Abstract
The exoenzyme S regulon is a set of coordinately regulated virulence genes of Pseudomonas aeruginosa. Proteins encoded by the regulon include a type III secretion and translocation apparatus, regulators of gene expression, and effector proteins. The effector proteins include two enzymes with ADP-ribosyltransferase activity (ExoS and ExoT) and an acute cytotoxin (ExoU). In this study, we identified ExoY as a fourth effector protein of the regulon. ExoY is homologous to the extracellular adenylate cyclases of Bordetella pertussis (CyaA) and Bacillus anthracis (EF). The homology among the three adenylate cyclases is limited to two short regions, one of which possesses an ATP-binding motif. In assays for adenylate cyclase activity, recombinant ExoY (rExoY) catalyzed the formation of cAMP with a specific activity similar to the basal activity of CyaA. In contrast to CyaA and EF, rExoY activity was not stimulated or activated by calmodulin. A 500-fold stimulation of activity was detected following the addition of a cytosolic extract from Chinese hamster ovary (CHO) cells. These results indicate that a eukaryotic factor, distinct from calmodulin, enhances rExoY catalysis. Site-directed mutagenesis of residues within the putative active site of ExoY abolished adenylate cyclase activity. Infection of CHO cells with ExoY-producing strains of P. aeruginosa resulted in the intracellular accumulation of cAMP. cAMP accumulation within CHO cells depended on an intact type III translocation apparatus, demonstrating that ExoY is directly translocated into the eukaryotic cytosol.
Pseudomonas aeruginosa, an opportunistic pathogen of humans, most commonly infects individuals with cystic fibrosis, impaired immune systems, or severe burns (1). Various virulence determinants have been shown to play a role in P. aeruginosa pathogenesis, including proteases, alginate, phospholipases, and toxins. Our analysis of P. aeruginosa virulence has focused on the exoenzyme S (ExoS) regulon, whose expression is correlated with the spread of the bacterium from epithelial colonization sites to the bloodstream (2). The members of the ExoS regulon are coordinately controlled at the transcriptional level by ExsA, an Ara C-family regulatory protein (3). ExsA regulates its own synthesis and a series of operons encoding proteins involved in the intoxication of eukaryotic cells by a process generally referred to as type III secretion or polarized translocation. Type III secretion and intoxication in P. aeruginosa are encoded by three classes of gene products that include components of a secretory apparatus (ExsD, PscB-L, and PscN-U), proteins mediating the translocation of effectors into the host cell cytoplasm (PopB and PopD), and effector proteins (ExoS, ExoT, and ExoU) that are postulated to disrupt normal cellular processes (2). Type III-mediated effector translocation is postulated to inhibit the phagocytic response to infection, allow bacterial replication, and promote epithelial injury.
Three effector proteins of the ExoS regulon have been identified (2). ExoS is an ADP-ribosyltransferase. Host target proteins modified by ExoS include members of the H- and K-Ras family of proteins, vimentin, the Fc region of IgG3, and apolipoprotein A1 (4, 5). Of these targets, ADP-ribosylation of Ras has been correlated to the uncoupling of Ras-mediated signal transduction in PC-12 neuronal cells (6). Two additional effector proteins, ExoT and ExoU, have been identified. ExoU expression is correlated with acute cytotoxicity and lung damage, but the role of ExoT remains elusive (7). Here, we report the identification of an extracellular adenylate cyclase, ExoY, a fourth effector protein of the ExoS regulon.
MATERIALS AND METHODS
Bacterial Strains and Growth Conditions.
P. aeruginosa strains were maintained on Vogel–Bonner medium supplemented with antibiotics as required (8). For transformation of P. aeruginosa, cells were made competent by using the MgCl2 method of Olsen et al. (9). As previously described, maximal induction of the ExoS regulon required growth in a dialysate of trypticase soy broth containing 10 mM nitrilotriacetic acid (10). Plasmids were maintained in Escherichia coli DH5α, and M13 bacteriophage clones were propagated in E. coli TG1 as described (11). Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; tetracycline, 25 μg/ml (E. coli) or 100 μg/ml (P. aeruginosa); and carbenicillin, 400 μg/ml.
Cloning and Nucleotide Sequence Analysis.
To clone exoY from P. aeruginosa strain 388, an exoY-specific probe was amplified with the primers 5′-ACCATGCGTATCGACGGTCATC and 5′-TTGCTGAGATGCTGGTCGACAC using strain 388 chromosomal DNA as a template. A bank of strain 388 KpnI chromosomal restriction fragments was cloned in pNOT19 and screened by hybridization with the exoY probe as described (11). One positive clone was mapped to position exoY by using restriction endonuclease digestion and Southern blot analysis. Subclones were generated in M13 vectors by using standard methods and subjected to nucleotide sequence analysis with an ALF automated DNA sequencer and reagents (Pharmacia). Analysis of nucleotide and protein sequences was performed using the Genetics Computer Group (Madison, WI) software.
Expression and Purification of recombinant ExoY (rExoY).
