Abstract
Kinetochores are complex macromolecular structures that link mitotic chromosomes to spindle microtubules. Although a small number of kinetochore components have been identified, including the kinesins CENP-E and XKCM1 as well as cytoplasmic dynein, neither how these and other proteins are organized to produce a kinetochore nor their exact functions within this structure are understood. For this reason, we have developed an assay that allows kinetochore components to assemble onto discrete foci on in vitro-condensed chromosomes. The source of the kinetochore components is a clarified cell extract from Xenopus eggs that can be fractionated or immunodepleted of individual proteins. Kinetochore assembly in these clarified extracts requires preincubating the substrate sperm nuclei in an extract under low ATP conditions. Immunodepletion of XKCM1 from the extracts prevents the localization of kinetochore-associated XKCM1 without affecting the targeting of CENP-E and cytoplasmic dynein or the binding of monomeric tubulin to the kinetochore. Extension of this assay for the analysis of other components should help to dissect the protein–protein interactions involved in kinetochore assembly and function.
Keywords: in vitro, mitosis, CENP-E, XKCMI, dynein
A key event during mitosis is the segregation of replicated chromosomes to daughter cells. During chromosome segregation, kinetochores play a central role by mediating chromosome–microtubule interactions. In mammalian cells, the kinetochore is a complex trilaminar structure assembled on the centromeric region of mitotic chromosomes (1–3). Molecules that may interact with microtubules are thought to localize to the fibrous corona and the outer plate—the two outermost regions of the kinetochore (3, 4).
Biochemical analysis of kinetochores has been limited by difficulties in purifying kinetochores and by their small size; kinetochore components constitute only a minor fraction of chromosomal proteins. Despite these drawbacks, a number of kinetochore components have been identified in mammals. These components fall into two classes: constitutive components, present at kinetochore regions throughout the cell cycle, and mitosis-specific components, targeted to the kinetochore region specifically during its mitotic maturation (1, 2).
The three known constitutive components, CENP-A, CENP-B, and CENP-C were identified in studies using autoimmune sera directed against human centromeres (5–7). Several mitosis-specific kinetochore components have also been identified. Included in this second group are three microtubule motor proteins: cytoplasmic dynein, CENP-E, and XKCM1. Cytoplasmic dynein, a minus end-directed microtubule motor, localizes to the corona which is the outer-most region of the kinetochore (4). CENP-E, a kinesin identified as a salt-resistant chromosomal scaffold component of mammalian tissue culture cell chromosomes, localizes to the kinetochore region of mitotic chromosomes (8). XKCM1 is a kinesin isolated from Xenopus that localizes to the centromeric region of mitotic chromosomes by light microscopy, but has yet to be localized at high resolution using immunoelectron microscopy. It has 55% overall amino acid identity with MCAK, a kinesin identified earlier in mammalian (CHO) cells that is similarly localized (9, 10).
The development of methods to isolate mitotic chromosomes from mammalian tissue culture cells has led to the in vitro study of kinetochore structure and function. Kinetochores can bind monomeric tubulin, nucleate microtubule assembly, capture and move microtubules, and remain attached to depolymerizing microtubules (refs. 11–15; reviewed in ref. 16). An alternative approach to the study of kinetochore structure and function has been the use of antibody microinjection into tissue culture cells (17, 18).
To extend the work on mammalian kinetochores to a biochemical level, it will be necessary to correlate specific molecules with specific functions. So far, little progress has been made in this direction. The major barriers to such progress are the difficulty in manipulating kinetochores on chromosomes purified from tissue culture cells and in interpreting the results of microinjection studies. Development of an in vitro system for kinetochore assembly would complement existing approaches by allowing the assembly of large numbers of kinetochores under reproducible conditions. In addition, the contribution of individual components could be tested by immunodepletion before assembly. Xenopus egg extracts provide an in vitro system for studying mitotic processes in which a pseudo-genetic approach is available using antibodies to inhibit or remove specific components (9).
To develop an in vitro kinetochore assembly assay, we first needed to establish markers for kinetochores in Xenopus. To this end, we demonstrate using antisera to three mitotic kinetochore markers—cytoplasmic dynein, CENP-E, and XKCM1—that Xenopus and mammalian kinetochores recruit similar mitosis-specific components. Using these proteins as markers, we have developed an in vitro reaction for kinetochore assembly on chromosomes condensed in clarified extracts of Xenopus eggs. Ideally, we would like to analyze kinetochore assembly in crude cycled extracts (extracts in which the sperm DNA has been replicated prior to assembling mitotic chromatin), so that the segregation competence of the kinetochores could be directly assayed following assembly. However, to date, it has been impossible to immunodeplete components from crude extracts while maintaining their ability to segregate chromosomes. In addition to ease of immunodepletion, clarified extracts have other advantages over crude extracts. Membranes and cytoskeletal elements do not assemble in clarified extracts facilitating identification of chromosomal components (18, 19), and clarified extracts are also more reproducible, more stable while frozen, and offer the potential for direct biochemical fractionation. Because of these technical advantages we have concentrated our efforts on clarified extracts.
