SUMMARY
Serving as the primary conduit for communication between the nucleus and the cytoplasm, nuclear pore complexes (NPCs) impact nearly every cellular process. The extent to which NPC composition varies and the functional significance of such variation in mammalian development have not been investigated. Here we report that a null allele of mouse nucleoporin Nup133, a structural subunit of the NPC, disrupts neural differentiation. We find that expression of Nup133 is cell type and developmental stage restricted, with prominent expression in dividing progenitors. Nup133-deficient epiblast and ES cells abnormally maintain features of pluripotency and differentiate inefficiently along the neural lineage. Neural progenitors achieve correct spatial patterning in mutant embryos; however, they are impaired in generating terminally differentiated neurons, as are Nup133-null ES cells. Our results reveal a role for structural nucleoporins in coordinating cell differentiation events in the developing embryo.
INTRODUCTION
Following implantation the mammalian embryo initiates a succession of morphogenetic events, coordinating growth with cell movement and differentiation to generate an organism comprised of correctly positioned and functional tissues. Key among these morphogenetic events is gastrulation. In mouse, the early gastrulating embryo consists of rapidly proliferating, pluripotent epiblast cells. These undergo various processes, including restriction of their differentiation potential, to form three distinct lineages of proliferating progenitor cells: ectoderm, mesoderm, and endoderm. The differentiation potential of these progenitors becomes further restricted as they give rise to different sub-lineages of dividing precursors, which ultimately will produce the non-proliferative, terminally differentiated cells present in all organ systems at birth (Tam et al., 2006). For example, upon receipt of signals emanating from extraembryonic tissues, and subsequently from embryonic mesendoderm, epiblast cells proliferate and undergo neural induction to generate the neuroepithelium, a population of neural-restricted progenitor cells, often referred to as the embryonic neural stem cell compartment (Gotz and Huttner, 2005; Stern, 2006). The neural progenitors continue to proliferate through successive stages of maturation, until they exit the cell cycle and generate several subtypes of terminally differentiated neurons and glia.
Interpretation of developmental signals requires nucleocytoplasmic transport: the import of transcriptional regulators and the export of mRNAs. All such exchanges between the cytoplasm and the nucleus occur through nuclear pore complexes (NPCs), macromolecular assemblies conserved in architecture from yeast to mammals. A NPC consists of approximately 30 distinct proteins (nucleoporins, Nups), each present in multiple copies (Tran and Wente, 2006). Current models of nucleocytoplasmic transport invoke interactions between mobile transport receptors (primarily the importin and exportin karyopherins) and a subset of nucleoporins, the FG-Nups which harbor repetitive stretches of Phe-Gly residues. Recent studies hint at distinct roles for different FG-Nups in protein and mRNA transport (Sabri et al., 2007) (Terry and Wente, 2007), as well as at possible non-transport-related functions of NPC constituents (Kalverda and Fornerod, 2007) (Taddei, 2007).
Current knowledge of NPC structure and function derives largely from cell biological, structural, and genetic studies in readily manipulated systems, such as yeast, Xenopus nuclear extracts, and cultured mammalian cells. To evaluate this knowledge in the context of a developing organism, targeted null mutations have been introduced into a small number of mouse nucleoporin genes. Loss-of-function mutations have been reported for six different nucleoporins and all resulted in embryonic lethality (Tran and Wente, 2006). Consistent with depletion of maternal stores of a protein essential for cell viability, null mutations in three of these Nups - CAN/Nup214, Rae1/Gle2, and Elys – led to developmental arrest at implantation (Babu et al., 2003; Okita et al., 2004; van Deursen et al., 1996). Loss of Nup98 arrested embryonic development during early gastrulation (Wu et al., 2001) and loss of Nup50 led to embryonic death during late gestation (Smitherman et al., 2000). The stage of embryonic death was not reported for the targeted disruption of Nup96 (Faria et al., 2006). It remains unknown why deficiencies in these individual components of the pore engendered such distinct developmental phenotypes.
We identified the mermaid (merm) mutant in an N-ethyl-N-nitrosourea (ENU) mutagenesis screen for recessive mutations disrupting mouse gastrulation (Garcia-Garcia et al., 2005). The merm mutation proved to be a functional null allele of nucleoporin Nup133, a constituent of the conserved Nup107-160 complex. We find that mouse embryos normally express Nup133 in a cell-type and stage-specific pattern and that NPCs can appropriately assemble in the absence of Nup133. However, under conditions that promote neural differentiation, embryonic stem (ES) and epiblast cells lacking Nup133 abnormally maintain features of an earlier progenitor cell and differentiate inefficiently. Our results uncover an unanticipated requirement for a distinct composition of the NPC during cell differentiation in the developing mouse embryo.
RESULTS
A null allele of nucleoporin Nup133 underlies the merm phenotype
The merm mutation resulted in lethality at mid-gestation (e9.5–e10.5), likely due to circulatory defects indicated by the presence of pericardial edema. At this stage merm embryos displayed a kinked and exencephalic neural tube, a shortened trunk region with irregularly segmented somites, and a thin, pointed primitive streak/tail bud (Figure 1A). Intrauterine growth retardation always accompanied the morphological phenotype. The expressivity of the phenotype was variable between mouse strains and within the same strain background (Figure 1B).
Figure 1. merm mutant phenotype and characterization of the merm allele.
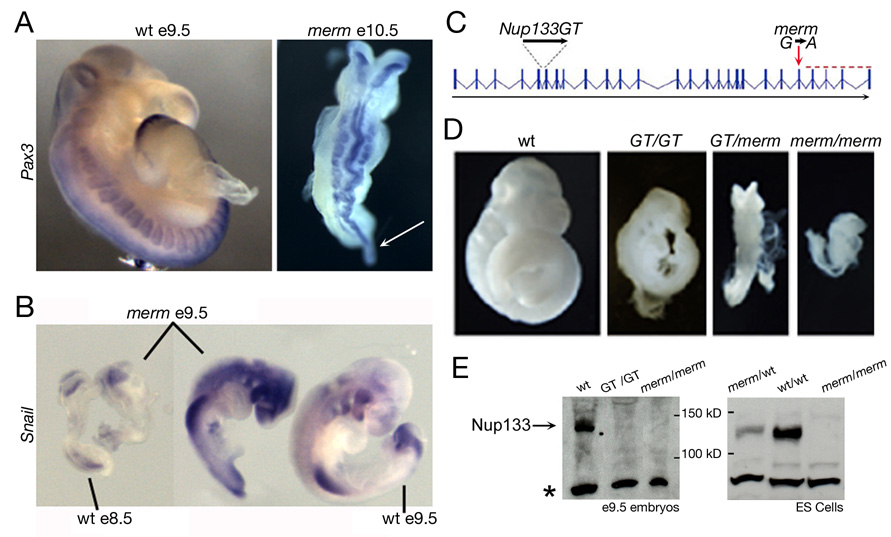
(A) Pax3 whole mount in situ hybridization revealed developmental delay and severely dysmorphic neural tube and somites in the merm e10.5 embryo compared to a wild-type embryo at e9.5. The arrow indicates the distended primitive streak/tail bud. (B) A comparison of e9.5 merm embryos to wild-type embryos at e8.5 and e9.5, hybridized to Snail - a marker of cephalic neural crest and limb mesenchyme - demonstrated the variability of the merm phenotype. (C) The merm mutation in intron 22 (red arrow) led to a C-terminal truncation of the Nup133 protein (red dashed line); the LacZ gene trap (RRK090; GT) inserted into intron 5 of Nup133. (D) The Nup133GT allele failed to complement the merm allele. All embryos are at e10.5. (E) Extracts prepared from e9.5 embryos and ES cells of the indicated genotypes were analyzed by Western blot using a polyclonal serum raised against human Nup133. Note the lack of detectable Nup133 in the merm embryos and ES cells. A faint band in the GT/GT embryos was consistent with low level expression of the full length Nup133 transcript. The non-specific lower band (*) showed comparable loading.
