β).
As expected pairwise linkage disequilibrium was significant for all observed major histocompatibility complex (MHC) loci (p value ≤ 0.001). Strongest disequilibria were found for HLA-DRB1:DQB1 (D' = 0.94), HLA-B:C (D' = 0.92), and HLA-B:DRB1 (D' = 0.68). Haplotype frequencies for HLA class I alleles with significant increases of anti-IFN-β reactivities in single test (Table S1 available online) do not indicate a common transmission with DRB1∗0401 and DRB1∗0408 (data not shown).
HLA-DRB1∗04 Promotes the Development of Binding and Neutralizing Antibodies Independent of the IFN-β Formulation
Whereas DRB1∗0401 and DRB1∗0408 are significantly associated with the presence of antibodies, HLA-DRB1∗0401 and ∗0408 frequencies were not different between patients who developed NABs or BABs (p value > 0.05) (Table 5), suggesting that the antibody development itself but not their neutralizing capacity is influenced by the HLA alleles. The rejection of the alternative hypothesis might, however, be caused by an insufficient power of the test.
Table 5.
Odds Ratios for HLADR∗0401- and HLADR∗0408-Positive Allele Carriers
|
Initial and Validation Study Group: Allele Carriers |
||||
|---|---|---|---|---|
| Treatment | AB Status | 0408+ and 0401+ | 0408− and 0401− | Odds Ratio (p Value) |
| All Products | ||||
| NABs | 37 | 107 | ||
| BABs |
23 |
46 |
0.7 (> 0.05) |
|
| ABs | 60 | 153 | ||
| no ABs | 21 | 276 | 5.2 (< 0.0001) | |
| IFN-β1a (i.m., Avonex) | ||||
| ABs | 3 | 7 | ||
| no ABs | 11 | 81 | 3.2 (> 0.1) | |
| IFN-β1b (s.c., Betaferon) | ||||
| ABs | 36 | 70 | ||
| no ABs | 5 | 93 | 9.6 (< 0.0001) | |
| IFN-β1a (s.c., Rebif 22 μg) | ||||
| ABs | 4 | 15 | ||
| no ABs | 1 | 30 | 8 (0.06) | |
| IFN-β1a (s.c., Rebif 44 μg) | ||||
| ABs | 17 | 61 | ||
| no ABs | 4 | 72 | 5 (< 0.005) | |
The difference of NABs and BABs cases among DRB1∗0401- and DRB1∗0408-positive patients was not significant in chi-square tests for the study (p = 0.19) as well as the validation group (p = 0.50). Odds ratios for the development of antibodies are increased irrespective of the type of the IFN-β preparation. This observation is true for the study as well as for the validation group. Overall, the relative risk (RR) for DRB1∗0401- and DRB1∗0408-positive carriers was 1.85 in the study and 2.43 in the validation group. Because of missing observations (no antibody-negative patients with HLA-DRB1∗0401 or HLA-DRB1∗0408) in the validation group, the exact odds ratio could not be calculated for IFN-β1a (s.c., Rebif 22 μg).
The odds ratios in the study and validation group for the development of antibodies to IFN-β were 5.15 for HLA-DRB1∗0401-positive patients and 9.6 for HLA-DRB1∗0408-positive patients. Odds ratios were increased for all IFN-β preparations, although they did not reach significance in the IFN-βa i.m.-treated and IFN-βa 22 μg s.c.-treated patients as a result of the low numbers of patients in these treatment groups (odds ratios were 3.2 and 8, respectively). The strongest effect was observed in patients who received IFN-β1b s.c. treatment (Table 5).
HLA Alleles Associated with Antibody Development Share a Similar Peptide-Binding Motif
To further investigate the molecular basis for HLA class II-restricted antibody development, we compared the sequences of the antibody-associated DRB1∗0401 and DRB1∗0408 to the DRB1∗0402, DRB1∗0403, and DRB1∗0404 alleles that are not associated with antibody development. DRB1∗0401 and DRB1∗0408 differ by one amino acid (71 K→R). DRB1∗0401 and DRB1∗0408 differ from DRB1∗0404, DRB1∗0402, and DRB1∗0403 at position 86, which forms the first pocket of the peptide-binding groove located at the terminal end of the same alpha-helix (Figure 3). Both alleles associated with antibody development share a glycine at position 86, whereas the nonassociated alleles have a valine at this position (86 G→V).
Figure 3.

Multiple Sequence Alignment of DR4 Alleles
HLA-DRB1∗0401 and DRB1∗0408 only differ in position 71 (site A). In contrast to protecting DR4 alleles, both sequences encode for a glycine in position 86 (site B). The residue is part of the C-terminal end of the epitope-binding alpha helix of the MHCII complex. The substitution of a large with a small hydrophobic amino acid at this position potentially increases the immunogenicity of IFN-β.