MATERIALS AND METHODS
Tissue Culture Cell Chromosome Isolation and Immunofluorescence.
Tissue culture cell chromosomes were purified from CHO cells and Xenopus A6 cells as described (4) with minor modifications for A6 cells.
For immunofluorescence, appropriate volumes of CHO and A6 chromosomes were diluted to 10 μl with PME buffer (10 mM K-Pipes, pH 7.2/5 mM MgCl2/1 mM EGTA) on ice and fixed by addition of 200 μl of PME plus 1% formaldehyde at room temperature for 10 min. Fixed chromosomes were layered onto a 5 ml cushion of 30% (vol/vol) glycerol in PME and pelleted onto polylysine-coated coverslips at 10,000 rpm for 15′ at 4°C in an HB-4 rotor. Pelleted chromosomes were postfixed in −20°C methanol for 3 min before processing for immunofluorescence.
In Vitro Kinetochore Assembly.
Clarified cytostatic factor (CSF)—arrested extracts and demembranated sperm nuclei—were prepared as described (20, 21) with minor modifications. Crude CSF extracts were prepared by crushing dejellied eggs exactly as described (20). The crude extract was supplemented with protease inhibitors, 10 μg/ml cytochalasin D, and 1× energy regeneration mix (added from a 20× stock; 1× = 7.5 mM creatine phosphate/1 mM ATP/1 mM MgCl2; see ref. 20 for details) then clarified at 50,000 rpm in a TLS55 rotor at 4°C for 2 h. The clarified extracts were recovered using a 20 gauge needle, respun at 55,000 rpm for 30 min in a TLS55 rotor at 4°C, frozen in liquid nitrogen in 50–100 μl aliquots, and stored at −80°C. Extracts prepared and stored in this manner retain the ability to reproducibly assemble kinetochores for at least 6 months.
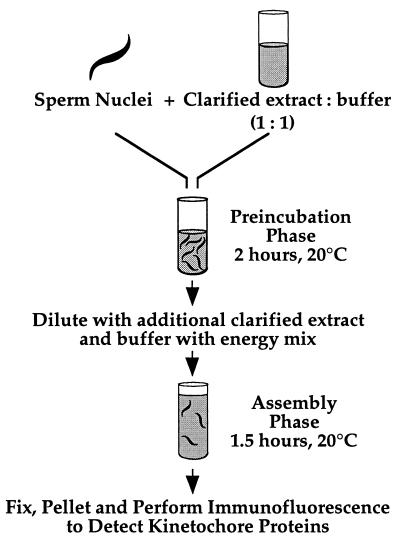
Kinetochore assembly was performed as diagrammed in Fig. 2. A total of 4 μl of 3 × 107 sperm nuclei/ml was mixed on ice with 8 μl of clarified extract and 8 μl of XBE5 (10 mM K-Hepes, pH 7.7/50 mM sucrose/100 mM KCl/0.1 mM CaCl2/5 mM EGTA/5 mM MgCl2) before incubation at 20°C for 2 h (preincubation). On ice, 5 μl of the preincubation reaction were mixed with 25 μl of XBE5 plus 2× energy mix (1× = 7.5 mM creatine phosphate/1 mM ATP/1 mM MgCl2/50 μg/ml creatine kinase) and 20 μl of clarified extract containing 10 μg/ml nocodazole, then incubated at 20°C for 1.5–2 h (assembly).
Figure 2.
Schematic of the two-step kinetochore assembly reaction in clarified Xenopus egg extracts (see text for details).
Immunofluorescence Analysis of in Vitro Kinetochore Assembly.
A total of 15 μl of the assembly reaction was fixed by pipeting in 300 μl of XBE2 (10 mM K-Hepes, pH 7.7/50 mM sucrose/100 mM KCl/0.1 mM CaCl2/5 mM EGTA/2 mM MgCl2) plus 1% formaldehyde and immediately mixing well with a cutoff pipet tip. After 12 min at room temperature the fixed reaction was pelleted onto polylysine-coated coverslips through an ice-cold 5 ml cushion of 30% (vol/vol) glycerol in XBE2. Coverslips were postfixed in −20°C methanol for 3 min and processed for immunofluorescence. Antibodies were used in the following order: 70.1 anti-dynein intermediate chain monoclonal ascites fluid, Texas Red goat anti-mouse IgM (Jackson ImmunoResearch), 3 μg/ml affinity-purified rabbit anti-human CENP-E (HX antibody raised to rod domain of CENP-E (22); gift of B. Schaar and T. Yen), or 0.5 μg/ml affinity-purified rabbit anti-XKCM1 (9), and fluorescein isothiocyanate donkey anti-rabbit (Jackson ImmunoResearch). Often, the dynein signal was amplified by use of a Texas Red-labeled tertiary antibody as the penultimate antibody (Texas Red donkey anti-goat, Jackson ImmunoResearch). Coverslips were rinsed with TBST containing 1 μg/ml Hoechst 33342 and mounted in 0.5% p-phenylenediamine (free base; Sigma) in 20 mM Tris⋅HCl, pH 9/90% glycerol.