In a backcross panel of 2051 recombination opportunities between C57Bl/6J and C3HeB/FeJ, we mapped the merm mutation to a 0.6 Mb interval on distal chromosome 8 that contained seven transcription units, including the Nup133 gene (Figure S1A). Nucleotide sequencing of the exons in the interval identified a G to A transition at the first base of intron 22 in Nup133 (Figure 1C). RT-PCR analysis (Figure S1B) showed that exon 21 was spliced directly onto exon 23, causing a frameshift that generated a premature stop codon (Figure S1C). A second mutant allele of Nup133, which carries a LacZ gene trap insertion in intron five, Nup133GT, failed to complement the Nup133merm allele in compound heterozygous Nup133merm/GT e9.5 embryos (Figure 1C, D), confirming that loss of Nup133 function caused the merm phenotype. Nup133GT/GT embryos exhibited a phenotype weaker than that of Nup133merm/merm mutants, while the Nup133merm/GT trans-heterozygotes showed an intermediate phenotype, suggesting that the Nup133GT allele supported the production of a small amount of protein. Consistent with this prediction, RT-PCR, using primers from exons flanking the gene trap, revealed a splice that deleted the LacZ insertion and generated a low level of wild-type transcript (Figure S1D).
The Nup133merm mutant allele was predicted to encode a truncated form of Nup133 (mNup133ΔC) lacking the COOH-terminal 162 amino acids. However, an anti-hNup133 antibody that recognizes a GFP-mNup133ΔC fusion transiently expressed in HeLa cells (Figure S1E), did not detect any signal on Western blots of e9.5 merm embryo and ES cell extracts (Figure 1E). In agreement with previous studies (Boehmer et al., 2003; Boehmer and Schwartz, 2007), this GFP-mNup133ΔC fusion neither interacted with Nup107 (Figure S1E), nor localized to NPCs in HeLa cells (Figure S1F). Together, these data argue that merm embryos expressed a truncated form of the Nup133 protein, which was mis-localized and degraded; thus Nup133merm behaved as a functional null allele.
merm embryos assemble nuclear pore complexes
merm mutants arrested their development after e9.5, indicating that Nup133 was dispensable for early post-implantation development. However, as far as is known, Nup133 functions only within the Nup107-160 complex (Belgareh et al., 2001; Loiodice et al., 2004; Vasu et al., 2001), which is essential for viability of HeLa cells and for NPC assembly upon nuclear reconstitution in Xenopus extracts (Boehmer et al., 2003; Harel et al., 2003; Walther et al., 2003). Therefore, we asked whether NPC formation was compromised in Nup133-deficient tissues. Figure 2A shows our results with MAb414, a diagnostic NPC assembly marker that detects a subset of FG-Nups; with antiserum to Nup153, a peripheral Nup on the nuclear side of the NPC, and with antiserum to Nup107, the direct binding partner of Nup133 in the Nup107-160 complex (Berke et al., 2004). Nuclei in both wild-type and merm neuroepithelium displayed punctate staining predominantly on the nuclear rim for the three different antibodies, indicating that NPCs were assembled in the nuclear envelope in the absence of Nup133. Antibodies specific for other individual nucleoporins (Nup98, CAN/Nup214, Tpr, and Nup50) gave similar staining patterns in wild-type and merm neural tissue (Figure S2). While Nup98, CAN/Nup214, and Tpr staining localized primarily to the nuclear membrane, Nup50 immunofluorescence was dispersed throughout the nucleus, consistent with its role as a mobile nucleoporin involved in transport (Moore, 2003). Thus, the absence of Nup133 did not manifestly impair recruitment of any of the tested nucleoporins to the nuclear envelope in mouse embryonic tissues.
Figure 2. NPC composition in merm neural tissue.

Immunofluorescence using anti-Nup133, anti-Nup107, anti-Nup153, or MAb414 antibodies as indicated: (A) sections of e9.5 neural tube imaged by widefield (Nup133 and MAb414) and confocal microscopy (Nup107 and Nup153); (B) undifferentiated ES cells photographed under a widefield microscope. The portion of the nuclear envelope enclosed within the white rectangle is shown at higher magnification above the panel. Despite the absence of Nup133 in the merm mutant, merm and wild-type cells exhibited similar anti-Nup107, anti-Nup153, and MAb414 punctate staining of nuclear pores. C and D, Western blot analysis detected comparable levels of several nucleoporins in extracts of wild-type and merm mutant cells. (C) wild-type (+/+) and two independent merm (−/−) e9.5 embryos; (D) wild-type (+/+), heterozygous (+/−) and merm (−/−) ES cells. In D, MAb414 antibody was used to detect both Nup153 and p62. In C, β-actin antibody staining served as a loading control.
To allow more detailed analysis of Nup133’s role in NPC assembly, we generated Nup133+/+, Nup133merm/+, and Nup133merm/merm ES cells. As in Nup133-deficient neuroepithelium, all nucleoporins analyzed localized properly in merm ES cells (Figure 2B and data not shown). Furthermore, Western blotting detected comparable levels of several nucleoporins in wild-type and merm e9.5 embryos (Figure 2C) and in ES cell extracts (Figure 2D), indicating that other nucleoporins were present at relatively normal ratios in the absence of Nup133. However, we cannot exclude the possibility that a subset of pores assembled nucleoporins at altered stoichiometries. When cultured under maintenance conditions, merm ES cells expanded at normal rates, and for at least 25 passages, appeared morphologically identical to wild-type ES cells (Figure S3A). Since any maternally-supplied Nup133 would have been exhausted before or shortly after the establishment of the merm ES cell lines, Nup133 functions are dispensable for NPC assembly and cell viability in mouse ES cells and early embryonic tissues. Consistent with their wild-type behavior in culture, merm ES cells exhibited no inherent chromosome instability compared to control ES cells. An examination of metaphase chromosome spreads found a modal chromosome number of 40 in three independent merm and two independent wild-type ES cell lines (Figure S3B; Table S1).
Nup133 expression is cell type-restricted in the mouse embryo
We examined Nup133 transcription in Nup133GT/+ embryos by whole-mount staining for β-galactosidase (β-Gal) activity (Figure 3) and corroborated the results by in situ hybridization to wild-type embryos (Figure S4). Early post-implantation embryos expressed Nup133 exclusively in the epiblast; we detected no expression in extraembryonic tissues (extraembryonic ectoderm and visceral endoderm; Figure 3A). Once gastrulation began, Nup133 expression became more complex. It varied not only between tissues, but also between developmental stages and axial positions within a single tissue (Figure 3B–F). Certain tissues, such as the node at e7.5–e8.0 (Figure 3B–C) and endocardium and hindgut endoderm at e9.5 (Figure 3D–F), lacked any detectable Nup133 expression. Tissues that were among the most dysmorphic in the merm mutant (Figure 1A), neuroepithelium and paraxial mesoderm/somites, expressed high levels of Nup133 (Figure 3D). Such a cell type and stage-specific expression pattern indicated that rather than acting as a core structural component, Nup133 functions as a modulator of NPC activity.
