Abstract
Enveloped viruses enter cells by protein-mediated membrane fusion. For influenza virus, membrane fusion is regulated by the conformational state of the hemagglutinin (HA) protein, which switches from a native (nonfusogenic) structure to a fusion-active (fusogenic) conformation when exposed to the acidic environment of the cellular endosome. Here we demonstrate that destabilization of HA at neutral pH, with either heat or the denaturant urea, triggers a conformational change that is biochemically indistinguishable from the change triggered by low pH. In each case, the conformational change is coincident with induction of membrane-fusion activity, providing strong evidence that the fusogenic structure is formed. These results indicate that the native structure of HA is trapped in a metastable state and that the fusogenic conformation is released by destabilization of native structure. This strategy may be shared by other enveloped viruses, including those that enter the cell at neutral pH, and could have implications for understanding the membrane-fusion step of HIV infection.
Keywords: membrane fusion, virus infection, protein folding, HIV
Specific control of membrane fusion is important in diverse biological functions such as fertilization, synaptic transmission, protein trafficking, and viral invasion. The mechanism of membrane fusion is best understood in the context of enveloped viruses, and studies of the influenza virus have been particularly informative (1–6). There is also interest in developing inhibitors of membrane fusion that may serve as pharmaceutical agents (7–14).
Viral infection requires fusion of the membrane surrounding the virus with the membrane surrounding a cell. Specialized viral proteins are responsible for promoting membrane fusion, a process that generally is otherwise very slow. Often, these membrane-fusion proteins also promote binding of the virus to specific cell-surface receptors. For example, the HIV envelope glycoprotein is a complex of two subunits—gp120, which mediates binding of the virus to specific cell-surface receptors, and gp41, which promotes membrane fusion.
In the influenza virus, the hemagglutinin (HA) protein mediates both binding of the virus to the cell surface and the subsequent fusion of viral and cellular membranes. HA is composed of a receptor-binding subunit, denoted HA1, and a fusogenic subunit, denoted HA2. The native HA1/HA2 complex, as found on the surface of the native virus, is fusion-inactive. After binding to its receptor (sialic acid) on the cell surface, the virus is endocytosed by the cell. During these processes, HA remains dormant for fusion. Initiation of fusion activity requires a decrease in pH that is provided within the cell as the endosome matures. As the conditions within the endosome approach pH 5, there is a conformational change in HA that induces the viral membrane to fuse with the cellular, endosomal membrane, permitting the nucleocapsid of the virus to be deposited into the cytoplasm of the cell. Thus, acidic pH is the physiological “trigger” of HA fusion activity (15–17).
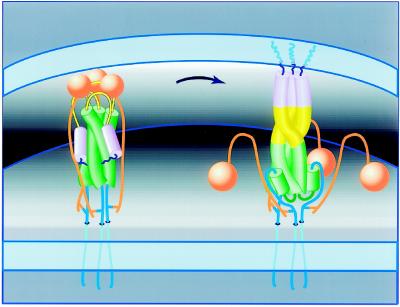
The acid-induced conformational change in HA is understood in considerable detail. In particular, the crystal structures of HA in the native (i.e., nonfusogenic) state and the low-pH converted state have been determined (18–20). Because low pH also activates influenza membrane fusion, the low-pH conformation of HA is generally regarded to be fusogenic. Comparison of the HA crystal structures indicates that a dramatic conformational change accompanies activation of HA fusion activity. This change was largely anticipated in the “spring-loaded” mechanism for activation of membrane fusion (21), in which a region folded as a long loop in native HA converts to a three-stranded coiled coil in the fusogenic state (Fig. 1).
Figure 1.
Model for the spring-loaded mechanism for membrane fusion (20, 21). Envelope of an influenza virus (bottom membrane) is juxtaposed to the target membrane of the cellular endosome (Top). In the native conformation (Left) an HA trimer facilitates viral attachment via the HA1 subunits (orange). In response to acidic pH (Right), the HA1 subunits dissociate, the loop regions (yellow) become helical and extend the HA2 coiled coil (green), and the fusion-peptide regions (blue) insert into the target membrane. Membrane fusion per se appears to require only the HA2 subunit (2). Figure by Jodi M. Harris (adapted from ref. 80).
Previously, we suggested that the native, nonfusogenic state of HA might have a metastable fold (21). According to this suggestion, the fusogenic state of HA is thermodynamically more stable than the native state of the protein, under native conditions (e.g., neutral pH). However, the native state is prevented from achieving the lower-energy fusogenic conformation by a kinetic barrier. This type of a barrier could be imposed during the folding of HA, for example, by factors that prevent direct acquisition of the fusogenic state from the unfolded state, or that actively direct the folding of HA to a metastable state. Notably, HA folds within the cell as the fusion-incompetent precursor, denoted HA0, which subsequently undergoes proteolytic cleavage to generate the mature, two-chain HA1/HA2 native state (22, 23).
Instead of postulating a metastable protein fold, a seemingly simpler model posits that HA folds to the thermodynamically most-stable state (i.e., the native state). At low pH, the fusogenic state becomes thermodynamically more favorable, and a conformational change ensues. Consistent with this model is the suggestion that low pH is required for the fusion activity of influenza HA. Short, synthetic peptides containing sequences from the HA fusion-peptide regions have substantially enhanced membrane-fusion ability at low pH, as compared with that at neutral pH (refs. 24–26; see, however, ref. 27). Furthermore, the coiled-coil region adjacent to the fusion-peptide region of HA displays substantial affinity for lipid bilayers only at acidic pH, a phenomenon postulated to facilitate membrane fusion (28).
The metastability model predicts that any destabilizing reagent will cause the same conformational change and membrane-fusion activity as acidic pH. The alternate model predicts that, although there may be some effect on structure by other destabilizing reagents, these effects will be unrelated to the acid-induced conformational changes that cause membrane fusion. Interestingly, Ruigrok et al. reported that heat can induce influenza fusion with target liposomes at neutral pH (ref. 29; see also ref. 30). However, these workers also found that heat treatment altered the biochemical properties of the HA ectodomain (i.e., the extra-viral portion) in a manner distinct from acid. They concluded that fusion at neutral pH occurs by a different mechanism, involving changes in HA that are more extensive and less specific than that of the acid-induced conformational change that induces membrane fusion (29).