Although both CENP-E and dynein were always detectable using a straight XBE2 plus 1% formaldehyde fixation, we observed some variation between extracts in our ability to detect XKCM1 using this fixation. We could eliminate this variability by diluting the reactions in 10 volumes of PME plus 0.2 M sucrose plus 0.1% Triton X-100 at 0°C for 10 min and then fixing by adding 10 volumes of PME plus 1% formaldehyde for 12 min at room temperature.
Assaying Tubulin Binding by Kinetochores.
To purify condensed sperm nuclei, 0.1 ml of 66% (wt/wt) sucrose in PME at the bottom of a 0.5 ml Eppendorf tube was overlaid with a 0.4 ml cushion of 30% (wt/vol) sucrose in PME. A total of 50–100 μl of the assembly reaction were layered on top of the cushion before spinning in an HB-4 rotor at 10,000 rpm for 15 min at 4°C. The cushion was washed several times while aspirating, and the interface between the cushion and the heavy sucrose pad was collected in the starting reaction volume. In case of salt extraction, the assembly reactions were diluted prior to purification with an equal volume of XBE5 plus 0.5× energy mix containing either 0 mM or 200 mM extra NaCl and incubated at room temperature for 10 min.
Tubulin binding assays were performed as described (14) with some modifications. Purified condensed sperm nuclei were diluted at least 10-fold into 50 mM K-Pipes, pH 7.2/1 mM EGTA/5 mM MgCl2/1 mM GTP with or without 10 μM bovine brain tubulin and incubated at 37°C for 10 min. The reaction mix was diluted with an equal volume of ice-cold PME plus 0.1% Triton X-100 and incubated on ice for 5 min to depolymerize any microtubules before being sedimented unfixed onto polylysine coated coverslips, fixed in PME plus 0.1% Triton X-100 plus 2% formaldehyde at 0°C for 10 min, postfixed in −20°C methanol for 3 min and processed for double label immunofluorescence for tubulin and CENP-E as described above. DM1α antibody was used to detect α-tubulin.
Transmission and Immunoelectron Microscopy.
For immunoelectron microscopy, fixed assembly reactions were sedimented onto Aclar (Ted Pella, Irvine, CA) coverslips as above, omitting methanol postfixation. Coverslips were blocked in EMBlock [PBS (137 mM NaCl/5.4 mM Na2HPO4/1.8 mM KH2PO4/2.7 mM KCl, pH 7.2) plus 0.1% Tween-20 plus 1.5% (wt/vol) BSA plus 0.5% (wt/vol) teleost fish gelatin (Sigma)] for 30 min, incubated in 3 μg/ml anti-CENP-E antibody diluted in EMBlock for 45 min, rinsed well in EMBlock, and then extensively in PBS plus 0.1% Tween (PBSTw), incubated for 45 min in 1/30 dilution in EMBlock of 6 nm colloidal gold-conjugated goat anti-rabbit secondary (Jackson ImmunoResearch), rinsed well in EMBlock, and extensively in PBSTw, rinsed with PBS, and fixed in 1% glutaraldehyde in 0.1 M Na-cacodylate (pH 7.4) for 30 min at room temperature. Coverslips were then rinsed in cacodylate buffer, osmicated in 1% reduced osmium in 1.6% potassium ferricyanide in 0.1 M Na-cacodylate (pH 7.4) for 15 min on ice, en bloc stained overnight at 4°C with 1% uranyl acetate, dehydrated, and embedded in Epon Araldite resin. Sections were cut on a Reichert Ultracut S ultramicrotome and observed with a Philips EM400 electron microscope.
For transmission electron microscopy XL177 Xenopus tissue culture cells were plated on polylysine-coated Aclar coverslips, simultaneously permeabilized, and fixed in EMB (80 mM K-Pipes, pH 6.9/3 mM MgCl2/5 mM EGTA/0.5% Triton X-100) plus 0.1% glutaraldehyde for 5 min at room temperature, and then fixed in EMB plus 1% glutaraldehyde for 60 min at room temperature. Subsequent processing steps were performed as described above.
RESULTS AND DISCUSSION
Reagents for Following Kinetochore Assembly in Xenopus Egg Extracts.