For expression and purification, exoY was cloned into the pET16b (pETexoY) and pET23b (pET23exoY) vectors (Novagen). The pETexoY expression plasmid encodes a fusion protein with 10 amino-terminal histidine residues, and pET23exoY encodes a fusion protein with 6 carboxy-terminal histidine residues. The purification of recombinant histidine fusion proteins from E. coli lysates was performed by nickel-affinity chromatography. ExoY was amplified incorporating XhoI restriction sites at both ends or with NheI and XhoI restriction sites at the 5′ and 3′ ends, respectively. The nucleotide sequence of amplified fragments was confirmed by sequence analysis. Clones, confirmed by restriction endonuclease mapping, were transformed into BL21(DE3)pLysS (Novagen).
Ni2+ affinity chromatography was performed as described (11). After elution from the Ni2+ resin, peak fractions were immediately pooled and dialyzed against a buffer [20 mM Tris⋅HCl(pH 8.0)/250 mM NaCl/6 mM MgCl2/0.2 mM CaCl2] containing 2 mM DTT at 4°C. Glycerol was added to 10%, and enzyme preparations were stored at −70°C. rExoY was quantitated by densitometry using BSA as a standard. Rabbit antisera specific for ExoY was generated using standard methodologies. Immunoblotting was performed as described (12). Bound IgG was detected with 125I-protein A.
Site-Directed Mutagenesis.
Site-directed mutations were introduced into exoY by using a Sculptor mutagenesis kit (Amersham). Primers K81M (5′-CGAGAAGCCC[A]TGGTCGGGAAAC), K88I (5′-GTTCGAGCTT[A]TCCCCTTCACC), D212N (5′-AGGTCATAAT[T]GGCGGTCATCG), and D214N (5′-AGGAAGAGGT[T]ATAATCGGCG), were annealed to single-stranded M13 templates, and reactions were performed according to the manufacturer’s instructions. Mutations were confirmed by nucleotide sequence analysis and returned to the pETexoY expression plasmid as a SacII–RsrII cassette.
Adenylate Cyclase Assays.
Reactions (100 μl) contained 10 mM Tris⋅HCl (pH 8.0), 6 mM MgCl2, 0.2 mM CaCl2, 2 mM ATP, 2 mM DTT, and rExoY. Reactions were incubated at 30°C, stopped with the addition of 186 μl of ethanol (65% final concentration), and incubated at room temperature for 5 min to precipitate protein. Where indicated, reactions were treated for 10 min with 5 μg of 3′:5′-cyclic nucleotide phosphodiesterase (Sigma) at 30°C before extraction with ethanol. Proteins were precipitated by centrifugation (14,000 × g) for 5 min, and supernatants were collected, lyophilized, and suspended in 50 μl of distilled water. cAMP was resolved from ATP and AMP and quantitated by using reverse-phase HPLC (13). Mobile phases consisted of 0.1 M potassium phosphate, pH 6.0 (buffer A) and 0.1 M potassium phosphate (pH 6.0) in 10% CH3OH (buffer B). Samples were injected into a C-18 column (0.46 cm × 15 cm) and resolved by washing for 5 min at 100% buffer A, followed by an 8-min gradient to 100% buffer B, held for 5 min at 100% buffer B, and returned to 100% buffer A for 5 min. Absorbance was monitored at 259 nm. Retention times were calculated using purified ATP, AMP, and cAMP as standards. Stimulation of ExoY activity was measured in the standard reaction containing bovine brain calmodulin (Sigma) or a postnuclear extract (PNE) from Chinese hamster ovary (CHO) cells (14).
Quantitation of cAMP in CHO cell lysates was performed as follows. Twelve-well plates were seeded with 2 × 105 CHO cells and cultured 18–24 hr in HAM’s F-12 nutrient mixture (supplemented with 10% newborn calf serum, 50 units/ml penicillin/streptomycin, 2 mM l-glutamine, 0.12% sodium bicarbonate, and 2.5 mM Hepes) at 37°C under 5% CO2. Confluent monolayers (>95%) were washed with Dulbecco’s phosphate-buffered saline, infected with 1 ml of medium (lacking newborn calf serum and antibiotics) containing 1 × 107 bacteria (multiplicity of infection = 25:1), and incubated at 37°C under 5% CO2. At the indicated times, cells were washed twice with phosphate-buffered saline, harvested with 150 μl of 4 mM EDTA, and heated to 100°C for 5 min. Lysates were extracted with ethanol, lyophilized, suspended in 50 μl of 50 mM Tris⋅HCl (pH 7.5)/4 mM EDTA, and assayed for cAMP using a BioTrak [3H] cAMP kit (Amersham) according to the manufacturer’s instructions. The protein content of the CHO cell lysates was determined with a BCA protein assay kit (Pierce). cAMP values were normalized to the protein content of the cell lysate.
RESULTS
Identification, Cloning, and Nucleotide Sequence Analysis of exoY.