Here we test the metastability model for membrane fusion by characterizing in detail the membrane-fusion activity of intact influenza virus at neutral pH that is induced by either heat or a chemical denaturant, urea. In parallel, we use proteolysis to assay the biochemical properties of HA, in the context of the intact virus, under these various conditions. Our results indicate that, at neutral pH, the native state of influenza HA is metastable.
MATERIALS
Concentrated samples of Influenza A/Beijing/32/92/X-117 H3N2 (1 mg/ml HA) were a gift from A. Donabedian of Parke–Davis, Rochester Operations. Lipids used to make synthetic vesicles were ordered from Avanti Polar Lipids: dioleoyl phosphatidylcholine (DOPC); N-(7-nitro-2–1, 3-benzoxadiazol-4-yl)-diacyl-phosphatidylethanolamine (NBD); and lissamine-rhodamine-phosphatidylethanolamine (rhodamine). As a source of the receptor sialic acid, bovine ganglioside (GD1a; Calbiochem–Nova Biochem) was added to the lipid mixture (32). Lipid quantitation was performed by using an inorganic phosphorus assay (Sigma). Recrystallized 5(6)-carboxyfluorescein (Kodak) and PD-10 (Pharmacia) G-25 columns were used for vesicle integrity assays. Polyclonal antisera against HA2 (238-3) was raised in rabbits with a keyhole-limpet hemocyanin-conjugate (Pierce) of LOOP-36 (21).
METHODS
Proteolysis Assay for the Conformational Change.
Influenza A samples were incubated for 15 min under experimental conditions, diluted and neutralized to standard conditions with 0.5 M Tris, pH 8, and digested with proteinase K (1:1 ratio of proteinase K to HA, by weight) for 1–4 hr at room temperature. Virus proteins and protein fragments were separated by 12% and 14% polyacrylamide gels, under nonreducing and reducing conditions, respectively. Proteins were either stained with Coomassie brilliant blue R250 or transferred to nitrocellulose for detection by immunoblot analysis by using polyclonal antiserum that detects the “loop region” of HA2 (21). Identity of the stable product was confirmed by five cycles of amino-terminal amino acid sequence analysis: intact HA begins at residue 1 of HA2 (sequence Gly-Ile-Phe-Gly-Ala) and the stable product begins at residue 28 of HA2 (sequence Asn-Ser-Glu-Gly-Thr).
Lipid-Mixing Assay for Membrane Fusion.
DOPC, GD1a, NBD, and rhodamine were mixed at molar ratios of 95.3:4.5:0.6:0.6 for preparation of large, unilamellar vesicles (31) in fusion buffer (50 mM Hepes, pH 7.4/150 mM sodium chloride/0.1 mM EDTA; ref. 32). Phospholipids were quantitated by using the inorganic phosphorus assay. Lipid bilayer integrity was confirmed for vesicles prepared in fusion buffer and for vesicles prepared in fusion buffer with 3.75 M urea by entrapping self-quenching concentrations of recrystallized 5(6)-carboxyfluorescein. Dilution of vesicles purified by gel filtration did not lead to dequenching of carboxyfluorescein. However, addition of 5 μl of 25% Triton X-100 to dilute vesicles led to rapid release of carboxyfluorescein and a color change to fluorescent green, indicating a breach of lipid bilayer integrity. Viral lipids were extracted (33) and quantitated by using the inorganic phosphorus assay.
Membrane-fusion activity was monitored by using a fluorescence resonance energy transfer assay (34). Fluorescence was monitored with an ISS PC Greg fluorescence spectrophotometer equipped with a thermostatted cuvette holder, with excitation at 465 nm and emission at 535 nm, using 16-nm slit widths for both. The lipid-mixing endpoint is defined as the fluorescence emission observed at 535 nm on addition of Triton X-100 to 1%. The fraction of lipid mixing (percent lipid mixing, Figs. 5a, 6a, and 7a) is expressed as a faction of the total lipid mixing (signal in 1% Triton X-100 minus initial signal) for each sample. Prebound virus and vesicles were added, to a final concentration of 50 μM in phospholipids, in a fluorescence cuvette containing fusion buffer and stirred at the temperature indicated. The pH of the sample was adjusted by addition of 10–30 μl of 0.1 M citric acid. For acid inactivation, virus was preincubated with citrate at pH 5.8 for 30 min at 37°C. For heat-induced fusion experiments, the initial baseline was estimated from the heat-inactivated control: virus was heated at the final temperature for 30 min and bound to vesicles, and the constant signal observed for the inactivation experiment was estimated as the initial baseline for calculation of percent fusion. Virus preincubated at 3.75 M urea for ≈1 hr at 37°C before addition of vesicles displayed no membrane-fusion activity, and the constant signal observed for the inactivation experiment was estimated as the initial baseline for calculation of percent fusion in the urea-induced fusion experiments. As a check for complete fusion, at the end of each experiment performed at neutral pH, 0.1 M citric acid was added to pH 5.8, and no change in signal was detected.
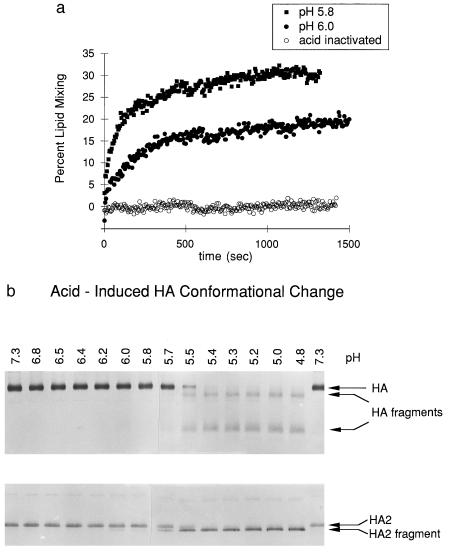
Figure 5.
Acid-induced membrane-fusion activation and the HA conformational change. (a) At 37°C, acid-induced activity is detected at pH 6.0 (solid circles), with optimal activity at pH 5.8 (solid squares) and no activity from acid-inactivated virus (open circles). (b) The conformational change detected by immunoblot analysis was induced by acid below pH 6.0, at 37°C.