Figure 3. Nup133 expression in the developing embryo.
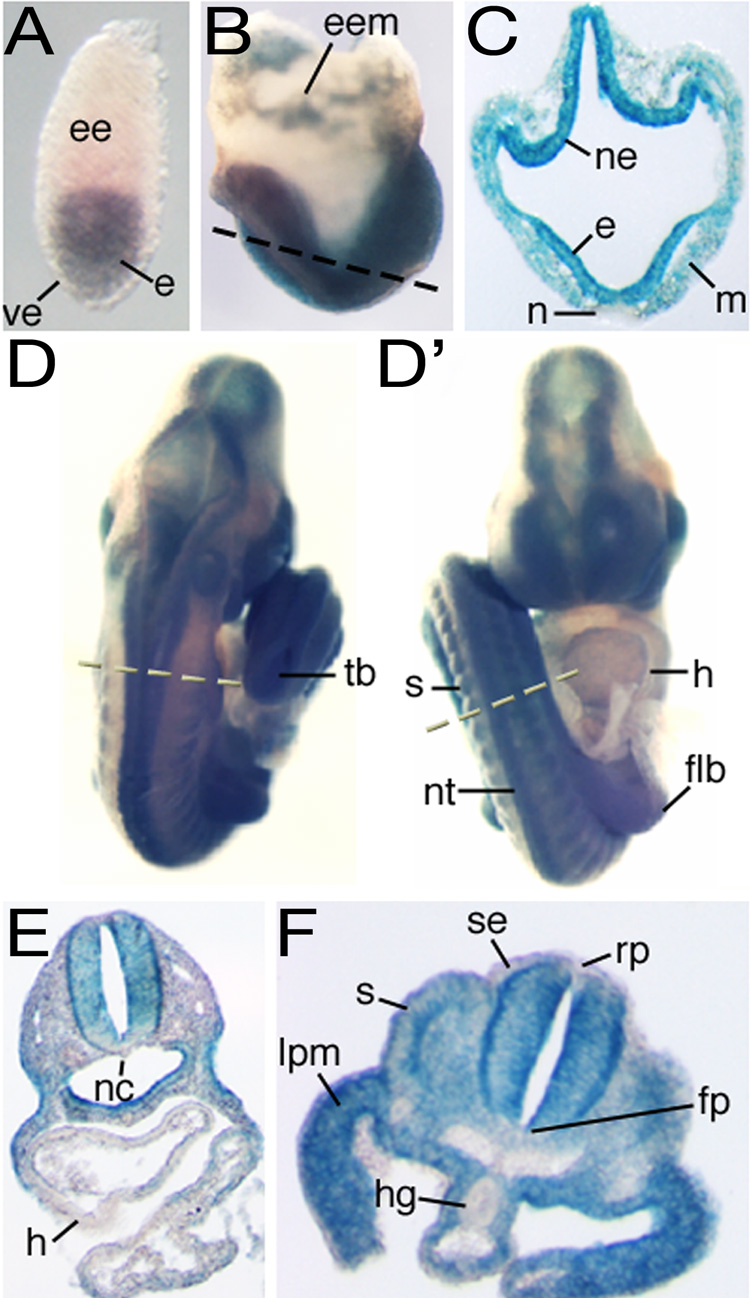
Staining for β-Gal activity in GT/+ embryos at (A) the egg cylinder stage, (B–C) the headfold (0-somite) stage, and (D–F) the forelimb bud stage. D and D' show, respectively, dorsal and ventral views of the same embryo. Dashed lines in B, D and D' indicate the level of the section shown in C, E and F, respectively. e, epiblast; ee, extraembryonic ectoderm; eem, extraembryonic mesoderm; flb, forelimb bud; fp, floor plate; h, heart; hg, hindgut endoderm; lpm, lateral plate mesoderm; m, mesoderm; n, node; nc, notochord; ne, neuroectoderm; nt; neural tube; rp, roof plate; s, somite; se, surface ectoderm; tb, tail bud; ve, visceral endoderm.
merm ES cells fail to contribute uniformly to tissues of chimeric embryos
We generated chimeric embryos containing both wild-type and merm cells to investigate the differentiation potential of Nup133-deficient cells in the developing mouse embryo. We injected LacZ-expressing ES cells into wild-type blastocysts, an experimental configuration in which the donor ES cells do not contribute to extraembryonic ectoderm and endoderm, but only to tissues in the embryo proper and to extraembryonic mesoderm (Bradley and Robertson, 1986; Tam and Rossant, 2003). We assessed the contribution of donor (Nup133+/+, Nup133merm/+, or Nup133merm/merm) ES cells in the resulting chimeric embryos by assaying for β-Gal activity. The findings shown in Figure 4 are representative of those obtained from blastocyst injections with one Nup133+/+, one Nup133merm/+, and two Nup133merm/merm ES cell lines.
Figure 4. merm ES cell contribution in chimeras.
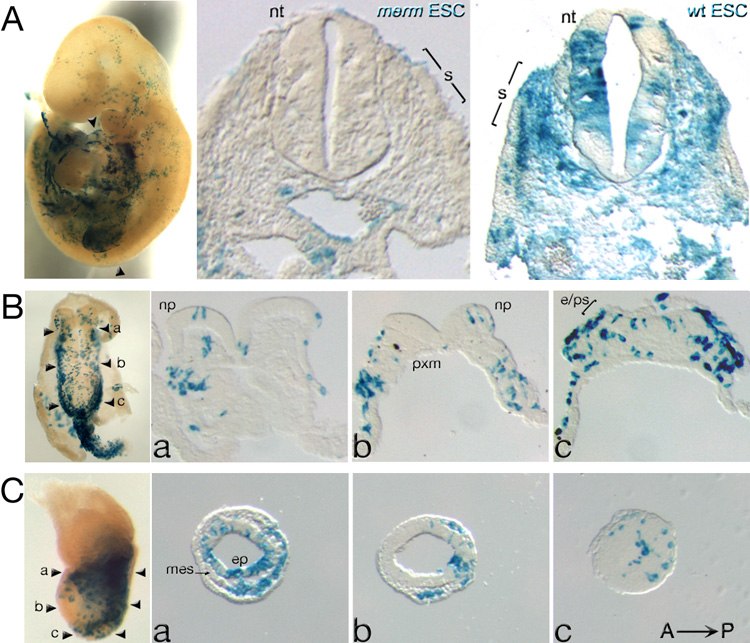
Chimeric embryos dissected at (A) e9.5; (B) e8.0- ventral view, and (C) e7.5. ES cell distribution detected by β-Gal activity (blue) in whole-mount preparations (left panels) and in sections (right panels). (A) Wild-type ES cells contributed uniformly to tissues of e9.5 chimeric embryos – far right panel, whereas merm ES cells were absent from the neural tube (nt) and somites (s) – middle panel. Arrowheads in the left panel indicate the level of the section in the middle panel. (B–C) In e8.0 and e7.5 chimeras, merm ES cells were detected in precursors to the neural tube - the neural plate (np) and anterior/distal epiblast (ep) – and in precursors to the somites – paraxial mesoderm (pxm). In the left most panels of (B) and (C) the arrowheads, labeled a, b, and c, indicate the level of the corresponding sections. mes, mesoderm.