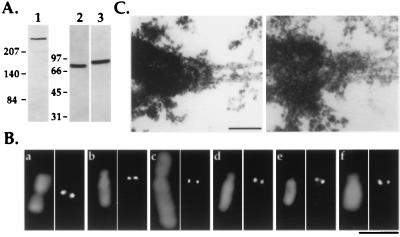
To develop an in vitro assay for kinetochore assembly using Xenopus egg extracts, we needed reagents directed against Xenopus kinetochore components. We analyzed four different human autoimmune sera (CREST sera; ref. 23) that recognize constitutive centromeric components in mammals, but failed to observe any localization of these sera to centromeric regions in Xenopus A6 tissue culture cells or on purified A6 chromosomes (not shown). We then tried antibodies to three different mitosis-specific kinetochore components—CENP-E, cytoplasmic dynein (intermediate chain of 78 kDa), and XKCM1—of which only XKCM1 is a Xenopus-specific protein. Antibodies to each of these three proteins recognize single bands of the expected sizes on immunoblots of Xenopus egg extracts (Fig. 1A) and localize identically to the kinetochore regions of both isolated A6 and CHO cell chromosomes (Fig. 1B), suggesting that Xenopus and mammalian kinetochores recruit similar mitosis-specific motor proteins. In addition, electron microscopy revealed that chromosomes in mitotic Xenopus tissue culture cells possess the characteristic trilaminar plate structure described for mammalian kinetochores (Fig. 1C).
Figure 1.
Reagents for following kinetochore assembly in Xenopus egg extracts. (A) Immunoblots of Xenopus extracts using 1 μg/ml affinity-purified anti-human CENP-E (lane 1), anti-chicken cytoplasmic dynein intermediate chain mAb 70.1 ascites fluid (lane 2), and 0.1 μg/ml affinity-purified anti-XKCM1 (lane 3) antibodies. Extracts were fractionated by 6% SDS/PAGE for CENP-E immunoblots and by 10% SDS/PAGE for dynein and XKCM1 immunoblots. Molecular mass markers (in kDa) are indicated to the left of each set of panels. (B) Comparative immunofluorescence of CHO (a, c, and e) and A6 (b, d, and f) cell chromosomes using antibodies to CENP-E (a and b), cytoplasmic dynein intermediate chain (c and d), and XKCM1 (e and f). Antibody localizations are shown in the right panel of each pair and DNA in the left panel. (Bar = 5 μm.) (C) Analysis of mitotic XL177 cells by electron microscopy demonstrating the presence of a trilaminar plate kinetochore structure at the point of interaction of kinetochore microtubules with the chromosome. Two separate examples are shown. (Bar = 0.2 μm.)
Kinetochore Assembly in Vitro Using Clarified Xenopus Egg Extracts.
To develop an assay for kinetochore assembly in vitro, we monitored the targeting of cytoplasmic dynein, CENP-E, and XKCM1 to discrete points on chromosomes condensed in clarified cytostatic factor-arrested Xenopus egg extracts. Using demembranated Xenopus sperm nuclei as substrates, we began our studies using the conditions described by Hirano and Mitchison (21), which were optimized for mitotic chromosome condensation. Under these conditions, we failed to detect targeting of any of the three kinetochore markers to discrete regions on the condensed chromosomes.
We found that modification of the chromosome assembly assay into a two-step reaction (diagrammed in Fig. 2) leads to efficient targeting of all three kinetochore markers to discrete dots on the condensed chromosomes. In the first step in this reaction, the “preincubation” phase, sperm nuclei are held for 2 h at 20°C in a 2-fold diluted clarified extract without addition of an energy regeneration system. At the end of this phase, the sperm nuclei are present as tightly clustered thin fibers of condensed chromatin, demonstrating maintenance of mitotic state. However, none of the kinetochore markers are detectable by immunofluorescence on the DNA at the end of the preincubation (data not shown). The second step of the reaction, the “assembly” phase, mimics the chromosome condensation reaction of Hirano and Mitchison (21). To initiate this phase of the reaction, additional clarified extract and buffer containing energy mix are added to the preincubation reaction. The assembly reaction is then incubated at 20°C for 1.5 h. Nocodazole is added in the second step of the reaction, but the results are essentially unchanged without it (there is no spindle assembly in these clarified extracts).