Discussion
As of December, 2006, 65 new biopharmaceuticals had been approved in the U.S., European Union, and Japan since 1999. Like IFN-β, 47 of these new biopharmaceuticals are derived from recombinant DNA technology.22 Applications range from the treatment of more prevalent rheumatic diseases (Ethanercept) to rare acquired or inherited disease with protein deficiencies (Galsulfase). It is predicted that many more therapeutic proteins will become available in the near future for an even wider range of indications.10 Despite their therapeutic potential, immunogenicity is a common problem to most, if not all, protein-based biopharmaceuticals and an important aspect of their safety. Antibodies might not only lead to the loss of drug efficacy but may also induce severe allergic reactions or neutralize the activity of endogenously produced protein.23 It has been argued that in the absence of resilient predictors of immunogenicity, patients might be put at a high risk.24 Preparation, handling, packaging, dose, frequency, and route of administration and treatment duration seem to affect immunogenicity of biopharmaceuticals.25–27 However, the role of genetic factors in immunogenicity to protein-based therapeutics has not been addressed. In the present study we show that—besides the influence of different preparations on immunogenicity—genetic factors play an important role in the development of antibodies against protein therapeutics. Patients who carry the HLA-DRB1∗0401 or HLA-DRB1∗0408 allele show a 5- and 14-fold increased risk to develop antibodies to IFN-β, respectively. This results in an average relative risk of 2.07 for DRB∗0401- and DRB∗0408-positive patients. Although the HLA-DRB1∗0401 or HLA-DRB1∗0408 alleles increase the risk of developing antibodies in response to all IFN-β formulations, it remains to be determined, in further studies and with a different study design, whether formulations with higher immunogenicity are more affected by the HLA haplotype than those with low immunogenicity.
These results are in line with the central role of T cells in eliciting B cell responses to protein targets: It seems that the peptide-binding motif of both alleles allows processing and binding of an IFN-β peptide that breaks tolerance by promoting a strong T helper cell response to the IFN-βprotein. This breakdown of tolerance promotes a B cell response that eventually leads to the development of an antibody production against the IFN-β protein. It is striking that this effect critically depends on a single amino acid in the HLA-DRB1∗04 allele sequence. The 86 G→V substitution in HLA-DRB1∗04 alleles is likely to interfere with elevated immunogenicity either by altering the binding affinity of the IFN-β peptide because of secondary structure changes of the epitope-binding alpha helix or by affecting the presentation of the IFN-β peptide itself. It seems that these two alleles influence the general likelihood of developing an antibody response independent of the IFN-β preparation. Although antibody development was strongly affected by the alleles, the neutralizing activity of the antibodies was not significantly influenced by these HLA alleles. This suggests that both HLA-DRB1∗04 alleles increase the likelihood of breaking tolerance to the protein by antibody production. Other factors, however, might determine whether the antibodies will neutralize IFN-β. One of those factors could be the intensity of an immune response to IFN-β. However, the rejection of the alternative hypothesis that the development of neutralizing antibodies is determined by HLA-DRB1∗04 alleles might be due to insufficient test power and remains to be clarified in further studies.
Interestingly, HLA-DRB1∗0401 has been associated with the development of autoantibodies against insulin. Congia et al. identified an immunodominant epitope in DRB1∗0401 transgenic mice that would have been proteolytically destroyed under normal conditions. Additionally, this epitope was processed and presented by human DRB1∗0401-positive Epstein-Barr virus-transformed B cells.28 First reports on the influence of HLA-DR4 serotypes in the development of porcine insulin antibodies date back to the early 1980s.29 The smaller glycine might endorse the binding of epitopes that contain larger amino acids in the vicinity of the substitution site or promote binding by a slight modification of the secondary structure. Although HLA-DRB1∗0101 is similar to HLA-DRB1∗0401 (both carry a glycine at position 86) and has analogous disease-modifying properties in MS, only HLA-DRB1∗0401 influences antibody development to IFN-β.30,31 Although not significant in single-allele tests, the explorative haplotype analysis based on a score test for unkown linkage phase reveals a significant reduction of MX1 production in DRB1∗0408-DQB1∗0301 carriers. Additionally, haplotype analysis showed a significant increase of anti-IFN-β reactivity and reduced MX1 mRNA expression in DRB1∗1601-DQB1∗0502 carriers, pointing to alleles other than DRB1∗0401 and DRB1∗0408 that might be associated with the development of binding and neutralizing antibodies. This is especially true for HLA-DRB1∗16 alleles that also potentially increase the risk to develop antibodies against IFN-β. On the other hand, DRB1∗1101 alleles seem to have a protective effect with respect to antibody production. It has been recently shown that trans interactions of HLA-DRB1 alleles play a significant role in the susceptibility to MS.32,33 Such trans interactions might also play a role in the immunogenicity of biotherapeutics. Hence, this study suggests that further genetic studies should be undertaken to clarify the influence of hereditary factors in immunogenicity of biotherapeutics and improve treatment decisions.