Consistent with Nup133 expression in epiblast but not in extraembryonic ectoderm and endoderm, chimeras with very high contribution of merm ES cells recapitulated the merm phenotype (data not shown). Chimeras with low ES cell contribution displayed distinct differences between the distributions of mutant and control ES cells. Nup133+/+ and Nup133merm/+ ES cells contributed to all tissues at all axial levels. In contrast, in e9.5–e10 chimeras, virtually no merm ES cell derivatives were present in the neural tube and somites (Figure 4A), tissues that normally expressed high levels of Nup133 (Figure 3D) and that were malformed in merm mutants (Figure 1A). Such an outcome suggested an intrinsic inability of merm ES cell derivatives to generate neural and somitic tissues. However, the presence of merm ES cell derivatives in neural tube precursors (the neural plate at e8.0, Figure 4B; anterior ectoderm at e7.5, Figure 4C), pointed instead to a gradual out-competition of mutant cells by wild-type cells as a more likely explanation for the absence of merm cells in the e9.5–e10 neural tube.
Differentiating merm ES cells retain progenitor cell features and fail to efficiently generate neurons in vitro
Of the 37 ES cell lines established from blastocysts isolated from Nup133merm/+ intercrosses, five were wild-type, 13 heterozygous, and 19 (51%) homozygous for the merm allele of Nup133. This was twice the number expected (9; 25%) from a Mendelian distribution of the Nup133merm and Nup133+ alleles. Thus, merm ES cells appeared to have a selective advantage under culture conditions designed to enrich for pluripotent cells. Consistent with this possibility, merm ES cells behaved comparably to normal ES cells under self-renewal conditions in culture, yet not in the context of a chimeric embryo. One possible explanation for these findings posits that differentiating merm ES cells tend to inappropriately maintain an earlier, pluripotent state. For example, in chimeric embryos merm ES cells contributed to early, but not to later stages of neural development which are incompatible with pluripotency. Therefore, we asked whether the lack of Nup133 compromised the ability of ES cells to differentiate along the neural pathway in vitro. When cultured in reduced levels of serum but at concentrations of LIF that support ES cell maintenance, wild-type ES cells produce a mixture of undifferentiated and neural progeny (Ying et al., 2003). Figure 5B shows that under similar culture conditions, merm ES cells generated undifferentiated colonies almost exclusively, based on (a–b) high levels of alkaline phosphatase activity - characteristic of germ cells and undifferentiated ES cells - and on the expression of two transcription factors - (c–d) Oct4 (Pou5f1) and (e–f) Nanog - required for maintenance of pluripotency of epiblast and ES cells (Tam and Loebel, 2007). For example, at day 12 of differentiation, two independent merm ES cell lines contained 80% Oct4-expressing cells (79.8 ± 1.2% & 79.7 ± 1%). In contrast, 14.4 (± 0.44)% of the cells in one wild-type line and 28.2 (± 0.74)% in a second line were Oct4-positive.
Figure 5. merm ES cell differentiation in culture.

(A) Upon LIF withdrawal and addition of retinoic acid (RA), merm colonies generated only small numbers of differentiated neurons marked by TuJ1 expression (green); these had short neurites compared to those in wild-type colonies. Gata4 (red) shows the presence of endodermal cells. (B) After growth in reduced levels of serum, merm ES cell colonies contained mostly alkaline phosphatase-positive (dark blue – b, higher magnification shown in lower panel), Oct4-positive (yellow, d), and Nanog-positive (magenta, f) cells. In contrast, wild-type ES cell colonies generated differentiated cells (unstained, a; green - phalloidin, c; and blue – DAPI, e).
LIF withdrawal and addition of retinoic acid (RA) promote the differentiation of wild-type ES cells into post-mitotic neurons (Ying et al., 2003). Compared to wild-type ES cells, merm ES cells generated approximately seven-fold fewer neurons under these conditions (Figure 5A). The merm ES cells, however, readily expressed Gata4, a marker of extraembryonic endoderm (Figure 5A) (Pfister et al., 2007), arguing that ES cell differentiation into the extraembryonic endoderm lineage does not require Nup133. To confirm impaired neural differentiation of merm ES cells, we applied a third differentiation protocol which involves the growth of ES cell derived embryoid bodies (EBs) in the presence of RA (Wichterle et al., 2002). merm ES cell differentiation was similarly impeded under these conditions (Figure S5A). In one experiment, wild-type EBs contained 23 ± 3% Oct4-positive and 17 ± 2% TuJ1-positive (neuron-specific β-III tubulin) cells; whereas merm EBs consisted of 50 ± 4% Oct4+ and 7 ± 2% TuJ1+ cells (T-test: p < 0.0001 for Oct4; p < 0.0015 for TuJ1). In a second experiment, using different wild type and merm ES cell lines, we detected 11 ± 2% Oct4+ and 25 ± 4% TuJ1 + cells in wild-type and 41 ± 5% Oct4+ and 11 ± 1% TuJ1+ cells in merm EBs (T-test p < 0.0001 for Oct4; p < 0.01 for TuJ1). Although unable to effectively execute terminal differentiation into post-mitotic neurons, the merm ES cells were able to form neural progenitors, identified by the expression and nuclear localization of the proneural transcription factors, Sox1 (Pevny et al., 1998) and Brn2 (Bertrand et al., 2002) (Figure S5B).
merm embryos exhibit defects in neural differentiation
To investigate whether the observed block in merm ES cell differentiation reflected similar defects in vivo in the developing merm embryo, we compared patterns of marker gene expression between mutant and control embryos. Identifying appropriate control embryos was complicated by the general developmental delay displayed by merm mutants (Figure 1B). Also, as somite formation was strongly perturbed in merm, we were unable to use somite number to select developmental stage-matched mutant and control embryos. Therefore, to evaluate whether altered expression of a marker reflected general developmental delay or a more specific impairment, we used both gestational age-matched and one-day earlier wild-type embryos as controls.
The e10.5 merm neural tube contained greatly reduced numbers of post-mitotic neurons expressing TuJ1, even relative to a one-day earlier e9.5 control (Figure 6A, a–b). A second pan-neuronal marker, Map2, verified this finding (Figure 6A, c–d). Expression of Sox1, the earliest known marker of neural precursors, established a neural identity for cells in the merm neuroepithelium (Figure 6A, e–f). The presence of Brn2+ cells indicated that the merm neural tube, like its wild-type counterpart, contained neural precursors at a more mature stage than that of the strictly Sox1+ cell (Figure 6A,c–d). To assess whether the observed defect in neuronal differentiation reflected perturbations in the specification of different subclasses of neural progenitors, we examined the expression of markers for dorsal-ventral patterning of the early neural tube. In most mutants at e10.5, the spatial distribution of markers for neural tube progenitors resembled that observed in age-matched controls: Msx1/2 for roof plate; Shh and Foxa2 for floor plate (and notochord); Pax6, Pax3, Pax7 and Nkx2.2 for dorsal and ventral neuroepithelium, respectively; and Olig2, a marker for precursors to motor neurons, the first neurons to differentiate in the developing neural tube (Jessell, 2000) (Figure 6A, g–h; 6B, a–h; and data not shown). Thus, once specified, merm neural progenitors were able to acquire largely normal positional identities along the dorsal-ventral axis of the neural tube. Next, we assessed the maturation of neural precursors by examining Id1 expression. Id1 is expressed in the epiblast and throughout the neural plate until e8.5. One day later, Id1-expressing neural progenitors segregate into dorsal and ventral domains. This regionalized expression persists until at least e12.5 (Jen et al., 1996, Jen et al., 1997). Accordingly, Id1 expression was already confined to its dorsal and ventral domains in a wild-type e9.5 neural tube (Figure 6A, g); however, it was detected throughout the neuroepithelium of merm embryos as late as e10.5 (Figure 6A, h). Thus, although neural progenitors in the merm embryo were correctly positioned, the persistence of Id1 expression indicated a defect in their maturation.