Previously, we generated and characterized transposon mutants of P. aeruginosa that possess a defective type III secretion apparatus (15). Comparison of the extracellular protein profiles from wild-type and mutant strains demonstrated that in addition to ExoS, several other proteins were absent in the secretion-deficient strains. By matching experimentally derived amino-terminal sequences of extracellular proteins with sequences deduced from cloned genes that were coordinately regulated with ExoS, we identified the major type III-secreted products, with the exception of a 42-kDa protein (16). To identify the 42-kDa protein, its amino-terminal sequence (MRIDGHRQVVSNATAQPGXLLR) was subjected to a tfasta search of sequences generated by the Pseudomonas Genome Project. This analysis revealed an ORF possessing 100% identity with the amino terminus of the 42-kDa protein. Homology to the calmodulin-activated adenylate cyclases of Bordetella pertussis and Bacillus anthracis was found immediately downstream of the amino-terminal sequence, in an alternative reading frame. The proximity of the 42-kDa amino-terminal sequence to the region encoding adenylate cyclase homology suggested that they were the same protein and that an error in this early release of genomic sequence may account for the difference in the reading frame. A chromosomal bank from P. aeruginosa strain 388 was constructed and screened with a specific probe amplified from genomic sequences. Clones that hybridized with the probe were subjected to nucleotide sequence analysis.
Nucleotide sequence analysis identified a 1,134-bp ORF (378 residues) encoding a protein with a predicted molecular mass of 41,673 Da, termed ExoY. The deduced amino-terminal sequence of ExoY and the amino-terminal sequence of the secreted 42-kDa protein were identical (data not shown). The lack of amino-terminal processing, the absence of an amino-terminal signal sequence, and the presence of an ExsA-binding site located immediately upstream of ExoY support the notion that ExoY is a member of the ExoS regulon and is coordinately regulated and secreted by a type III mechanism (2, 16, 17).
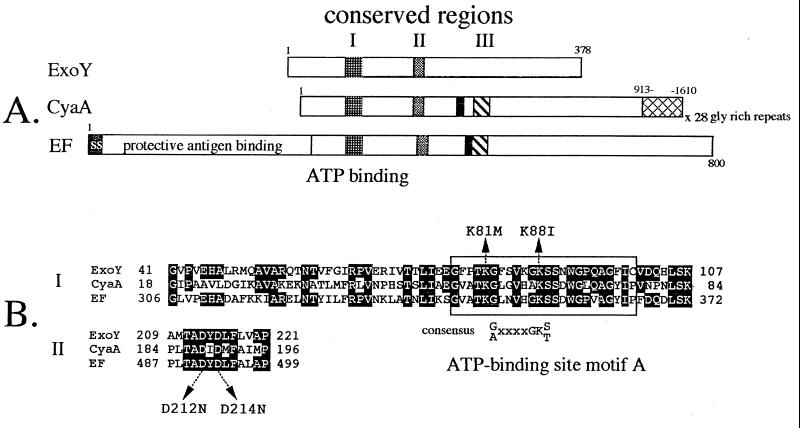
Nucleotide sequence analysis confirmed that the amino terminus of ExoY and the region of adenylate cyclase homology were in the same reading frame. bestfit and pileup alignments of ExoY with the Bordetella pertussis (CyaA) and Bacillus anthracis (EF) adenylate cyclases identified homologous regions (conserved regions I and II) extending from residues 41–107 and 209–221 of ExoY (Fig. 1). Both of these regions have previously been identified as areas of high homology between CyaA and EF (18, 19). Conserved region I contains an ATP/GTP-binding site A motif, which is thought to play a role in contacting the α-phosphate of bound nucleotide (20). Conserved region II, common to ExoY, CyaA, and EF, shares identity with a wide range of nucleotide-binding proteins including phosphofructose kinase, pyruvate kinase, and thymidylate kinase and is proposed to participate in contacting the β- and γ-phosphates of bound nucleotide (20). A third short stretch of homology, conserved region III, which has been described for CyaA and EF, is not present in ExoY (Fig. 1A). CyaA and EF each possess a binding site for calmodulin, a eukaryotic protein that stimulates (CyaA) or is required to activate (EF) adenylate cyclase activity. Significant homology to the calmodulin-binding domains of CyaA and EF was not detected in ExoY.
Figure 1.
(A) Schematic representation of the ExoY, CyaA, and EF adenylate cyclases. The positions of conserved regions I, II, and III are shown. Solid boxes represent the calmodulin-binding domains of CyaA and EF. The glycine-rich repeats of CyaA (hatched), signal sequence (ss), and protective antigen-binding site of EF are labeled. (B) pileup alignment of conserved regions I and II. Residues within CyaA and EF that are homologous to ExoY are shaded. The position of ATP-binding site motif A and its consensus sequence are shown. Residues of ExoY altered by site-directed mutagenesis are indicated by arrows.