Supplemental Data
Supplemental Data include one table and can be found with this article online at http://www.ajhg.org/.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
IMGT/HLA database, http://www.ebi.ac.uk/imgt/hla
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
Acknowledgments
This study was supported by the European Union (NABinMS), Deutsche Forschungsgemeinschaft (He2386/7-1), and BiogenIdec.
References
- 1.Sawcer S., Goodfellow P.N., Compston A. The genetic analysis of multiple sclerosis. Trends Genet. 1997;13:234–239. doi: 10.1016/S0168-9525(97)01140-2. [DOI] [PubMed] [Google Scholar]
- 2.Oksenberg J.R., Baranzini S.E., Barcellos L.F., Hauser S.L. Multiple sclerosis: Genomic rewards. J. Neuroimmunol. 2001;113:171–184. doi: 10.1016/s0165-5728(00)00444-6. [DOI] [PubMed] [Google Scholar]
- 3.Dyment D.A., Ebers G.C., Sadovnick A.D. Genetics of multiple sclerosis. Lancet Neurol. 2004;3:104–110. doi: 10.1016/s1474-4422(03)00663-x. [DOI] [PubMed] [Google Scholar]
- 4.Hafler D.A., Compston A., Sawcer S., Lander E.S., Daly M.J., De Jager P.L., de Bakker P.I., Gabriel S.B., Mirel D.B., Ivinson A.J. Risk alleles for multiple sclerosis identified by a genomewide study. N. Engl. J. Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 5.Olerup O., Hillert J. HLA class II-associated genetic susceptibility in multiple sclerosis: A critical evaluation. Tissue Antigens. 1991;38:1–15. doi: 10.1111/j.1399-0039.1991.tb02029.x. [DOI] [PubMed] [Google Scholar]
- 6.Fogdell-Hahn A., Ligers A., Gronning M., Hillert J., Olerup O. Multiple sclerosis: A modifying influence of HLA class I genes in an HLA class II associated autoimmune disease. Tissue Antigens. 2000;55:140–148. doi: 10.1034/j.1399-0039.2000.550205.x. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi Y., Watanabe S., Konishi M., Yokoi M., Kakehashi R., Kaito M., Kondo M., Hayashi Y., Jomori T., Suzuki S. Quantitation and typing of serum hepatitis C virus RNA in patients with chronic hepatitis C treated with interferon-beta. Hepatology. 1993;18:1319–1325. [PubMed] [Google Scholar]
- 8.Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Treatment of SARS with human interferons. Lancet. 2003;362:293–294. doi: 10.1016/S0140-6736(03)13973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sainz B., Mossel E.C., Peters C.J., Garry R.F. Interferon-beta and interferon-gamma synergistically inhibit the replication of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) Virology. 2004;329:11–17. doi: 10.1016/j.virol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schellekens H. Immunogenicity of therapeutic proteins: clinical implications and future prospects. Clin. Ther. 2002;24:1720–1740. doi: 10.1016/s0149-2918(02)80075-3. [DOI] [PubMed] [Google Scholar]
- 11.Sorensen P.S., Ross C., Clemmesen K.M., Bendtzen K., Frederiksen J.L., Jensen K., Kristensen O., Petersen T., Rasmussen S., Ravnborg M. Clinical importance of neutralising antibodies against interferon beta in patients with relapsing-remitting multiple sclerosis. Lancet. 2003;362:1184–1191. doi: 10.1016/S0140-6736(03)14541-2. [DOI] [PubMed] [Google Scholar]
- 12.Bertolotto A., Deisenhammer F., Gallo P., Solberg S.P. Immunogenicity of interferon beta: differences among products. J. Neurol. 2004;251(Suppl 2):II15–II24. doi: 10.1007/s00415-004-1204-7. [DOI] [PubMed] [Google Scholar]
- 13.Hemmer B., Stuve O., Kieseier B., Schellekens H., Hartung H.P. Immune response to immunotherapy: the role of neutralising antibodies to interferon beta in the treatment of multiple sclerosis. Lancet Neurol. 2005;4:403–412. doi: 10.1016/S1474-4422(05)70117-4. [DOI] [PubMed] [Google Scholar]
- 14.Vinuesa C.G., Tangye S.G., Moser B., Mackay C.R. Follicular B helper T cells in antibody responses and autoimmunity. Nat. Rev. Immunol. 2005;5:853–865. doi: 10.1038/nri1714. [DOI] [PubMed] [Google Scholar]