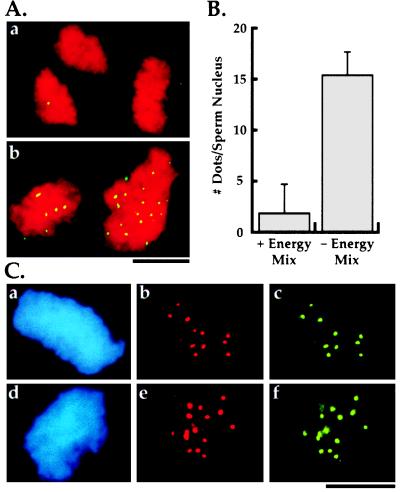
An example of the products from this reaction scheme and the inhibitory effect of energy regeneration mix in the preincubation buffer is shown in Fig. 3A; quantitation of this effect is shown in Fig. 3B. The requirement for a preincubation under low ATP conditions is specific to clarified extracts and is not necessary in low speed crude Xenopus extracts. We do not understand the function of the preincubation phase; it could be necessary to overcome an imbalance in cell cycle state regulators generated by centrifugation (24, 25) or for some prerequisite alteration in centromeric chromatin structure. Absence of sperm nuclei in the preincubation phase of the two-step reaction, followed by the addition of sperm nuclei for a brief period prior to the assembly phase, does not result in kinetochore assembly (data not shown), suggesting that the preincubation phase is priming the sperm chromatin for kinetochore assembly.
Figure 3.
Characterization of in vitro kinetochore assembly. (A) Effect of presence (a) or absence (b) of energy mix in preincubation buffer on kinetochore assembly. DNA is in red and CENP-E is in green. The same result is obtained with anti-dynein and anti-XKCM1 antibodies. There is no striking difference in the morphology of the condensed DNA at the end of the assembly reaction between the two conditions. (B) Quantitation of kinetochore assembly and effect of energy mix in preincubation buffer. The bars represent the mean number of dots of kinetochore marker proteins per sperm nucleus ± 1 SD. For the left column (+ Energy Mix in preincubation buffer) n = 103 sperm nuclei from two experiments on two different extract preparations. For the right column (− Energy Mix in preincubation buffer; standard kinetochore assembly reaction) n = 303 sperm nuclei from four experiments on three different extract preparations. The absence of energy mix in the preincubation buffer results in >90% of the sperm nuclei demonstrating colocalization of all three markers at the reported density. The residual sperm nuclei show no localization of any of the three markers. (Bars = 10 μm.) (C) Double label immunofluorescence of in vitro assembled kinetochores demonstrating colocalization of all three mitotic markers to discrete foci on the DNA. Panels show 4′,6′diamidino-2-phenylindole (DAPI)-labeled sperm nucleus DNA (a and d) and corresponding double label localization of dynein and CENP-E (b and c) or dynein and XKCM1 (e and f). Dots associated with the chromosome clusters are distributed over several focal planes; the images shown are photomicrographs of single focal planes and do not account for all dots present on these chromosomal clusters.
As shown in Fig. 3C, the structures obtained in our in vitro assembly reaction colocalize all three mitotic kinetochore markers to the same discrete foci on the DNA. In addition, the number of dots of kinetochore markers associated with each sperm nucleus is consistent with the number expected. Xenopus sperm nuclei have 18 chromosomes (26) and, because there is no DNA replication in clarified extracts, we expect one kinetochore per chromosome at the end of the assembly reaction. Counting the number of dots of kinetochore marker proteins associated with each condensed chromosomal cluster, we find on average 16 dots (Fig. 3B), close to the expected number of 18. Unlike the reaction conditions used by Hirano and Mitchison (21), our assay conditions do not result in separated single chromosomes; instead, we obtain clusters of tightly intertwined, condensed chromosomes presumably derived from single sperm nuclei.
The targeting of mitosis-specific kinetochore components such as cytoplasmic dynein and CENP-E in our assay suggests that underlying more chromatin-proximal structures may also be present on the in vitro assembled kinetochores. Confirming this will require Xenopus-specific antibodies to constitutive kinetochore components such as CENP-A and CENP-C.
Morphology of in Vitro-Assembled Kinetochores.
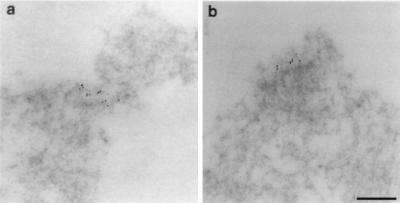
In previous studies, electron microscopy was used on spindles formed in crude Xenopus extracts to characterize the structures at the point where spindle microtubules intersect the mitotic chromatin (27, 28). Even though these chromosomes are segregation-competent and were fixed using optimal fixation methods while undergoing anaphase (28), no trilaminar plate-like structures were observed. Because sperm nuclei are converted to large clusters of condensed chromosomes in our assay, with the domains of interest forming only a minute subset of the total condensed chromatin, we decided to use immunoelectron microscopy to examine the morphology of our in vitro-assembled kinetochores. Our fixation conditions were based on those used previously to preserve the trilaminar plate morphology of CHO chromosomes for immunoelectron microscopy (4, 14). Two examples using the anti-CENP-E antibody are shown in Fig. 4. We observe very specific localization of gold particles to small regions of the mitotic chromatin. These regions had on average seven tightly clustered gold particles with a diameter of about 0.2 μm (range, 0.1–0.3 μm; n = 24 gold clusters) and typically stained more lightly than the surrounding chromatin. The gold clusters were usually, but not exclusively, observed near the edge of the chromatin. The absence of any background gold particles strongly suggests that these clusters represent the dots observed by light microscopy. We did not observe a striking lamellar morphology associated with any of the gold clusters, although we did observe some degree of chromatin differentiation relative to the surrounding chromatin at all of the examined clusters. Although we cannot exclude the possibility that our experimental manipulations damaged existing lamellar kinetochore structure, our results are consistent with the results obtained for segregation-competent chromosomes in crude extracts.