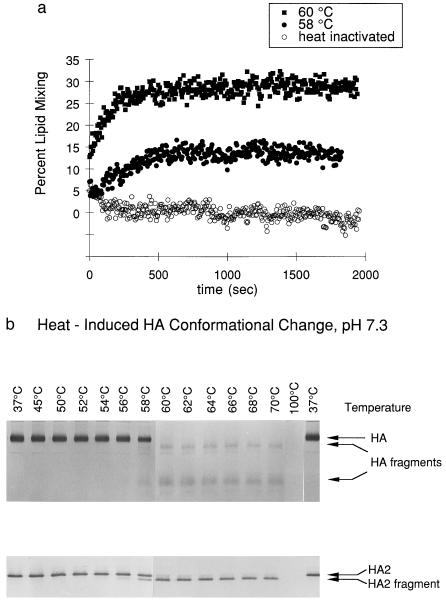
Figure 6.
Heat-induced membrane-fusion activation and the HA conformational change. (a) At pH 7.3, heat-induced activity is detected at 58°C (solid circles), with optimal activity at 60°C (solid squares) and no activity from heat-inactivated virus (open circles). (b) The conformational change detected by immunoblot analysis was induced by heat above 58°C at pH 7.3.
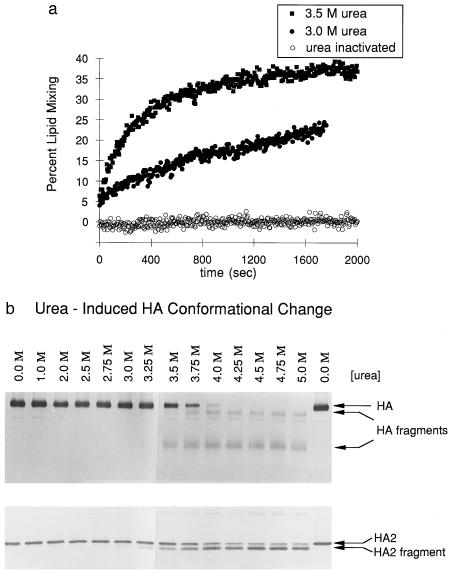
Figure 7.
Denaturant-induced membrane-fusion activation and the HA conformational change. (a) At pH 7.3 and 37°C, denaturant-induced activity is detected at 3.0 M urea (solid circles), with optimal activity at 3.5 M urea (solid squares) and no activity from urea-inactivated virus (open circles). (b) The conformational change detected by immunoblot analysis was induced by urea above 3.5M at 37°C, pH 7.3.
RESULTS
Acid-Induced Conformational Change Is also Induced at Neutral pH.
We evaluated the conformational change in HA by using a biochemical proteolysis assay (35). In the native conformation, HA is resistant to digestion by proteinase K, but in the acid-induced conformation, HA is digested to a specific, protease-resistant product (Fig. 2). These species are readily resolved by PAGE and detected by immunoblot analysis (Fig. 3), providing a convenient assay for the conformational change in HA. Because the transition to the fusogenic state is irreversible (16, 35, 36), the proteolysis assay is performed under standard conditions, following incubation of the virus under the conditions to be evaluated. The transition from a protease-resistant, native conformation to the protease-sensitive, acid-induced state is sharply defined at a specific pH for each strain of Influenza A. The transition pH of the intact, X-117 influenza A/Beijing/H3/N2 utilized in these studies is 5.8.
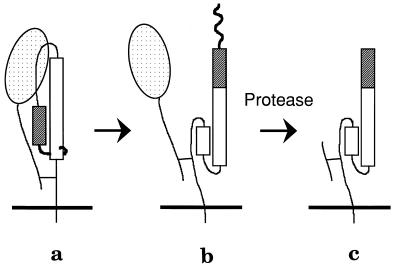
Figure 2.
Schematic of the protease assay for the conformational change of HA. In one monomer of the HA trimer, the HA1 polypeptide (stippled ball and string) is connected to the HA2 polypeptide by a disulfide bond. The HA2 polypeptide spans the viral membrane (Bottom) once and includes the fusion peptide region. (a) Native HA is resistant to proteolysis. (b) The conformational change: HA1 dissociates from the top of HA2, and the fusion peptide is propelled to the top of the molecule. (c) Proteolysis of the fusogenic conformation: some of HA1 and the fusion peptide of HA2 are proteolytically degraded, leaving the majority of HA2 disulfide-bonded to a fragment of HA1. As a result, both the HA1 and the HA2 polypeptides are smaller, and this difference can be detected by SDS/PAGE and immunoblot analysis.
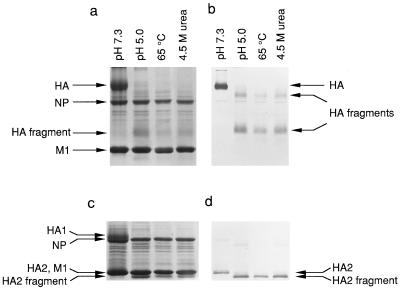
Figure 3.
Conditions that induce the HA conformational change. The protease assay is used to monitor the conformation of HA on virus exposed to experimental conditions (indicated above the gel lanes). (a) Viral polypeptides are separated by SDS/PAGE and detected by Coomassie blue staining of viral proteins separated under nonreducing conditions. (b) Immunoblot analysis of samples used for a with anti-HA2 antibodies reveals disulfide-bonded HA and HA fragments, separated by SDS/PAGE under nonreducing conditions. (c) Samples identical to those in a are separated by SDS/PAGE under reducing conditions and stained with Coomassie blue (lanes are as in a). (d) Immunoblot analysis of samples used for c with anti-HA2 antibodies reveals HA2 and HA2 fragments, separated by SDS/PAGE under reducing conditions (lanes are as in b). Identity of the HA2 fragment was confirmed for all four samples (pH 7.3, pH 5.0, 65°C, and 4.5 M urea) by five cycles of amino-terminal amino acid sequence analysis (see text). HA, an HA1 polypeptide disulfide bonded to an HA2 polypeptide. HA fragments, disulfide-bonded HA1 and HA2 proteolytic fragments. HA2, intact HA2 polypeptide. HA2 fragment, proteolytic fragment of HA2. M1, matrix protein. NP, nucleoprotein.