- 15.Reveille J.D. The genetic basis of autoantibody production. Autoimmun. Rev. 2006;5:389–398. doi: 10.1016/j.autrev.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 16.Sundberg E.J., Deng L., Mariuzza R.A. TCR recognition of peptide/MHC class II complexes and superantigens. Semin. Immunol. 2007;19:262–271. doi: 10.1016/j.smim.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bein G., Glaser R., Kirchner H. Rapid HLA-DRB1 genotyping by nested PCR amplification. Tissue Antigens. 1992;39:68–73. doi: 10.1111/j.1399-0039.1992.tb01909.x. [DOI] [PubMed] [Google Scholar]
- 18.Pachner A.R. An improved ELISA for screening for neutralizing anti-IFN-beta antibodies in MS patients. Neurology. 2003;61:1444–1446. doi: 10.1212/01.wnl.0000094198.37489.11. [DOI] [PubMed] [Google Scholar]
- 19.Pachner A., Narayan K., Price N., Hurd M., Dail D. MxA gene expression analysis as an interferon-beta bioactivity measurement in patients with multiple sclerosis and the identification of antibody-mediated decreased bioactivity. Mol. Diagn. 2003;7:17–25. doi: 10.1007/BF03260016. [DOI] [PubMed] [Google Scholar]
- 20.Schaid D.J., Rowland C.M., Tines D.E., Jacobson R.M., Poland G.A. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum. Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J.H., Tan Q. Integrated analysis of genetic data with R. Hum. Genomics. 2006;2:258–265. doi: 10.1186/1479-7364-2-4-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsuji K., Tsutani K. Approval of new biopharmaceuticals 1999–2006: Comparison of the US, EU and Japan situations. Eur. J. Pharm. Biopharm. 2008;68:496–502. doi: 10.1016/j.ejpb.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 23.Sharma B. Immunogenicity of therapeutic proteins. Part 3: Impact of manufacturing changes. Biotechnol. Adv. 2007;25:325–331. doi: 10.1016/j.biotechadv.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 24.Schellekens H. When biotech proteins go off-patent. Trends Biotechnol. 2004;22:406–410. doi: 10.1016/j.tibtech.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Cleland J.L., Powell M.F., Shire S.J. The development of stable protein formulations: A close look at protein aggregation, deamidation, and oxidation. Crit. Rev. Ther. Drug Carrier Syst. 1993;10:307–377. [PubMed] [Google Scholar]
- 26.Schellekens H. Bioequivalence and the immunogenicity of biopharmaceuticals. Nat. Rev. Drug Discov. 2002;1:457–462. doi: 10.1038/nrd818. [DOI] [PubMed] [Google Scholar]
- 27.Schellekens H. Factors influencing the immunogenicity of therapeutic proteins. Nephrol. Dial. Transplant. 2005;20(Suppl 6):vi3–vi9. doi: 10.1093/ndt/gfh1092. [DOI] [PubMed] [Google Scholar]
- 28.Congia M., Patel S., Cope A.P., De V.S., Sonderstrup G. T cell epitopes of insulin defined in HLA-DR4 transgenic mice are derived from preproinsulin and proinsulin. Proc. Natl. Acad. Sci. USA. 1998;95:3833–3838. doi: 10.1073/pnas.95.7.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludvigsson J. Insulin antibodies in diabetic children treated with monocomponent porcine insulin from the onset: relationship to B-cell function and partial remission. Diabetologia. 1984;26:138–141. doi: 10.1007/BF00281121. [DOI] [PubMed] [Google Scholar]
- 30.Jones E.Y., Fugger L., Strominger J.L., Siebold C. MHC class II proteins and disease: a structural perspective. Nat. Rev. Immunol. 2006;6:271–282. doi: 10.1038/nri1805. [DOI] [PubMed] [Google Scholar]
- 31.DeLuca G.C., Ramagopalan S.V., Herrera B.M., Dyment D.A., Lincoln M.R., Montpetit A., Pugliatti M., Barnardo M.C., Risch N.J., Sadovnick A.D. An extremes of outcome strategy provides evidence that multiple sclerosis severity is determined by alleles at the HLA-DRB1 locus. Proc. Natl. Acad. Sci. USA. 2007;104:20896–20901. doi: 10.1073/pnas.0707731105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dyment D.A., Herrera B.M., Cader M.Z., Willer C.J., Lincoln M.R., Sadovnick A.D., Risch N., Ebers G.C. Complex interactions among MHC haplotypes in multiple sclerosis: Susceptibility and resistance. Hum. Mol. Genet. 2005;14:2019–2026. doi: 10.1093/hmg/ddi206. [DOI] [PubMed] [Google Scholar]
- 33.Ramagopalan S.V., Morris A.P., Dyment D.A., Herrera B.M., DeLuca G.C., Lincoln M.R., Orton S.M., Chao M.J., Sadovnick A.D., Ebers G.C. The inheritance of resistance alleles in multiple sclerosis. PLoS Genet. 2007;3:1607–1613. doi: 10.1371/journal.pgen.0030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