Figure 4.
Morphological analysis of in vitro assembled kinetochores using immunoelectron microscopy. Anti-CENP-E antibody was used for the immunoelectron microscopy. Two examples of gold clusters (a and b) show the specificity of labeling, the absence of a striking lamellar morphology and differentiation of the chromatin at the region of labeling. The cluster in a appears to show a slight degree of constriction; the cluster in b shows clear organization of the underlying chromatin fibers. (Bar = 0.2 μm.)
Analysis of Kinetochore Assembly.
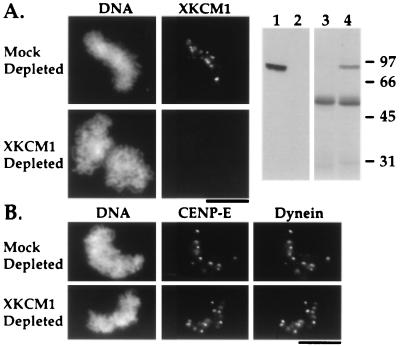
As antibodies to kinetochore components become available, our system can be used to dissect dependency relationships in kinetochore assembly. To demonstrate this potential, we have used our assay to determine if prior assembly of XKCM1 is required for targeting of either CENP-E or cytoplasmic dynein to kinetochores. Immunodepletion of XKCM1 from extracts prior to kinetochore assembly eliminates the observed localization of XKCM1 on chromosomes but has no effect on the targeting of either CENP-E or cytoplasmic dynein (Fig. 5). Therefore, we can conclude that assembly of CENP-E and cytoplasmic dynein onto kinetochores is independent of the assembly of XKCM1. We could not perform the equivalent experiment for CENP-E or dynein because neither the anti-human CENP-E nor the anti-dynein antibody are capable of immunodepleting their Xenopus antigens to a sufficient extent to prevent kinetochore targeting (data not shown). UV-vanadate cleavage of dynein heavy chain in extracts also does not affect the targeting of dynein to kinetochores (data not shown).
Figure 5.
XKCM1 depletion does not affect targeting of dynein and CENP-E to kinetochores. (A) XKCM1 localization to condensed chromosomes is abolished by XKCM1 depletion. Right two panels show an immunoblot (lanes 1 and 2) of equal amounts of mock-depleted (lane 1) and XKCM1-depleted (lane 2) extracts probed with anti-XKCM1 antibody, and a Coomassie-stained gel (lanes 3 and 4) of the corresponding immunoprecipitates: control IgG (lane 3) and anti-XKCM1 (lane 4). (B) XKCM1 depletion does not affect CENP-E and dynein localization. CENP-E and dynein targeting to kinetochores was compared in XKCM1-depleted vs. mock-depleted extracts. Quantitation of number of dots of CENP-E and dynein per sperm nucleus show no difference between mock- and XKCM1-depleted extracts (n = 100 sperm nuclei from two experiments on two different extract preparations). (Bars = 10 μm.)
In addition to a requirement for reagents capable of immunodepleting other kinetochore components from extracts, this type of analysis will also depend on the contribution of pre-established centromere structure present in the substrate sperm chromatin to kinetochore assembly in our assay. The presence of constitutive kinetochore components on mature sperm chromatin in mice and amphibians (29, 30) suggests that the substrate sperm nuclei do posses some amount of pre-established centromere structure. This structure may play an important role in kinetochore assembly in our assay. However, experiments in several organisms have shown that sperm nuclei undergo a dramatic reorganization of their chromatin structure upon entry into eggs (31). This reorganization may include exchange of pre-existing constitutive kinetochore components with components present in the extract, thus making it feasible to assay the role of such components in kinetochore assembly. Defining the role of pre-established centromere structure in our assay will require reagents to follow constitutive kinetochore components in Xenopus.
In Vitro-Assembled Kinetochores Bind Monomeric Tubulin.