The stable, protease-resistant product of acid-induced HA corresponds closely to the extended coiled-coil core of the fusogenic conformation (20, 21). This stable product begins at residue 28 of HA2, as determined by amino-terminal sequence analysis, indicating that the fusion-peptide region of HA2 has been removed. Based on the apparent molecular weight of the product, and the observation that this molecular weight decreases on exposure to reducing agents (Fig. 3), we conclude that the stable product extends at least to Cys-138 of HA2, which is disulfide bonded to a fragment of HA1. Thus, the stable product corresponds well to the fragment of the fusogenic state that was crystallized for x-ray diffraction analysis (20).
Exposure of the virus, at neutral pH, to either heat (65°C) or urea (4.5 M) results in a state that gives rise to the same specific proteolytic pattern as that observed for the acid-induced conformation of HA (Fig. 3). Amino-terminal sequencing indicates that, in all three cases, the stable product begins at residue 28 of HA2. In a previous study of high-temperature, HA-induced fusion at neutral pH, it was found that the proteolysis pattern for HA after high-temperature treatment was different from that observed after low-pH treatment. The authors thus concluded that the mechanism of fusion at neutral pH is distinct from that at acid pH and involves more extensive and less specific changes in HA structure (29). These previous proteolysis studies were performed on HA ectodomain samples that were exposed to high temperature, subsequent to cleavage from the surface of the virus with the protease bromelain. In contrast, we have studied proteolysis of HA in the context of the intact virus. One possible explanation for the discrepancy between our studies and previous studies is that the soluble, bromelain-cleaved HA ectodomain is more labile to thermal denaturation than HA within the intact virus, complicating interpretation of the earlier results. Our results indicate that the temperature-induced conformational change in HA at neutral pH is indistinguishable, as judged by the proteolysis assay, from the low-pH induced conformational change, or from the urea-induced conformational change at neutral pH.
Membrane Fusion Induced at Neutral pH.
We utilized a fluorescence lipid-mixing assay that has been used in earlier studies to monitor the fusion of viral and vesicle membranes, especially for HA-mediated membrane fusion (37, 38). The assay monitors changes in fluorescence as the lipids of the viral envelope mix with fluorescent donor and acceptor lipids of the target-vesicle membrane, thereby decreasing the amount of resonance-energy transfer in the target vesicle (34). Membrane-fusion activity is measured as an increase in donor fluorescence that results from the loss of energy transfer (Fig. 4). This assay does not measure directly the mixing of viral and vesicle contents and therefore cannot distinguish hemifusion (39) from complete fusion.
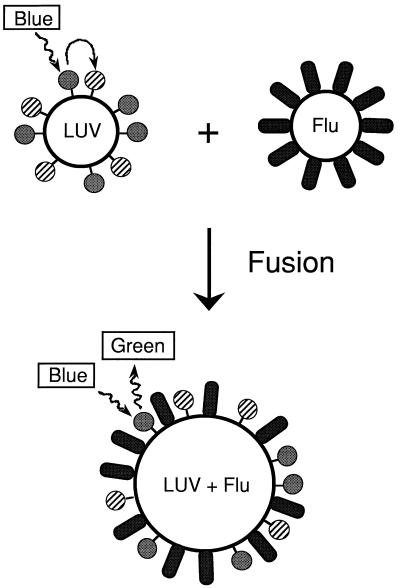
Figure 4.
Schematic of the assay for membrane-fusion activity. Large, unilamellar vesicles (LUV) are prepared with fluorescent donor and acceptor lipids (solid and striped spheres, respectively). On excitation of the donor fluorophore (blue light, 465 nm), there is substantial resonance energy transfer to the acceptor fluorophore, so that little donor fluorescence is observed (green light, 535 nm). After membrane fusion, the lipids of the viral envelope (Flu) mix with the vesicle, separating the donor from the acceptor (LUV + Flu), leading to an increase in observed donor fluorescence.
We monitored lipid mixing of fluorescently labeled vesicles with intact X-117 virus, the same virus that was used in the proteolysis assay. Membrane-fusion activity is observed on lowering the pH to 6.0; the activity is more pronounced at pH 5.8 (Fig. 5a). The pH of onset of fusion activity coincides with the conformational changes leading to the appearance of the stable HA2 fragment (the extended coiled-coil core of the fusogenic conformation) in the proteolysis assay (Fig. 5b). Thus, in agreement with earlier reports (35, 36, 40, 41), we find that acid causes both the HA conformational change and membrane fusion at the same pH.
Membrane fusion is also observed at neutral pH on addition of either heat or urea. Strikingly, the onset of membrane-fusion activity induced by either heat (Fig. 6a) or urea (Fig. 7a) is concurrent with the appearance of the HA2 fragment corresponding to the extended coiled-coil core of the fusogenic conformation, as monitored with the protease assay (Fig. 6b and Fig. 7b). Thus, heat and urea induce a conformational change, at neutral pH, to the same fusogenic state as that induced by low pH.
Viral Inactivation.
In the absence of target membranes, acidic conditions irreversibly inactivate the membrane-fusion activity of HA (16, 35, 36, 42). Electron micrographs of inactivated virus preparations depict virus aggregates with entangled HA molecules in which the fusion-peptide regions are thought to be inserted into viral membranes (6, 36, 43, 44). Therefore, it is likely that viral inactivation results from insertion of the fusion peptide into the same or other viral membranes and/or aggregation of virus. The rate of membrane fusion matches the rate of inactivation of HA in the absence of target membranes, and both rates are retarded at lower temperatures, leading to the conclusion (45) that the fusogenic state of HA can also lead to viral inactivation (Fig. 8).
Figure 8.
A model for the mechanism of the HA conformational change. The metastable native state (N) undergoes an irreversible transition to the more stable fusogenic state (F). The fusogenic state induces membrane-fusion activity in the presence of target membranes and causes viral inactivation in the absence of target membranes.