Expression and Purification of rExoY.
rExoY was expressed as a histidine-tagged fusion protein in E. coli pETexoY and purified from bacterial lysates by using Ni2+ affinity chromatography. Following elution from the Ni2+ affinity column, however, rExoY had a tendency to aggregate. In contrast to other secreted effectors of the ExoS regulon (ExoS, ExoT, and ExoU), which encode no cysteine residues, ExoY possesses five cysteines. rExoY aggregated when dialyzed in the absence of reducing agents, but remained soluble when dialysis was performed in the presence of DTT or 2-mercaptoethanol (data not shown). rExoY and all site-directed mutant proteins were stored and assayed for adenylate cyclase activity in the presence of 2 mM DTT.
rExoY Possesses Adenylate Cyclase Activity.
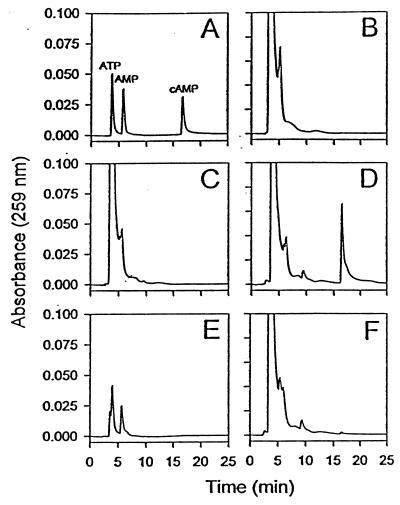
To determine whether rExoY possessed adenylate cyclase activity, purified rExoY (1 μM) was incubated in the presence of ATP and assay buffer at 30°C for 4 hr. Formation of cAMP was monitored by reverse-phase HPLC and calculated as a percentage of the original ATP substrate (Fig. 2). Standard solutions containing ATP, AMP, or cAMP were used to calculate retention times of 3.6, 5.7, and 16.7 min., respectively (Fig. 2A). Reaction mixtures containing rExoY catalyzed the formation of a product that eluted with authentic cAMP (Fig. 2D). This product was not detected when rExoY was heated to 100°C for 5 min prior to the assay (data not shown) or when reactions were treated with cAMP-5′:3′-phosphodiesterase (Fig. 2F). Treatment of ATP and cAMP standards with phosphodiesterase resulted in the conversion of cAMP to AMP (Fig. 2E). Formation of cAMP was not detected in control reactions lacking enzyme (Fig. 2B) or containing a vector control extract from BL21pLysS (Fig. 2C). When GTP was substituted for ATP in the standard reaction, formation of cGMP was not detected, suggesting that under the standard assay conditions, ATP is the preferred substrate for ExoY (data not shown). These data confirm that rExoY possesses adenylate cyclase activity.
Figure 2.
Reverse-phase HPLC analysis of cAMP production. The elution profile of ATP, AMP, and cAMP (30 nmol) is shown in A, standard reactions lacking enzyme in B, reactions containing a control extract from BL21pLysS in C, and 1.0 μM rExoY in D. ATP and cAMP (30 nmol) treated with 3′:5′-cyclic nucleotide phosphodiesterase is shown in E. A reaction identical to that seen in D treated with 5 μg of 3′:5′-cyclic nucleotide phosphodiesterase for 10 min at 30°C is shown in F.
Several residues have been shown to be essential for the adenylate cyclase activity of CyaA (21–24). Two residues (Lys-81 and Lys-88) located within conserved region I are predicted to be directly involved in ATP binding (Fig. 1B). Two additional residues (Asp-212 and Asp-214), also thought to play a role in contacting bound nucleotide, are located within conserved region II (Fig. 1B) (20). All four residues are conserved among EF, CyaA, and ExoY. To determine whether these four residues were required for the adenylate cyclase activity of ExoY, each was altered by site-directed mutagenesis. Lys-81 was changed to Met (K81M), Lys-88 to Ile (K88I), and Asp-212 and -214 were changed to asparagines (D212N and D214N). An additional clone was constructed to move the histidine tag from the amino-terminal position (rExoY) to a carboxyl-terminal position (r23ExoY). Of all of the recombinants tested, only rExoY and r23ExoY possessed detectable adenylate cyclase activity (6.7 ± 2.7 and 4.5 ± 0.9 nmol of cAMP/min⋅mg−1 of ExoY, respectively). These data indicate that each of the four conserved residues is essential for the adenylate cyclase activity of ExoY.
rExoY Adenylate Cyclase Activity Is Stimulated by a Eukaryotic Factor.
Calmodulin is absolutely required for EF catalytic activity (25). Furthermore, the basal adenylate cyclase activity of CyaA is stimulated 500- to 1000-fold by calmodulin (26). To determine whether calmodulin stimulated the adenylate cyclase activity of ExoY, 1 μM rExoY or r23ExoY was incubated in the absence or presence of 0.1 μM, 1 μM, or 10 μM calmodulin in a standard assay. Stimulation of adenylate cyclase activity was undetectable (<2-fold) at all three concentrations of calmodulin tested (data not shown). These data suggest that calmodulin does not play a role in the stimulation of ExoY adenylate cyclase activity.