Figure 6. Neural differentiation in merm embryos.

(A) The e10.5 merm neural tube (b, d) contained fewer post-mitotic neurons (TuJ1- positive, a–b; Map2-positive, c–d), even compared to an e9.5 control (a, c). Wild-type and merm neural tubes expressed comparable levels of Sox2 (a–b), a marker of both epiblast and neural progenitor cells, and of Sox1 (e–f), a marker of neural progenitor cells. The persistence of Id1-positive cells throughout the merm neuroepithelium (h) indicated that the progenitors were more immature than those in the e9.5 wild-type neuroepithelium (g). (B) The patterning of neural progenitors was comparable between wild-type and merm neural tubes at e10.5 (a–h). (C) Markers of the G1 phase of the cell cycle were mostly absent in the merm neural tube (cyclin D1, a–b; cyclin D2, c–d; and p27Kip1, e–f), whereas expression of phosphorylated histone H3 (phospho-H3) indicated that merm and wild-type cells were undergoing mitosis at similar levels (g–h). Immunofluorescence with the indicated antibodies was performed on cryosections of neural tubes prepared from e9.5 or e10.5 merm and wild-type embryos and imaged by confocal or widefield microscopy.
Id1 is expressed in mouse ES cells, where it prevents differentiation in the presence of LIF (Ruzinova and Benezra, 2003; Ying et al., 2003). Additionally, Id gene expression blocks the differentiation of many cell lines in culture by promoting the G1/S transition (Ruzinova and Benezra, 2003). The G1 phase of the ES cell cycle, in contrast to that of most types of dividing cells, is very short and cyclin D-independent. ES cells express minimal levels of cyclin D and p27Kip1, a CDK inhibitor (Burdon et al., 2002; Savatier et al., 1996). Similarly, cyclin D1 and D2 are not expressed in the epiblast of the early embryo until the start of gastrulation. Thereafter cyclin D1 and D2 continue to be expressed in neural progenitors (Wianny et al., 1998), consistent with a function in the transition from epiblast to lineage- restricted progenitor cells. The merm neuroepithelium expressed very little cyclin D1, cyclin D2, and p27Kip1 (Figure 6C a–f), despite exhibiting appreciable levels of mitotic cells (phospho-Histone H3 staining; Figure 6C, g–h). Thus, although able to proliferate, merm progenitor cells could not readily establish a cyclin D-dependent G1 phase. Taken together, the results in Figure 6 indicate that merm neural progenitors were unable to fully transition into the neural lineage and that they abnormally maintained features of their pluripotent epiblast precursors.
merm embryonic neural precursors maintain features of earlier pluripotent epiblast cells
We examined marker gene expression in merm mutants at e8.5. Figure 7A shows results for two genes, Otx2 and Fgf5, whose epiblast expression pattern changes in parallel with lineage commitment during gastrulation. Otx2 is expressed throughout the epiblast at the pre and early primitive streak stages but becomes progressively restricted to the anterior third of the embryo by the headfold stage (Ang et al., 1994). Fgf5 is expressed in the epiblast just prior to gastrulation and its expression completely disappears by the late streak stage (Hebert et al., 1991). merm mutants abnormally maintained Otx2 and Fgf5 expression in the epiblast at e8.5, whereas wild-type embryos had already confined or extinguished epiblast expression of these genes by e7.5, the late streak stage (Figure 7A, a–d). These data were indicative of a significant developmental delay in the epiblast. In contrast, based on unperturbed marker gene expression in merm mutants, development of extraembryonic ectoderm and endoderm proceeded appropriately, consistent with the normal lack of Nup133 expression in these tissues. Otx2 was expressed correctly in e8.5 merm anterior visceral endoderm (arrowheads in Figure 7A, a–b; (Rhinn et al., 1998) and Bmp4 was expressed correctly in merm extraembryonic ectoderm (arrowheads in Figure 7A, e–f; (Winnier et al., 1995). These marker expression assays demonstrated that differentiation of the Nup133-deficient epiblast was out of sync with other morphogenetic events in the gastrulating embryo; notably the late streak/early headfold merm epiblast retained features characteristic of precursors normally present in the pre- and early streak embryo.
Figure 7. Epiblast differentiation in merm embryos.
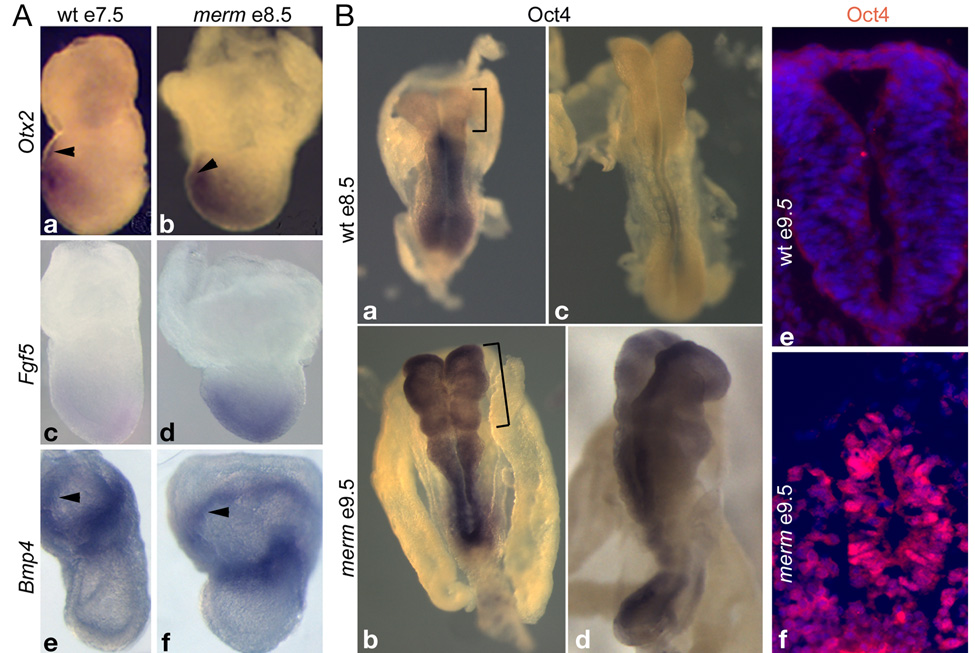
(A) Patterning of merm e8.5 extraembryonic tissues was similar to that of an e7.5 wild-type embryo: Otx2, anterior visceral endoderm – arrowheads (a–b); Bmp4, extraembryonic ectoderm – arrowheads (e–f). Patterning of the merm epiblast was delayed by more than 24 hours: Otx2 (a–b); Fgf5 (c–d). Whole-mount in situ hybridization was performed on wild-type and merm embryos on the C3HeB/FeJ strain background; anterior is to the left. (B) Whole-mount in situ hybridization with an Oct4 probe to e8.5 wild-type (a, c) and e9.5 merm embryos (b, d) and anti-Oct4 immuno-labeling on e9.5 neural tube sections (e–f) revealed delayed extinction of Oct4 transcription in the differentiating merm epiblast/neural plate and the persistence of Oct4 nuclear localization in the merm neural tube. DAPI nuclear staining is shown in blue.