As expected, we find that the X-117 virus is inactivated after incubation at acidic pH, as evaluated with the fluorescence lipid-mixing assay (Fig. 5a). At neutral pH, viral HA molecules are also inactivated by pretreatment at either high temperature (Fig. 6a) or moderate concentrations of urea (Fig. 7a). Thus, the parallels between acid activation and neutral-pH activation by either heat or urea extend to the phenomenon of viral inactivation. Specifically, each condition that induces membrane-fusion activity also inactivates the virus in the absence of target membranes.
Competition between inactivation and membrane fusion (Fig. 8) may be responsible for differences that we observe in the efficiency of fusion under suboptimal conditions. At conditions of pH or temperature that stimulate the conformational change to the fusogenic state for only a small fraction of HA molecules, membrane fusion is slow and incomplete (Figs. 5a and 6a). Curiously, however, membrane fusion at suboptimal urea concentrations, although slow, is efficient (Fig. 7a). This result could be explained if urea disfavored inactivation, for example, by retarding virus aggregation.
DISCUSSION
Influenza Fusion at Neutral pH.
Enveloped viruses can be classified according to whether they fuse directly with the cell membrane at neutral pH or are first endocytosed and then fuse with the cellular membrane in response to acidic conditions. Our results indicate that influenza virus, which enters cells via the endocytic route, can fuse with target membranes at neutral pH. This confirms the observation made by Rugroik et al. that influenza can fuse at neutral pH and elevated temperatures (29). These workers concluded that neutral pH fusion involves a distinct mechanism from that at low pH, perhaps explaining why there has not been significant follow-up of this early observation. In contrast, our results strongly suggest that there is a common mechanism of membrane-fusion activation, involving destabilization of the native, spring-loaded state of HA, that can be triggered by either acid pH, heat, or chemical denaturant at neutral pH. Importantly, models for influenza fusion that stipulate a low-pH requirement for membrane fusion are untenable with our results. We conclude that, in vivo, the mildly acidic environment of the endosome acts, in a general manner, to destabilize the native state of HA, unleashing the more-stable, fusogenic conformation.
Metastable Protein Folding.
It is commonly accepted that most proteins fold to their thermodynamically most-stable state, as proposed in the Anfinsen hypothesis (46). Our results indicate that the native state of HA is folded, at neutral pH, into a conformation that is metastable. This conclusion is based on a rather rigorous criterion: destabilizing conditions at neutral pH, such as the addition of denaturant, induce a conformational change to a second, stably folded, unique state, which persists after removal of the destabilizing conditions.
The conclusion that HA is metastable is consistent with other observations. First, activation of HA to the fusogenic state is irreversible (16, 35, 36). (Note, however, that although irreversibility is one of the criteria for metastability, the irreversibility of the HA conformational change could be a consequence of other properties of the protein, such as interaction of the fusion-peptide region with membranes). Second, recombinant models of the coiled-coil core of the fusogenic conformation of HA are exceedingly stable to thermal denaturation (47, 48), with thermal-unfolding temperatures much higher than the transition temperature for fusion activation at neutral pH. Finally, for mutants of HA that have an altered pH of activation, there is a striking correlation between the increased pH of fusion and the decreased temperature of fusion at neutral pH (29), consistent with metastable folding of HA. Moreover, these same mutations have very little effect on the thermal unfolding temperature of a recombinant model of the coiled-coil core of the fusogenic conformation of HA (C.M.C., M. A. Milhollen & P.S.K., unpublished results), indicating that the mutations selectively destabilize the native, metastable state, as opposed to stabilizing the fusogenic state.
The existence of a metastable state for HA raises an intriguing question: how does HA fold into a metastable conformation? The mechanism of in vivo protein folding and the three-dimensional structure of the HA precursor (HA0) are relevant to this question. Folding of HA0 in vivo involves chaperonins (49–51), which may facilitate formation of a metastable HA0 trimer. Alternatively, the HA0 precursor polypeptide may fold into a thermodynamically most-stable conformation, with proteolytic cleavage yielding the metastable, native HA1/HA2 complex. In this regard, it is noteworthy that HA0 must undergo maturation cleavage to become fusion-competent (22, 23). After cleavage of the peptide bond, there is substantial structural rearrangement: the newly created amino terminus of HA2 and carboxyl terminus of HA1 in mature HA are ≈22 Å apart in the native structure (18). Thus, it is possible that HA0 cannot topologically access the fusogenic conformation until maturation cleavage separates HA1 from HA2, trapping HA in a metastable conformation. Whether HA0 folds directly into a metastable state, or whether maturation cleavage of a thermodynamically most-stable state of HA0 produces the metastable HA1/HA2 state, remains to be determined.
Metastable protein folding has been suggested for a few other proteins, including α-lytic protease, subtilisin, luciferase, and the serpin family of protease inhibitors (52–54). Metastable folding has also been detected in the disulfide-bonded intermediates that are populated in the oxidative folding of bovine pancreatic trypsin inhibitor (BPTI), by using the rigorous criterion that destabilizing conditions can induce a transition to a second, unique, more-stable state that persists after removal of the destabilizing conditions. Addition of heat, high concentrations of urea, or the enzyme protein disulfide isomerase to kinetically trapped BPTI intermediates increases the rate of folding to the final native protein (55–57).
Implications.
One can envision reasons why membrane-fusion proteins might have evolved to utilize metastability for membrane fusion. First, coupling the energetically expensive membrane-fusion reaction to an energetically favorable conformational change may help to drive the reaction toward complete membrane fusion. Second, the irreversibility of a conformational change to a state that is fusion-active in the presence, and fusion-inactivated in the absence, of a target membrane (Fig. 8) may prevent indiscriminate and toxic fusion events.
Regardless of whether the native state is metastable, it seems likely that membrane fusion proteins generally will prove capable of folding into two states with substantially different conformations. As in influenza HA and HIV gp120/gp41, many membrane-fusion proteins function by exposing a hydrophobic, glycine-rich stretch of ≈25 residues, termed the “fusion-peptide” region, which inserts into the target membrane (38, 58–61). In the native, dormant state of HA, the fusion-peptide region is buried in the hydrophobic core of the protein, presumably to avoid either fusion-inactivation or indiscriminate membrane-fusion activity. It is reasonable to expect that fusion-peptide regions of other proteins will also be buried in the native state. It is unlikely, however, that the conformation of the cores of these proteins will remain intact after removal of the fusion-peptide regions from a buried location. Native proteins generally are only marginally stable, and creating a void in the hydrophobic core of a protein is expected to have major destabilizing consequences (62, 63). Accordingly, it seems likely that membrane-fusion proteins have evolved the capacity to adopt a second conformation, with a stable core that is packed in a completely different manner, as has been established for the case of influenza HA (18–20).