Assays testing kinetochore functions require reisolation of the condensed chromosomes after kinetochore assembly. Therefore, we devised a method to purify the end products of our assembly reaction in native form by trapping condensed chromosome clusters at the interface between a sucrose cushion and a heavy sucrose pad. We obtain 20–30% recovery of the starting number of sperm nuclei using this procedure.
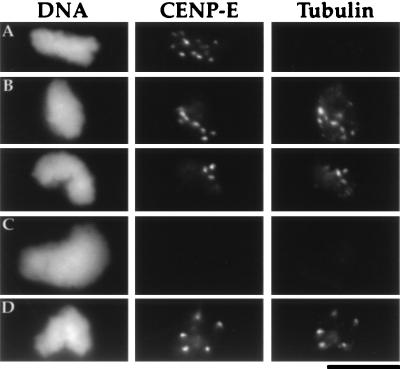
Earlier studies demonstrated that CHO chromosomes, when purified from cells arrested with high concentrations of microtubule depolymerizing drugs, lack any kinetochore-associated monomeric tubulin. When mixed with pure tubulin, these purified chromosomes bind monomeric tubulin at their kinetochore region (11, 14). To mimic the conditions used in the isolation of tissue culture cell chromosomes, and to permit direct functional comparisons between our in vitro assembled chromosomes and tissue culture cell chromosomes, the assembly phase of our two-step reaction is performed in the presence of nocodazole. Therefore, we predicted that there would be no monomeric tubulin associated with the in vitro assembled kinetochores and, as shown in Fig. 6A, we found this to be the case. Furthermore, when purified chromosome clusters were mixed with monomeric tubulin, we detected binding of the tubulin to the kinetochore regions (Fig. 6B). Extraction with 0.1 M NaCl after assembly abolishes the tubulin binding (Fig. 6C), but XKCM1 depletion does not affect the tubulin binding of in vitro-assembled kinetochores (Fig. 6D). These results suggest that the tubulin binding activity of the kinetochore is due to a salt-sensitive kinetochore component that is not XKCM1.
Figure 6.
Tubulin binding by in vitro-assembled kinetochores. Kinetochores were identified using the anti-CENP-E antibody. (A) Purified condensed chromosome clusters lack associated monomeric tubulin. (B) In vitro-assembled kinetochores bind exogenously added monomeric tubulin. Purified condensed chromosome clusters were incubated with 10 μM tubulin and then assayed for binding of monomeric tubulin. As shown here for two separate sperm nuclei, every CENP-E dot was associated with monomeric tubulin (100 sperm nuclei, two experiments). The tubulin also shows faint punctate background staining of the DNA and, occasionally, a few larger dots which do not correspond to CENP-E dots. (C) In vitro-assembled kinetochores fail to bind monomeric tubulin after salt extraction. Assembled kinetochores were extracted using 100 mM extra NaCl before being purified and assayed for binding of monomeric tubulin. This extraction removes kinetochore-associated CENP-E, XKCM1, and dynein but does not remove chromosomal histones (data not shown). (D) XKCM1 depletion does not affect tubulin binding by in vitro-assembled kinetochores. XKCM1 depletion was assayed in parallel by immunoblots and immunofluorescence as described in Fig. 5. (Bar = 10 μm.)
Future Directions.
The primary application of our assay will be in the dissection of protein–protein interactions involved in kinetochore assembly. Doing this will require antibodies capable of immunodepleting different kinetochore components from Xenopus egg extracts, a task likely to be aided by the current emphasis on genome sequencing. Once such reagents are available, our assay will be useful for a systematic dissection of the dependency relationships in kinetochore assembly.
An interesting avenue for future exploration will be to assay different kinetochore functions following biochemical manipulation of kinetochore assembly. We have shown that kinetochores assembled in our assay will bind exogenously added monomeric tubulin. Tubulin binding is independent of XKCM1 but is dependent on components removed by salt extraction of assembled kinetochores. It may be possible to add back this tubulin binding component by incubating salt-extracted kinetochores with extract, and use this as an assay to identify the component responsible for the tubulin binding activity. Additional functional assays will become possible if the technical problem of separating single chromosomes from the clusters formed in our system can be solved.
Finally, our assay may be of some use in the definition of vertebrate centromeric DNA sequences and in the identification of novel kinetochore components. This will require that pure DNA, as opposed to sperm chromatin, is able to recruit kinetochore components when used in our assay.
Acknowledgments
We thank Bruce Schaar and Tim Yen for their gift of anti-CENP-E antibody, Peg Siebert for help with electron microscopy, and Linda Wordeman and Tatsuya Hirano for advice and encouragement during the early phases of this work. Sarita Jain, Matt Welch, Karen Oegema, Jody Rosenblatt, and Doug Kellogg provided valuable comments on the manuscript. This work was supported by a National Institutes of Health Grant (to T.J.M.); C.E.W. is a National Institutes of Health postdoctoral fellow; A.D. is a Howard Hughes Medical Institute predoctoral fellow.