In normal embryos, restriction to the neural lineage largely coincides, spatially and temporally, with the loss of Oct4 expression, a marker of the undifferentiated, self-renewing state of pluripotent ICM and early epiblast cells (Chambers and Smith, 2004; Ralston and Rossant, 2005; Scholer et al., 1990). We examined Oct4 expression in merm embryos to assess whether Nup133-deficient epiblast appropriately restricted pluripotency during formation of the neural lineage. In wild-type embryos (Figure 7B, a; c) Oct4 down-regulation proceeded in an anterior to posterior manner, such that by the 6-somite-stage (e8.5) Oct4 expression was restricted to primordial germ cells. In contrast, the down-regulation of Oct4 was markedly delayed in merm embryos. For example, the mutant in Figure 7B (b) continued to express Oct4 at e9.5 even though, based on anterior neural plate morphology, it had developed beyond the e8.5 wild-type embryos (Figure 7B, a; c). In extreme cases, Oct4 was still present, albeit at reduced levels, throughout the neuroepithelium of mutants with more than 6 somites (Figure 7B, d). These results, confirmed by anti-Oct4 antibody staining (Figure 7B, e–f), indicated that merm cells maintained features of the pluripotent epiblast long after restriction to the neural lineage should have occurred.
DISCUSSION
Here we show that the conserved nucleoporin Nup133 performs an unsuspected role during mouse embryonic development. Since NPCs can assemble in its absence, Nup133 likely serves a modulatory rather than a structural function. We demonstrate a specific requirement for this nucleoporin during the transition of a pluripotent cell into the neural–restricted lineage. Neural progenitors generated in the absence of Nup133 abnormally maintain features of pluripotent early epiblast/ES cells and exhibit impaired neuronal differentiation.
Nup133 is not an obligate component of the NPC in the mouse embryo
Nup133 was originally identified as a component of the evolutionarily conserved Nup107-160 complex, a major NPC structural subunit that resides on both sides of the nuclear envelope (Alber et al., 2007; Hetzer et al., 2005). siRNA depletion in HeLa cells of several constituents of the Nup107-160 complex, including Nup133, provided strong evidence that this complex is essential for NPC assembly in vertebrates (Harel et al., 2003; Walther et al., 2003). In contrast, studies in whole organisms (S. cerevisiae and S. pombe, A. nidulans, Lotus japonicus and C. elegans) found that Nup133 is dispensable for cell viability (Bai et al., 2004; Doye et al., 1994; Galy et al., 2003; Kanamori et al., 2006; Osmani et al., 2006). Our characterization of the mermaid mutant revealed that mouse embryos lacking a functional Nup133 allele developed through mid-gestation. Additionally, merm blastocysts gave rise to stable ES cell lines. These findings demonstrate that in mouse, as in other organisms, Nup133 is not essential for cell viability.
We did not observe destabilization or mis-localization of any other nucleoporin in merm ES cells and embryonic tissues (Figure 2), indicating that NPC assembly can proceed in the absence of Nup133. Most significantly, Nup107, the direct binding partner of Nup133 in this sub-complex, did localize appropriately to the membrane of merm nuclei. However, due to the lack of suitable antibodies to other components of the Nup107-160 sub-complex, mis-localization and potential dysfunction of its other members cannot be excluded in merm nuclei.
NPC subunit composition varies among different embryonic cell types
The data in Figure 3 established that Nup133 is not ubiquitously transcribed in all cell types during mouse development. The expression of only one other nucleoporin has been examined in the mouse embryo, that of Nup50 (Guan et al., 2000). Although ubiquitously expressed, levels of Nup50 varied significantly among different tissues, with the highest levels found in the neural tube, the most severely compromised tissue in Nup50-deficient embryos (Smitherman et al., 2000). Similar to the merm allele of Nup133, a null mutation in Nup50 did not generate a cell-lethal phenotype. Moreover, absence of Nup50 did not perturb NPC formation, at least in primary embryonic fibroblasts.
A comparison of the phenotypes displayed by merm and Nup50 deficient embryos points to different requirements for the two nucleoporins during development. In both mutants morphological abnormalities became apparent after e8.5 and in both, these abnormalities included growth retardation, a kinked neural tube, and exencephaly. However, whereas merm embryos died at mid-gestation, Nup50−/− embryos survived up to birth, suggesting that Nup133 performs essential functions in a wider variety of cell types than does Nup50. The expression of p27Kip1 was perturbed in both nucleoporin mutants. In the Nup50−/− e10.5 neural tube, cells expressing p27Kip1 were scattered throughout the neuroepithelium; in the merm e10.5 neural tube p27Kip1 expressing cells were located properly at the outer edge of the neuroepithelium, but in greatly reduced numbers. These results are consistent with different cellular defects leading to a similar disruption of neural tube morphology: delayed or blocked differentiation of neural progenitors in merm and possibly abnormal migration or premature differentiation of Nup50−/− neural progenitors.
The distinct neural progenitor differentiation defects in merm mutants suggest that cell-type and stage-specific structures of NPCs confer distinct functional capabilities upon the pore. Interestingly, Nup96+/− mice, although viable, exhibit a selective impairment of the immune system, indicating that a given stoichiometry of nucleoporins differentially impacts NPC function in different tissues (Faria et al., 2006).
merm cells retain precursor characteristics and display impaired differentiation
During normal gastrulation, restriction of epiblast cell differentiation potential is accompanied by spatial and temporal alterations in gene expression. As shown in Figure 7, merm mutants exhibited a marked delay in such shifts in gene expression, suggesting an impediment to development beyond an early, pluripotent state.
Neural progenitor cells lacking Nup133 abnormally retained features of their epiblast precursors and had a diminished capacity to generate post-mitotic neurons in ES cell culture and in the developing embryo, pointing to a developmental requirement for Nup133 in establishing the neural lineage. The execution of neuronal differentiation may depend on concurrent activities of Nup133 and/or its functions at earlier steps in the neural lineage. Since dorsal-ventral patterning of neural progenitors occurred appropriately in merm mutants, the lack of Nup133-containing pores does not completely block epiblast cell differentiation at the uncommitted, pluripotent state of a pre-streak embryo. Instead, our findings suggest that Nup133 regulates only a subset of events required for correct differentiation, potentially related to the cell cycle, and that these events must be in sync with other activities in differentiating progenitors for the formation of functional neurons.
merm mutants displayed abnormalities in a number of tissues in addition to the neural tube, indicating that cell differentiation in other epiblast-derived lineages likely requires Nup133. For example, the irregular somites in merm embryos (Figure 1A) suggest that Nup133 is also pivotal for efficient differentiation of their precursors in the paraxial mesodermal lineage. Wild-type paraxial mesoderm cells expressed high levels of Nup133 (Figure 3), while Nup133-deficient ES cells did not contribute to somites in e9.5 chimeric embryos (Figure 4). Conversely, the existence of tissues such as node, notochord, and hindgut that, although epiblast-derived, normally express little or no Nup133 (Figure 3) implies that down-regulation of Nup133 in certain cell types is functionally important for proper development.