For HA, fusion activation is accompanied by exposure of the fusion-peptide regions and concurrent formation of a new hydrophobic core at the base of an extended coiled-coil structure in the low pH-induced conformation (20). A remarkably similar conformation is seen in the x-ray crystal structures of the cores of the fusogenic subunits from the murine leukemia virus (MLV) and from HIV (14, 64, 65). Moreover, residues in the core of the MLV structure are conserved among many members of the retrovirus and filovirus families including Ebola (64). For HIV, it is likely that the native gp120/gp41 complex will be folded into a conformation that is substantially different from that present in the core gp41 structures (14, 65). Peptides that make up the core of gp41 are themselves effective inhibitors of HIV infection and appear to work in a dominant-negative manner by binding to viral gp41 (66–68), suggesting strongly that these regions are available transiently during the conformational change to the fusogenic state.
The native state of the HIV membrane-fusion protein complex, gp120/gp41, may also be metastable, like that of HA. The core of the fusogenic conformation of gp41 is exceedingly stable to thermal denaturation (66), with an apparent melting temperature of approximately 90°C. In contrast, the native HIV gp120/gp41 complex is labile, as indicated by the ease with which gp120 dissociates in virus preparations (69, 70). Binding of HIV to its cellular receptor (CD4) and coreceptors (e.g., CCR5 or CXCR4) is thought to destabilize the native state of the viral envelope protein, triggering a conformational change to a fusion-active state (refs. 71 and 72; see also ref. 73). Moreover, the core of the fusogenic conformation of the transmembrane (TM) subunit from the MLV retrovirus is very thermostable (64, 74). Recent studies indicate that the native glycoprotein complex is less stable than the TM subunit core, strongly suggesting a metastable native fold (D. Fass & P.S.K., unpublished data).
An interesting phenomenon referred to as “enhancement” has been observed for HIV, in which agents that inactivate HIV at high concentrations, such as soluble CD4 (sCD4) or some neutralizing antibodies, increase infectivity at low concentrations, presumably by assisting or inducing membrane-fusion activity (75, 76). The apparent conflict between neutralization and enhancement could be reconciled by analogy to current thinking about HA-mediated membrane fusion: virus that is prevented from binding to cell-surface receptors by high concentrations of neutralizing agents will be rapidly inactivated, whereas low concentrations of these agents might allow some cell-surface binding to occur and promote the conformational change by destabilizing a metastable native state.
Our results suggest that the envisioned differences in mechanism between viral fusion proteins that are activated at low and neutral pH may actually be nonexistent or less pronounced than previously thought. Viruses that normally fuse at neutral pH could use binding of the envelope protein by cellular receptors to destabilize a metastable, native envelope structure. Viruses that enter cells via an endocytic pathway could utilize acid as a general destabilizing agent to trigger the conformational change from a metastable state. Thus, metastability may be a common feature of viral membrane-fusion proteins, regardless of whether they promote membrane fusion at the cell surface or within acidic endosomal compartments.
Finally, however, we note that metastability per se does not appear to be absolutely required for viral membrane fusion. Activation of the membrane-fusion proteins of rhabdoviruses [e.g., Rabies virus, Vesicular Stomatitis virus (VSV)] involves a reversible, pH-dependent conformational change (77–79). In particular, it has been shown that the glycoprotein (G) from VSV undergoes a pH-dependent conformational change to a fusogenic state that reverses back to the native, nonfusogenic state on reneutralization (79). Thus, although metastability may prove to be a common theme among fusion proteins, evolution has provided alternative solutions to the problem of virus invasion.
Acknowledgments
We especially thank A. Donabedian of Parke–Davis, Rochester Operations, for his enthusiasm and generosity in supplying concentrated influenza virus; strain A/Beijing/32/92/X-117 (H3N2); X. Xu and M. Shaw of the Influenza Branch, Centers for Disease Control, for the HA nucleotide sequence from the X-117 strain; R. Rutkowski, D. Fass, and L. Wu for critical editing and members of the Kim lab for helpful comments on the manuscript; J. McKnight for initial help with fluorescence assays; P. Petillo for help with the carboxyfluorescein recrystallization; and T. Stegmann, J. Lear, and J. Bentz for helpful discussions about the fusion assay. This research was supported by the Howard Hughes Medical Institute.