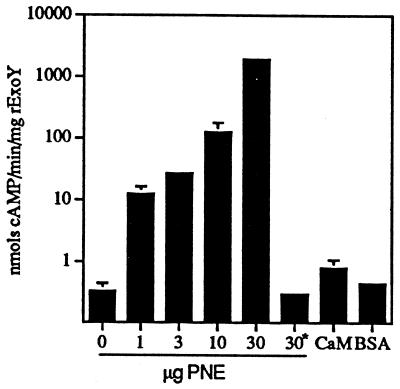
To determine whether another eukaryotic protein stimulated the adenylate cyclase activity of ExoY, a crude extract of cytosolic proteins (PNE) was prepared from CHO cells. In the absence of rExoY, PNE possessed no detectable adenylate cyclase activity (data not shown). In contrast, the addition of PNE (1–30 μg) to rExoY (1 μM) stimulated adenylate cyclase activity in a dose-dependent manner (Fig. 3). PNE heated to 100°C for 5 min possessed no stimulatory activity, suggesting that the factor is proteinaceous in nature. An irrelevant protein, BSA, did not stimulate adenylate cyclase activity (Fig. 3). Comparison of the stimulatory activity of three independently prepared extracts indicated that adenylate cyclase activity was increased at least 500-fold. The catalytic activity of the site-specific mutants was not improved by the addition of PNE, except for rExoYK88I, which now expressed 0.1% the PNE-stimulated activity of rExoY.
Figure 3.
A eukaryotic factor stimulates rExoY adenylate cyclase activity. rExoY (1.0 μM) was assayed for adenylate cyclase activity in the presence or absence of PNE from CHO cells for 30 min under standard conditions. ∗ indicates that PNE was heated to 100°C for 5 min prior to addition to the assay mixture. Calmodulin (CaM, 10 μM) or 50 μg/ml BSA did not stimulate rExoY adenylate cyclase activity.
In Vivo Effect of ExoY.
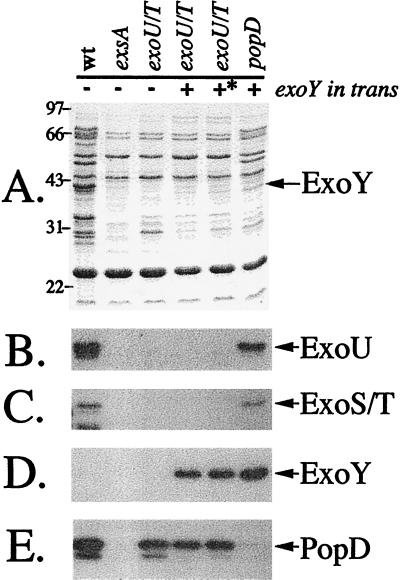
Based on the model of type III secretion, ExoY is predicted to be directly translocated by adherent P. aeruginosa to the cytosol of target cells, resulting in the elevation of intracellular cAMP levels. To address this hypothesis, we took advantage of P. aeruginosa strain PA103, which naturally lacks ExoS and ExoY expression (ref. 27; data not shown). To eliminate effects from other known secreted effector proteins, targeted mutations were introduced into the exoU and exoT loci, resulting in expression strain PA103ΔexoUexoT∷Tc. An independent chromosomal mutation was also introduced into a gene encoding the putative translocator protein PopD, PA103popD∷Ω (16). Plasmids expressing native ExoY or a site-directed mutant of ExoYK81M, which possessed no detectable adenylate cyclase activity, were constructed and transformed into PA103ΔexoUexoT∷Tc or PA103popD∷Ω. Transformants were cultured under inducing conditions for ExoS production, and extracellular protein profiles were examined by SDS/PAGE (Fig. 4A) and Western blot analysis (Fig. 4 B–E). Wild-type PA103 produces and secretes ExoU, ExoT, and PopD, but not ExoY. Insertional inactivation of the transcriptional activator exsA results in a generalized repression of the entire type III system (absence of ExoU, ExoT, and PopD). The specific mutations in the effector proteins ExoU and ExoT eliminate their expression but have no effect on the translocation protein PopD. ExoY and the catalytically inactive mutant K81M (∗) are synthesized and secreted from PA103ΔexoUexoT∷Tc. The translocation mutant PA103popD∷Ω makes no PopD but is competent for synthesis and secretion of ExoU, ExoT, and ExoY (in trans).
Figure 4.
Extracellular protein profiles of ExoY-expressing strains. P. aeruginosa strains (as in Table 1; wt, PA103; exsA, PA103exsA∷Ω; exoU/T, PA103ΔexoUexoT∷Tc; popD, PA103popD∷Ω) were grown to an OD540 between 4.0 and 5.0, and extracellular fractions were prepared. A is a Coomassie-stained gel of concentrated supernatants. Molecular-mass markers (in kDa) are labeled on the left of A. B–E are immunoblots of gels identical to A probed with antisera against ExoU, ExoS/T, ExoY, and PopD, respectively. For ExoY expression in strain PA103, ExoY was amplified with its native promoter and cloned into pUCP18. ∗ denotes the expression of a catalytically (K81M) inactive form of ExoY.