A prominent feature of both differentiating merm ES cells and the merm neural tube was the prolonged expression of Oct4, a marker and regulator of the undifferentiated pluripotent progenitor cell state. Forced expression of Oct4 in adult intestinal epithelium inhibits progenitor cell differentiation and results in dysplasia. Thus, adult progenitor cells are able to respond to embryonic signals and failure to extinguish such signals can promote tumorigenesis (Hochedlinger et al., 2005). Interestingly, NUP133 point mutations were recently identified in human breast cancers (Sjöblom et al., 2006; Wood et al., 2007), suggesting that perturbation of Nup133 function can contribute to oncogenesis.
Nup133 function during development
Several studies indicate that the Nup107-160 complex participates in more than one cellular process: in interphase nuclei as a NPC component and in mitosis for spindle assembly and/or proper kinetochore function (Boehmer et al., 2003; Harel et al., 2003; Orjalo et al., 2006; Vasu et al., 2001; Walther et al., 2003; Zuccolo et al., 2007). The long term viability, normal rate of expansion, and chromosome stability of merm ES cells demonstrate that Nup133 is dispensable for any essential structural function of the Nup107-160 complex.
The smaller size of merm embryos might reflect a separate function of Nup133 during cell division. However, it might also represent a complex secondary effect of inefficient differentiation, as proliferation and differentiation are normally tightly coordinated during development.
Studies in HeLa cells identified a contribution of Nup133 to mRNA export (Vasu et al., 2001; Walther et al., 2003). However, as merm embryos develop through mid-gestation, bulk mRNA export likely proceeds normally in the absence of Nup133. In agreement, in situ hybridization detected comparable levels of nuclear and cytoplasmic polyA+ RNA in wild-type and merm cells present in RA-differentiated embryoid bodies (Figure S5C).
Studies based on classical NLS- and NES-mediated transport did not reveal an involvement of Nup133 in nuclear protein transport in HeLa cells (Vasu et al., 2001; Walther et al., 2003). Consistent with this finding, a number of transcription factors localized properly to the nuclei of merm neural progenitor cells (Figure 6A–B; Figure 7B, f, and Figure S5), including Oct4, Nanog, and Brn2, whose nuclear import is NLS-dependent (Pan et al., 2004) (Do et al., 2007) (Yasuhara et al., 2007). A recent study, using gain and loss of function assays in ES cells, found that importin-α subtype switching triggers Brn2 nuclear localization and subsequent neuronal differentiation (Yasuhara et al., 2007). Both over-expression of importin α1 and knockdown of importin α5 in differentiating ES cells leads to a phenotype comparable to that of merm ES cells in culture. Our preliminary experiments detected equivalent patterns of expression and localization for Brn2, importin α1, and importin α5 in the neural tube of e9.5 merm and wild-type embryos (Figure 6; Figure S6). Thus, although we cannot exclude a subtle defect in importin α-mediated transport, Nup133 likely acts either downstream or in parallel with the importin class switch in this neural tissue. Additionally, it is conceivable that Nup133 regulates the transport of a subset of proteins, perhaps using less canonical, karyopherin-independent (and thus NLS-independent) nuclear transport pathways, such as those uncovered for a number of transcriptional regulators (Xu and Massague, 2004).
Nup133’s position at the periphery of the Nup107-160 complex, with its NH2-terminal β-propeller domain free for interaction, strongly argues that Nup133 mediates its developmental role through contact with other proteins or protein complexes (Berke et al., 2004). Thus, in addition to, or instead of, participating in nuclear transport and mitosis, Nup133-containing pores may anchor specific proteins or protein-RNA complexes in the nucleus to mediate as yet unknown functions of the NPC in vertebrate cells. Potential interesting candidates include components of chromatin remodeling complexes and repressor/activator complexes with histone-modifying activities (Kouzarides, 2007). Consistent with this idea is the finding that ES cell differentiation requires inhibition of histone deacetylation (Lee et al., 2004). Recent studies in yeast and Drosophila point to a contribution of the NPC in the transcriptional regulation of a number of genes. For example, upon induction maximally transcribed yeast genes are found associated with nuclear pores. Similarly, the Drosophila dosage compensation complex, which promotes hyper-transcription of the male X chromosome, interacts with components of the NPC (Akhtar and Gasser, 2007) (Taddei, 2007). Analyses of oncogenic fusion proteins containing Nup sub-regions strongly hint at transport-independent functions in transcription for mammalian nucleoporins (Kalverda and Fornerod, 2007) and interestingly, both the MHC-Oct4 region and the proneural gene Mash1 appear to be regulated by changes in chromatin structure and location during neural differentiation of ES cells (Aoto et al., 2006; Williams et al., 2006).
EXPERIMENTAL PROCEDURES
Mouse Strains and Genotyping
merm mice were genotyped based on linkage to flanking SSLP markers developed during positional cloning of the Nup133merm allele. Phenotypic analysis was performed in a congenic 129Sv/ImV strain, unless stated otherwise. The RRK090 ES cell line carrying a gene trap insertion in Nup133 was obtained from BayGenomics. Nup133GT (GT) mice derived from this ES cell line were genotyped by PCR using primers for LacZ or primers that distinguish between the wild-type and Nup133GT alleles. See Supplemental Data for primer sequences. All animal experiments were performed following protocols and procedures approved by the MSKCC IACUC.
Analysis of merm Embryos
In situ hybridization, immunofluorescence and β-Gal staining were carried out as described (Eggenschwiler and Anderson, 2000). Unless otherwise stated, photos of wild-type and merm embryos and tissues were taken at the same magnification.
Chimeras and ES Cell Culture
ES cells were generated from blastocysts obtained from merm/+; Rosa26 X merm/+ crosses according to (Hogan et al., 1994). Chimeric embryos were produced using early passage (passage 3) ES cells according to standard protocols. The first two ES cell differentiation protocols were adapted from (Ying et al., 2003). Briefly, ES cells were maintained under self-renewal conditions (ESC medium supplemented with 1000U/ml ESGRO-LIF and 15% FBS). For differentiation at normal LIF but reduced serum levels, ES cells were cultured for 12 days in ES cell medium + 1000U/ml ESGRO-LIF + 5% serum. For RA-induced differentiation, ES cells were cultured for two days in ES cell medium + 5% serum + 1µM RA, followed by 10 days in ES cell medium + 5% serum. For counting, cells were trypsinized and resuspended in PBS, fixed in 4% para-formaldehyde, then smeared on a slide and stained with antibodies. For each marker cell counts from ten fields were averaged per line. ES cell differentiation into embryoid bodies (EBs) was performed as described in (Wichterle et al., 2002). For each marker cells counts were averaged from 14–15 sections per line. To obtain growth curves, ES cells were seeded in 24-well plates and 4 wells were counted per time point per line. Alkaline phosphatase activity was detected using NBT/BCIP (Roche). At least two wild-type and two merm ES cell lines, at three different passage numbers, were used in each experiment. Photographs were taken with a Nikon E800 compound fluorescent microscope using a digital camera (Princeton Instruments) and Metamorph imaging software (Molecular devices).