ABBREVIATION
- HA
hemagglutinin
References
- 1.Wiley D C, Skehel J J. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 2.Schoch C, Blumenthal R, Clague M J. FEBS Lett. 1992;311:221–225. doi: 10.1016/0014-5793(92)81107-w. [DOI] [PubMed] [Google Scholar]
- 3.Stegmann T, Helenius A. In: Viral Fusion Mechanisms. Bentz J, editor. Boca Raton, FL: CRC; 1993. pp. 89–111. [Google Scholar]
- 4.Hughson F M. Curr Opin Struct Biol. 1995;5:507–513. doi: 10.1016/0959-440x(95)80036-0. [DOI] [PubMed] [Google Scholar]
- 5.Tatulian S A, Hinterdorfer P, Baber G, Tamm L K. EMBO J. 1995;14:5514–5523. doi: 10.1002/j.1460-2075.1995.tb00238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanaseki T, Kawasaki K, Murata M, Ikeuchi Y, Ohnishi S. J Cell Biol. 1997;137:1041–1056. doi: 10.1083/jcb.137.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wild C T, Oas T, McDanal C B, Bolognesi D, Matthews T J. Proc Natl Acad Sci USA. 1992;89:10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodian D L, Yamasaki R B, Buswell R L, Stearns J F, White J M, Kuntz I D. Biochemistry. 1993;32:2967–2978. doi: 10.1021/bi00063a007. [DOI] [PubMed] [Google Scholar]
- 9.Jiang S, Lin K, Strick N, Neurath A R. Nature (London) 1993;365:113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- 10.Mammen M, Dahmann G, Whitesides G M. J Med Chem. 1995;38:4179–4190. doi: 10.1021/jm00021a007. [DOI] [PubMed] [Google Scholar]
- 11.Rapaport D, Ovadia M, Shai Y. EMBO J. 1995;14:5524. doi: 10.1002/j.1460-2075.1995.tb00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lambert D M, Barney S, Lambert A L, Guthrie K, Medinas R, Davis D E, Bucy T, Erickson J, Merutka G, Petteway S R., Jr Proc Natl Acad Sci USA. 1996;93:2186–2191. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao O, Compans R W. Virology. 1996;223:103–112. doi: 10.1006/viro.1996.0459. [DOI] [PubMed] [Google Scholar]
- 14.Chan D C, Fass D, Berger J M, Kim P S. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 15.Maeda T, Ohnishi S. FEBS Lett. 1980;122:283–287. doi: 10.1016/0014-5793(80)80457-1. [DOI] [PubMed] [Google Scholar]
- 16.Maeda T, Kawasaki K, Ohnishi S. Proc Natl Acad Sci USA. 1981;78:4133–4137. doi: 10.1073/pnas.78.7.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang R T, Rott R, Klenk H-D. Virology. 1981;110:243–247. doi: 10.1016/0042-6822(81)90030-1. [DOI] [PubMed] [Google Scholar]
- 18.Wilson I A, Skehel J J, Wiley D C. Nature (London) 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 19.Weiss W I, Brünger A T, Skehel J J, Wiley D C. J Mol Biol. 1990;212:737–761. doi: 10.1016/0022-2836(90)90234-D. [DOI] [PubMed] [Google Scholar]
- 20.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Nature (London) 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 21.Carr C M, Kim P S. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 22.Klenk H D, Rott R, Orlich M, Blödorn J. Virology. 1975;68:426–439. doi: 10.1016/0042-6822(75)90284-6. [DOI] [PubMed] [Google Scholar]
- 23.Lazarowitz S G, Choppin P W. Virology. 1975;68:440–454. doi: 10.1016/0042-6822(75)90285-8. [DOI] [PubMed] [Google Scholar]
- 24.Murata M, Sugahara Y, Takahashi S, Ohnishi S. J Biochem. 1987;102:957–962. doi: 10.1093/oxfordjournals.jbchem.a122137. [DOI] [PubMed] [Google Scholar]
- 25.Wharton S A, Martin S R, Ruigrok R W H, Skehel J J, Wiley D C. J Gen Virol. 1988;69:1847–1857. doi: 10.1099/0022-1317-69-8-1847. [DOI] [PubMed] [Google Scholar]
- 26.Rafalski M, Ortiz A, Rockwell A, van Ginkel L C, Lear J D, DeGrado W F, Wilschut J. Biochemistry. 1991;30:10211–10220. doi: 10.1021/bi00106a020. [DOI] [PubMed] [Google Scholar]
- 27.Lear J D, DeGrado W F. J Biol Chem. 1987;262:6500–6505. [PubMed] [Google Scholar]
- 28.Yu Y G, King D S, Shin Y-K. Science. 1994;266:274–276. doi: 10.1126/science.7939662. [DOI] [PubMed] [Google Scholar]
- 29.Ruigrok R W H, Martin S R, Wharton S A, Skehel J J, Bayley P M, Wiley D C. Virology. 1986;155:484–497. doi: 10.1016/0042-6822(86)90210-2. [DOI] [PubMed] [Google Scholar]
- 30.Huang R T C, Wahn K, Klenk H-D, Rott R. Virology. 1980;104:294–302. doi: 10.1016/0042-6822(80)90334-7. [DOI] [PubMed] [Google Scholar]
- 31.Mayer L D, Hope M J, Cullis P R. Biochim Biophys Acta. 1986;858:161–168. doi: 10.1016/0005-2736(86)90302-0. [DOI] [PubMed] [Google Scholar]
- 32.Alford D, Ellens H, Bentz J. Biochemistry. 1994;33:1977–1987. doi: 10.1021/bi00174a002. [DOI] [PubMed] [Google Scholar]
- 33.Bligh E G, Dyer W J. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 34.Struck D K, Hoekstra D, Pagano R E. Biochemistry. 1981;20:4093–4099. doi: 10.1021/bi00517a023. [DOI] [PubMed] [Google Scholar]
- 35.Skehel J J, Bayley P M, Brown E B, Martin S R, Waterfield M D, White J M, Wilson I A, Wiley D C. Proc Natl Acad Sci USA. 1982;79:968–972. doi: 10.1073/pnas.79.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato S B, Kawasaki K, Ohnishi S-I. Proc Natl Acad Sci USA. 1983;80:3153–3157. doi: 10.1073/pnas.80.11.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stegmann T, Hoekstra D, Sherphof G, Wilschut J. Biochemistry. 1985;24:3107–3113. doi: 10.1021/bi00334a006. [DOI] [PubMed] [Google Scholar]
- 38.Stegmann T, Delfino J M, Richards F M, Helenius A. J Biol Chem. 1991;266:18404–18410. [PubMed] [Google Scholar]
- 39.Kemble G W, Danieli T, White J M. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 40.Yewdell J W, Gerhard W, Bachi T. J Virol. 1983;48:239–248. doi: 10.1128/jvi.48.1.239-248.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doms R W, Helenius A, White J. J Biol Chem. 1985;260:2973–2981. [PubMed] [Google Scholar]