To determine the in vivo effect of ExoY expression, CHO cells were infected with P. aeruginosa strains for 225 min. CHO cells were lysed and assayed for cAMP. Comparison of uninfected CHO cells to those infected with the vector control strain (PA103ΔexoUexoT∷Tc, pUCP18) demonstrated that the parental strain has no significant effect on the morphology of CHO cells or levels of cAMP (Table 1). In contrast, following infection with an ExoY-expressing strain, CHO cells showed a rounded morphology that correlated to increased cAMP levels. The changes in morphology and cAMP levels were not detected following infection with the catalytically inactive K81M mutant of ExoY (Fig. 5). As a positive control, the morphological responses of the CHO cells to heat-labile enterotoxin (LT) and pertussis toxin (PT) were determined (28). Intoxication with LT or PT elicited an elongation response or a clustering response, respectively (data not shown). The differential morphological response of CHO cells to ExoY relative to LT or PT is unclear, but may involve the magnitude of the change in intracellular cAMP levels or may be related to differences in the subcellular distribution of each toxin.
Table 1.
Elevation of cAMP levels in P. aeruginosa-infected CHO cells
| P. aeruginosa strain | Plasmid | cAMP, pmol* | Morphology change† |
|---|---|---|---|
| Uninfected | NA | <0.5 | − |
| PA103ΔexoUexoT∷Tc | pUCP | <0.5 | − |
| PA103ΔexoUexoT∷Tc | pUCPexoY | 136.9 ± 4.5 | + |
| PA103ΔexoUexoT∷Tc | pUCPexoY K81M‡ | <0.5 | − |
| PA103popD∷Ω | pUCPexoY | <0.5 | − |
| PA103ΔexoUexoT∷Tc | pUCPexoY | 95.8 ± 16.8§ | NA |
| PA103ΔexoUexoT∷Tc | pUCPexoY | <0.5¶ | + |
NA, not applicable.
Data are reported as pmol of cAMP normalized to 417 μg of cellular protein per well.
CHO cell morphology change is defined as the change from an elongated triangular shape to a rounded morphology.
K81M is a catalytically inactive mutant of ExoY.
Cells were pretreated with cytochalasin D for 30 min prior to infection.
CHO cell lysate was treated with 5′:3′-cAMP phosphodiesterase prior to assaying for cAMP.
Figure 5.
Kinetics of cAMP accumulation in infected CHO cells. CHO cells were infected with PA103ΔexoUexoT∷Tc expressing either native ExoY or catalytically inactive ExoYK81M. At the indicated times, lysate fractions were prepared and assayed for cAMP. Values are the average of three wells and reported as pmols of cAMP per 144 μg of cellular protein per well.
After infection with the popD∷Ω-expressing ExoY, no changes in CHO cell morphology or cAMP levels were detected, indicating that delivery of ExoY depended on type III-mediated translocation. Pretreatment of CHO cells with cytochalasin D, which prevents bacterial invasion, followed by infection with an ExoY-expressing strain resulted in elevated cAMP levels, suggesting that extracellular bacteria are able to mediate ExoY intoxication. Treatment of the CHO cell lysate with phosphodiesterase prevented detection of cAMP, which confirmed the product specificity of ExoY. A kinetic analysis of cAMP accumulation in CHO cells demonstrated a rapid increase in cAMP after 100 min of infection with P. aeruginosa expressing ExoY (Fig. 5). These data suggest that ExoY is directly translocated from the bacterial cell to the eukaryotic cytosol and that elevated cAMP levels are associated with the rounded morphology of the CHO cells. Furthermore, these data demonstrate that the elevation of cAMP is directly attributable to the adenylate cyclase activity of ExoY.
DISCUSSION
The identification of ExoY represents the discovery of a unique bacterial protein possessing adenylate cyclase activity. Two properties distinguish ExoY from the adenylate cyclases of Bordetella pertussis and Bacillus anthracis. In this article, we demonstrate that ExoY is an adenylate cyclase delivered into eukaryotic cells via a type III translocation mechanism. Evidence to support this conclusion includes the coordinate regulation and secretion of ExoY by the ExoS regulon of P. aeruginosa. In previous studies, this regulon has been shown to encode type III secretion, translocation, and effector proteins (2, 15, 16). The amino-terminal region in ExoY is not processed during secretion, and the carboxyl-terminal region does not contain a glycine-rich repeat region; these data eliminate type II and type I mechanisms of secretion. Infection of eukaryotic cells with P. aeruginosa strains, producing catalytically active ExoY, results in an elevation of intracellular cAMP and cell-morphology changes. Cell-morphology changes or cAMP accumulation were not observed when the type III translocation apparatus was compromised by mutation (popD∷Ω), when a catalytically inactive form of ExoY was provided in trans, or when rExoY was added directly to cells (data not shown). These data are consistent with a type III translocation mechanism mediating the intoxication of cells by ExoY.