References
- 1.Brinkley B R, Ouspenski I, Zinkowski R P. Trends Cell Biol. 1992;2:15–21. doi: 10.1016/0962-8924(92)90139-e. [DOI] [PubMed] [Google Scholar]
- 2.Earnshaw W C, Tomkiel J E. Curr Opin Cell Biol. 1992;4:86–93. doi: 10.1016/0955-0674(92)90063-i. [DOI] [PubMed] [Google Scholar]
- 3.Rieder C L. Int Rev Cytol. 1982;79:1–58. doi: 10.1016/s0074-7696(08)61672-1. [DOI] [PubMed] [Google Scholar]
- 4.Wordeman L, Steuer E R, Sheetz M P, Mitchison T. J Cell Biol. 1991;114:285–294. doi: 10.1083/jcb.114.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmer D K, O’Day K, Wener M H, Andrews B S, Margolis R L. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Earnshaw W C, Sullivan K F, Machlin P S, Cooke C A, Kaiser D A, Pollard T D, Rothfield N F, Cleveland D W. J Cell Biol. 1987;104:817–829. doi: 10.1083/jcb.104.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saitoh H, Tomkiel J, Cooke C A, Ratrie H D, Maurer M, Rothfield N F, Earnshaw W C. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- 8.Yen T J, Li G, Schaar B T, Szilak I, Cleveland D W. Nature (London) 1992;359:536–539. doi: 10.1038/359536a0. [DOI] [PubMed] [Google Scholar]
- 9.Walczak C E, Mitchison T J, Desai A. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- 10.Wordeman L, Mitchison T J. J Cell Biol. 1995;128:95–104. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balczon R D, Brinkley B R. J Cell Biol. 1987;105:855–862. doi: 10.1083/jcb.105.2.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coue M, Lombillo V A, McIntosh J R. J Cell Biol. 1991;112:1165–1175. doi: 10.1083/jcb.112.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyman A A, Mitchison T J. Nature (London) 1991;351:206–211. doi: 10.1038/351206a0. [DOI] [PubMed] [Google Scholar]
- 14.Mitchison T J, Kirschner M W. J Cell Biol. 1985a;101:755–765. doi: 10.1083/jcb.101.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitchison T J, Kirschner M W. J Cell Biol. 1985b;101:766–777. doi: 10.1083/jcb.101.3.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desai A, Mitchison T J. J Cell Biol. 1995;128:1–4. doi: 10.1083/jcb.128.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernat R L, Delannoy M R, Rothfield N F, Earnshaw W C. Cell. 1991;66:1229–1238. doi: 10.1016/0092-8674(91)90045-z. [DOI] [PubMed] [Google Scholar]
- 18.Tomkiel J, Cooke C A, Saitoh H, Bernat R L, Earnshaw W C. J Cell Biol. 1994;125:531–545. doi: 10.1083/jcb.125.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirano T, Mitchison T J. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 20.Murray A W. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 21.Hirano T, Mitchison T J. J Cell Biol. 1993;120:601–612. doi: 10.1083/jcb.120.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lombillo V A, Nislow C, Yen T J, Gelfand V I, McIntosh J R. J Cell Biol. 1995;128:107–115. doi: 10.1083/jcb.128.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Earnshaw W C, Rothfield N. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 24.Felix M A, Pines J, Hunt T, Karsenti E. EMBO J. 1989;8:3059–3069. doi: 10.1002/j.1460-2075.1989.tb08457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leiss D, Felix M A, Karsenti E. J Cell Sci. 1992;102:285–297. doi: 10.1242/jcs.102.2.285. [DOI] [PubMed] [Google Scholar]
- 26.Graf J D, Kobel H R. Methods Cell Biol. 1991;36:19–34. doi: 10.1016/s0091-679x(08)60270-8. [DOI] [PubMed] [Google Scholar]
- 27.Sawin K E, Mitchison T J. J Cell Biol. 1991;112:925–940. doi: 10.1083/jcb.112.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shamu C E, Murray A W. J Cell Biol. 1992;117:921–934. doi: 10.1083/jcb.117.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brinkley B R, Brenner S L, Hall J M, Tousson A, Balczon R D, Valdivia M M. Chromosoma. 1986;94:309–317. doi: 10.1007/BF00290861. [DOI] [PubMed] [Google Scholar]
- 30.Haaf T, Grunenberg H, Schmid M. Exp Cell Res. 1990;187:157–161. doi: 10.1016/0014-4827(90)90130-3. [DOI] [PubMed] [Google Scholar]
- 31.Poccia D. In: The Molecular Biology of Fertilization. Schatten H, Schatten G, editors. San Diego: Academic; 1989. pp. 115–135. [Google Scholar]