- 42.White J, Kartenbeck J, Helenius A. EMBO J. 1982;1:217–222. doi: 10.1002/j.1460-2075.1982.tb01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weber T, Paesold G, Galli C, Mischler R, Semenza G, Brunner J. J Biol Chem. 1994;269:18353–18358. [PubMed] [Google Scholar]
- 44.Wharton S A, Calder L J, Ruigrok R W H, Skehel J J, Steinhauer D A, Wiley D C. EMBO J. 1995;14:240–246. doi: 10.1002/j.1460-2075.1995.tb06997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramalho-Santos J, Nir S, Düzgünes N, Pato de Carvalho A, da Conçeicão Pedroso de Lima M. Biochemistry. 1993;32:2771–2779. doi: 10.1021/bi00062a006. [DOI] [PubMed] [Google Scholar]
- 46.Anfinsen C B. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 47.Chen J, Wharton S A, Weissenhorn W, Calder L J, Hughson F M, Skehel J J, Wiley D C. Proc Natl Acad Sci USA. 1995;92:12205–12209. doi: 10.1073/pnas.92.26.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carr C M. Ph.D. thesis. Cambridge, MA: Massachusetts Institute of Technology; 1995. [Google Scholar]
- 49.Gething M-J, McCammon K, Sambrook J. Cell. 1986;46:939–950. doi: 10.1016/0092-8674(86)90076-0. [DOI] [PubMed] [Google Scholar]
- 50.Segal M S, Bye J M, Sambrook J F, Gething M-J. J Cell Biol. 1992;118:227–244. doi: 10.1083/jcb.118.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hebert D N, Foellmer B, Helenius A. Cell. 1995;81:425–433. doi: 10.1016/0092-8674(95)90395-x. [DOI] [PubMed] [Google Scholar]
- 52.Baker D, Agard D A. Biochemistry. 1994;33:7505–7509. doi: 10.1021/bi00190a002. [DOI] [PubMed] [Google Scholar]
- 53.Sinclair J F, Ziegler M M, Baldwin T O. Nat Struct Biol. 1994;1:320–326. doi: 10.1038/nsb0594-320. [DOI] [PubMed] [Google Scholar]
- 54.Lee K N, Park S D, Yu M-H. Nat Struct Biol. 1996;3:497–500. doi: 10.1038/nsb0696-497. [DOI] [PubMed] [Google Scholar]
- 55.Weissman J S, Kim P S. Science. 1991;253:1386–1393. doi: 10.1126/science.1716783. [DOI] [PubMed] [Google Scholar]
- 56.Weissman J S, Kim P S. Nature (London) 1993;365:185–188. doi: 10.1038/365185a0. [DOI] [PubMed] [Google Scholar]
- 57.Weissman J S, Kim P S. Nat Struct Biol. 1995;2:1123–1130. doi: 10.1038/nsb1295-1123. [DOI] [PubMed] [Google Scholar]
- 58.Tsurudome M, Glück R, Graf R, Falchetto R, Schaller U, Brunner J. J Biol Chem. 1992;267:20225–20232. [PubMed] [Google Scholar]
- 59.Durrer P D, Gaudin Y, Ruigrok R W H, Graf R, Brunner J. J Biol Chem. 1995;270:17575–17581. doi: 10.1074/jbc.270.29.17575. [DOI] [PubMed] [Google Scholar]
- 60.Rafalski M, Lear J, DeGrado W. Biochemistry. 1990;29:7917–7922. doi: 10.1021/bi00486a020. [DOI] [PubMed] [Google Scholar]
- 61.Pereira F B, Goñi F M, Muga A, Nieva J L. Biophys J. 1997;73:1977–1986. doi: 10.1016/S0006-3495(97)78228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lim W A, Sauer R T. J Mol Biol. 1991;219:359–376. doi: 10.1016/0022-2836(91)90570-v. [DOI] [PubMed] [Google Scholar]
- 63.Matthews B W. Adv Prot Chem. 1995;46:249–278. doi: 10.1016/s0065-3233(08)60337-x. [DOI] [PubMed] [Google Scholar]
- 64.Fass D, Harrison S C, Kim P S. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 65.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Nature (London) 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 66.Lu M, Blacklow S C, Kim P S. Nat Struct Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 67.Chen C H, Matthews T J, McDanal C B, Bolognesi D P, Greenberg M L. J Virol. 1995;69:3771–3777. doi: 10.1128/jvi.69.6.3771-3777.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wild C, Greenwell T, Shugars D, Rimsky-Clarke L, Matthews T. AIDS Res Hum Retroviruses. 1995;11:323–325. doi: 10.1089/aid.1995.11.323. [DOI] [PubMed] [Google Scholar]
- 69.Kalyanaraman V S, Rodriguez V, Veronese F, Rahman R, Lusso P, DeVico A L, Copeland T, Oroszlan S, Gallo R C, Sarngadharan M G. AIDS Res Hum Retroviruses. 1990;6:371–380. doi: 10.1089/aid.1990.6.371. [DOI] [PubMed] [Google Scholar]
- 70.Helseth E, Olshevsky U, Furman U, Sodroski J. J Virol. 1991;65:2119–2123. doi: 10.1128/jvi.65.4.2119-2123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sattentau Q J, Moore J P. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fauci A. Nature (London) 1996;384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 73.Gilbert J M, Hernandez L D, Balliet J W, Bates P, White J M. J Virol. 1995;69:7410–7415. doi: 10.1128/jvi.69.12.7410-7415.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fass D, Kim P S. Curr Biol. 1995;5:1377–1383. doi: 10.1016/s0960-9822(95)00275-2. [DOI] [PubMed] [Google Scholar]
- 75.Allan J S, Strauss J, Buck D W. Science. 1989;247:1084–1088. doi: 10.1126/science.2309120. [DOI] [PubMed] [Google Scholar]
- 76.Sullivan N, Sun Y, Li J, Hofmann W, Sodroski J. J Virol. 1995;69:4413–4420. doi: 10.1128/jvi.69.7.4413-4422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gaudin Y, Tuffereau C, Segretain D, Dnossow M, Flamand A. J Virol. 1991;65:4853–4859. doi: 10.1128/jvi.65.9.4853-4859.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blumenthal R, Bali-Puri A, Walter A, Covell D, Eidelman O. J Biol Chem. 1987;262:13614–13619. [PubMed] [Google Scholar]
- 79.Pak C C, Puri A, Blumenthal R. Biochemistry. 1997;36:8890–8896. doi: 10.1021/bi9702851. [DOI] [PubMed] [Google Scholar]
- 80.Carr C M, Kim P S. Science. 1994;265:234–236. doi: 10.1126/science.7939658. [DOI] [PubMed] [Google Scholar]