Each of the known adenylate cyclase toxins is secreted from the bacterium and enters host cells by a distinct mechanism. ExoY enters cells by a translocation mechanism that is coupled to type III secretion from P. aeruginosa. CyaA of Bordetella pertussis is secreted by a type I mechanism for which α-hemolysin of E. coli serves as the prototypical protein. Like α-hemolysin, CyaA possesses glycine-rich carboxy-terminal repeat sequences that are required for binding to host cells, hemolytic activity, and subsequent translocation of the catalytic domain to the eukaryotic cytosol (29). Bacillus anthracis-encoded EF is secreted by the general secretory pathway (type II) and enters cells by receptor-mediated endocytosis (30). The entry of EF depends on the expression and secretion of protective antigen, which serves as the binding subunit of this toxin. Unlike CyaA and EF, which are secreted and may exert their effects at sites distal from the site of infection, the mechanism of delivery to host cells limits ExoY intoxication to sites of colonization. The production of ExoY may play a role in the protection of the bacterium from local phagocytic cells. Both CyaA and EF have been shown to suppress immune function (31–34).
A second property of ExoY is the stimulation of adenylate cyclase activity by a eukaryotic factor that is distinct from calmodulin. Calmodulin is absolutely required for the detection of adenylate cyclase activity from EF and stimulates the basal activity of CyaA 500- to 1000-fold (25, 26). Both CyaA and EF possess domains that are required for calmodulin binding, and peptides derived from these regions have been shown to directly bind calmodulin (35, 36). In contrast, ExoY lacks homology with the CyaA/EF calmodulin-binding domains, and the addition of calmodulin to enzyme assays has only a marginal effect on activity. We found that the basal activity of ExoY was stimulated at least 500-fold by the addition of PNE from CHO cells and when heated, this extract failed to activate ExoY. In addition, high levels of cAMP were detected when CHO cells were infected with P. aeruginosa strains expressing ExoY. Taken together, these observations indicate that a eukaryotic protein distinct from calmodulin stimulates the adenylate cyclase activity of ExoY. The stimulation of ExoY adenylate cyclase activity by a eukaryotic cofactor may represent a regulatory strategy to prevent the generation of cAMP within the bacterium before intoxication of host cells. This regulatory strategy can also be applied to ExoS and ExoT, which demonstrate an absolute requirement for FAS to express ADP ribosyltransferase activity under physiological conditions. The discovery of ExoY and the eventual identification of the eukaryotic activator will aid in the understanding of the relationship between type III secretion, epithelial damage, and bacterial dissemination.
Acknowledgments
We are grateful to the Pseudomonas Genome Project for the release of sequence information critical to the discovery of ExoY. This work was supported by Grants AI31665 (to D.W.F), AI01289 (to D.W.F.), and AI30162 (to J.T.B.) from the National Institute of Allergy and Infectious Diseases.
ABBREVIATIONS
- CyaA
adenylate cyclase from Bordetella pertussis
- EF
edema factor adenylate cyclase from Bacillus anthracis
- CHO
Chinese hamster ovary
- ExoS
exoenzyme S
- PNE
postnuclear extract
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF061745).
References
- 1.Bodey G P, Bolivar R, Fainstein V, Jadeja L. Rev Infect Dis. 1983;5:279–313. doi: 10.1093/clinids/5.2.279. [DOI] [PubMed] [Google Scholar]
- 2.Frank D W. Mol Microbiol. 1997;26:621–629. doi: 10.1046/j.1365-2958.1997.6251991.x. [DOI] [PubMed] [Google Scholar]
- 3.Yahr T L, Frank D W. J Bacteriol. 1994;176:3832–3838. doi: 10.1128/jb.176.13.3832-3838.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coburn J. Curr Top Microbiol Immunol. 1992;175:133–143. doi: 10.1007/978-3-642-76966-5_7. [DOI] [PubMed] [Google Scholar]
- 5.Knight D A, Barbieri J T. Infect Immun. 1997;65:3304–3309. doi: 10.1128/iai.65.8.3304-3309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganesan A K, Frank D W, Misra R P, Schmidt G, Barbieri J T. J Biol Chem. 1998;273:7332–7337. doi: 10.1074/jbc.273.13.7332. [DOI] [PubMed] [Google Scholar]
- 7.Finck-Barbancon V, Goranson J, Zhu L, Sawa T, Wiener-Kronish J P, Fleiszig S M J, Wu C, Mende-Mueller L, Frank D W. Mol Microbiol. 1997;25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 8.Vogel H J, Bonner D M. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 9.Olsen R H, Debusscher G, McCombie W R. J Bacteriol. 1982;150:60–69. doi: 10.1128/jb.150.1.60-69.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank D W, Iglewski B H. J Bacteriol. 1991;173:6460–6468. doi: 10.1128/jb.173.20.6460-6468.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yahr T L, Barbieri J T, Frank D W. J Bacteriol. 1996;178:1412–1419. doi: 10.1128/jb.178.5.1412-1419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank D W, Nair G, Schweizer H P. Infect Immun. 1994;62:554–563. doi: 10.1128/iai.62.2.554-563.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stocchi V, Cucchiarini L, Magnani M, Chiarantini L, Palma P, Crescentini G. Anal Biochem. 1985;146:118–124. doi: 10.1016/0003-2697(85)90405-1. [DOI] [PubMed] [Google Scholar]
- 14.Xu Y, Barbieri J T. Infect Immun. 1995;63:825–832. doi: 10.1128/iai.63.3.825-832.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yahr T L, Goranson J, Frank D W. Mol Microbiol. 1996;22:991–1003. doi: 10.1046/j.1365-2958.1996.01554.x. [DOI] [PubMed] [Google Scholar]
- 16.Yahr T L, Mende-Mueller L M, Friese M B, Frank D W. J Bacteriol. 1997;179:7165–7168. doi: 10.1128/jb.179.22.7165-7168.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hovey A K, Frank D W. J Bacteriol. 1995;177:4427–4436. doi: 10.1128/jb.177.15.4427-4436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Escuyer V, Duflot E, Sezer O, Danchin A, Mock M. Gene. 1988;71:293–298. doi: 10.1016/0378-1119(88)90045-5. [DOI] [PubMed] [Google Scholar]
- 19.Mock M, Labruyere E, Glaser P, Danchin A, Ullmann A. Gene. 1988;64:277–284. doi: 10.1016/0378-1119(88)90342-3. [DOI] [PubMed] [Google Scholar]
- 20.Glaser P, Munier H, Gilles A-M, Krin E, Porumb T, Barzu O, Sarfati R, Pellecuer C, Danchin A. EMBO J. 1991;10:1683–1688. doi: 10.1002/j.1460-2075.1991.tb07692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glaser P, Elmaoglou-Lazaridou A, Krin E, Ladant D, Barzu O, Danchin A. EMBO J. 1989;8:967–972. doi: 10.1002/j.1460-2075.1989.tb03459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labruyere E, Mock M, Surewicz W K, Mantsch H H, Rose T, Munier H, Sarfati R S, Barzu O. Biochemistry. 1991;30:2619–2624. doi: 10.1021/bi00224a008. [DOI] [PubMed] [Google Scholar]
- 23.Xia Z G, Storm D R. J Biol Chem. 1990;265:6517–6520. [PubMed] [Google Scholar]
- 24.Munier H, Bouhss A, Krin E, Danchin A, Gilles A M, Glaser P, Barzu O. J Biol Chem. 1992;267:9816–9820. [PubMed] [Google Scholar]
- 25.Leppla S H. Proc Natl Acad Sci USA. 1982;79:3162–3166. doi: 10.1073/pnas.79.10.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolff J, Cook G H, Goldhammer A R, Berkowitz S A. Proc Natl Acad Sci USA. 1980;77:3841–3844. doi: 10.1073/pnas.77.7.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fleiszig S M J, Wiener-Kronish J P, Miyazaki H, Vallas V, Mostov K E, Kanada D, Sawa T, Benedict Yen T S, Frank D W. Infect Immun. 1997;65:579–586. doi: 10.1128/iai.65.2.579-586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hewlett E L, Sauer K T, Myers G A, Cowell J L, Guerrant R L. Infect Immun. 1983;40:1198–1203. doi: 10.1128/iai.40.3.1198-1203.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon V M, Leppla S H, Hewlett E L. Infect Immun. 1988;56:1066–1069. doi: 10.1128/iai.56.5.1066-1069.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon V M, Young W W, Jr, Lechler S M, Gray M C, Leppla S H, Hewlett E L. J Biol Chem. 1989;264:14792–14796. [PubMed] [Google Scholar]
- 31.Confer D L, Eaton J W. Science. 1982;217:948–950. doi: 10.1126/science.6287574. [DOI] [PubMed] [Google Scholar]
- 32.O’Brien J, Friedlander A, Dreier T, Ezzell J, Leppla S. Infect Immun. 1985;47:306–310. doi: 10.1128/iai.47.1.306-310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wade B H, Wright G G, Hewlett E L, Leppla S H, Mandell G L. Proc Soc Exp Biol Med. 1985;179:159–162. doi: 10.3181/00379727-179-42078. [DOI] [PubMed] [Google Scholar]
- 34.Pearson R D, Symes P, Conboy M, Weiss A A, Hewlett E L. J Immunol. 1987;139:2749–2754. [PubMed] [Google Scholar]
- 35.Craescu C T, Bouhuss A, Mispelter J, Diesis E, Popescu A, Chiriac M, Barzu O. J Biol Chem. 1995;270:7088–7096. doi: 10.1074/jbc.270.13.7088. [DOI] [PubMed] [Google Scholar]
- 36.Munier H, Blanco F J, Precheur B, Diesis E, Nieto J L, Craescu C T, Barzu O. J Biol Chem. 1993;268:1695–1701. [PubMed] [Google Scholar]