Antibodies and Reagents
For a list of primary antibodies, see Supplemental Data. Fluorescent secondary antibodies were from Jackson Laboratories and Molecular Probes. HRP-conjugated secondary antibodies were from Amersham. Filamentous actin was detected with AlexaFluor-conjugated phalloidin (Molecular Probes) and nuclei were visualized with DAPI (Sigma).
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Burke, B. Clurman, V. Cordes, G. Grosveld, B. Fontoura, R. Benezra, and J. van Deursen for antibodies; the MSKCC Mouse Genetics Core for the blastocyst injections; and the MSKCC Molecular Cytology Core for guidance on confocal microscopy. We are grateful to A. K. Hadjantonakis and L. Selleri for valuable discussions and to J. Eggenschwiler, A. Koff, N. Matova, and L. Studer for insightful comments on the manuscript. This research was supported by NRSA HD42355 (F.L.), RO1 GM58726 (E.L.) 1UO1 HD043478 (K.V.A.), the Ligue Nationale contre le Cancer (équipe labellisée 2006, V.D.), the Association pour la Recherche contre le Cancer (ARC, V.D.), the Ministère de l’Education Nationale, de la Recherche et de l’Enseignement Supérieur (A.C.I. “Jeunes chercheurs”, V.D.), the Ministère délégué à la Recherche et aux nouvelles Technologies fellowship (A.A.). The authors declare no competing financial interests related to this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Akhtar A, Gasser S. The nuclear envelope and transcriptional control. Nature Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff L, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait B, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Ang S, Conlon R, Jin O, Rossant J. Positive and negative signals from mesoderm regulate the expression of mouse Otx2 in ectoderm explants. Development. 1994;120:2979–2989. doi: 10.1242/dev.120.10.2979. [DOI] [PubMed] [Google Scholar]
- Aoto T, Saitoh N, Ichimura T, Niwa H, Nakao M. Nuclear and chromatin reorganization in the MHC-Oct3/4 locus at developmental phases of embryonic stem cell differentiation. Dev Biol. 2006;298:354–367. doi: 10.1016/j.ydbio.2006.04.450. [DOI] [PubMed] [Google Scholar]
- Babu J, Jeqanathan K, Baker D, Wu X, Kang-Decker N, van Deursen J. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol. 2003;160:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S, Rouquette J, Umeda M, Faigle W, Loew D, Sazer S, Doye V. The fission yeast Nup107-120 complex functionally interacts with the small GTPase Ran/Spi1 and is required for mRNA export, nuclear pore distribution, and proper cell division. Mol Cell Biol. 2004;24:6379–6392. doi: 10.1128/MCB.24.14.6379-6392.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N, Rabut G, Bai S, van Overbeek M, Beaudouin J, Daigle N, Zatsepina O, Pasteau F, Labas V, Fromont-Racine M, et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke I, Boehmer T, Blobel G, Schwartz T. Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J Cell Biol. 2004;167:591–597. doi: 10.1083/jcb.200408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N, Castro D, Guillemot F. Proneural genes and the specification of neural cell types. Nature Reviews Neuroscience. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Boehmer T, Enninga J, Dales S, Blobel G, Zhong H. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc Natl Acad Sci U S A. 2003;100:981–985. doi: 10.1073/pnas.252749899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer T, Schwartz T. Purification, crystallization and preliminary X-ray analysis of a Nup107–Nup133 heterodimeric nucleoporin complex. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:816–818. doi: 10.1107/S1744309107040523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley A, Robertson E. Embryo-derived stem cells: a tool for elucidating the developmental genetics of the mouse. Curr Top Dev Biol. 1986;20:357–371. doi: 10.1016/s0070-2153(08)60675-4. [DOI] [PubMed] [Google Scholar]
- Burdon T, Smith A, Savatier P. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 2002;12:432–438. doi: 10.1016/s0962-8924(02)02352-8. [DOI] [PubMed] [Google Scholar]
- Chambers I, Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene. 2004;23:7150–7160. doi: 10.1038/sj.onc.1207930. [DOI] [PubMed] [Google Scholar]
- Crawford K, Flick R, Shelly D, Paul R, Bove K, Kumar A, Lessard J. Mice lacking skeletal muscle actin show reduced muscle strength and growth deficits and die during the neonatal period. Mol Cell Biol. 2002;22:5887–5896. doi: 10.1128/MCB.22.16.5887-5896.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do H-J, Lim H-Y, Kim J-H, Song H, Chung H-M, Kim J-H. An intact homeobox domain is required for complete nuclear localization of human Nanog. Biochem Biophys Res Commun. 2007;353:770–775. doi: 10.1016/j.bbrc.2006.12.100. [DOI] [PubMed] [Google Scholar]
- Doye V, Wepf R, Hurt E. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J. 1994;13:6062–6075. doi: 10.1002/j.1460-2075.1994.tb06953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenschwiler J, Anderson K. Dorsal and lateral fates in the mouse neural tube require the cell-autonomous activity of the open brain gene. Dev Biol. 2000;227:648–660. doi: 10.1006/dbio.2000.9918. [DOI] [PubMed] [Google Scholar]
- Faria A, Levay A, Wang Y, Kamphorst A, Rosa M, Nussenzveig D, Balkan W, Y M, Levy D, Fontoura B. The nucleoporin Nup96 is required for proper expression of interferon-regulated proteins and functions. Immunity. 2006;24:283–290. doi: 10.1016/j.immuni.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Galy V, Mattaj I, Askjaer P. Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell. 2003;14:5104–5115. doi: 10.1091/mbc.E03-04-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Garcia M, Eggenschwiler J, Caspary T, Alcorn H, Wyler M, Huangfu D, Rakeman A, Lee J, Feinberg E, Timmer J, et al. Analysis of mouse embryonic patterning and morphogenesis by forward genetics. Proc Natl Acad Sci U S A. 2005;102:5913–5919. doi: 10.1073/pnas.0501071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz M, Huttner W. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Guan T, Kehlenbach R, Schirmer A, Kehlenbach A, Fan F, Clurman B, Arnheim N, Gerace L. Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol Cell Biol. 2000;20:5619–5630. doi: 10.1128/mcb.20.15.5619-5630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A, Orjalo A, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes D. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- Hebert J, Boyle M, Martin G. mRNA localization studies suggest that murine FGF-5 plays a role in gastrulation. Development. 1991;112:407–415. doi: 10.1242/dev.112.2.407. [DOI] [PubMed] [Google Scholar]
- Hetzer M, Walther T, Mattaj I. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu Rev Cell Dev Biol. 2005;21:347–380. doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Habor: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Jen Y, Manova K, Benezra R. Expression patterns of Id1, Id2, and Id3 are highly related but distinct from that of Id4 during mouse embryogenesis. Dev Dyn. 1996;207:235–252. doi: 10.1002/(SICI)1097-0177(199611)207:3<235::AID-AJA1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Jen Y, Manova K, Benezra R. Each member of the Id gene family exhibits a unique expression pattern in mouse gastrulation and neurogenesis. Dev Dyn. 1997;208:92–106. doi: 10.1002/(SICI)1097-0177(199701)208:1<92::AID-AJA9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Jessell T. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Kalverda B, Fornerod M. The Nuclear Life of Nucleoporins Dev Cell. 2007;13:164–165. doi: 10.1016/j.devcel.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Kanamori N, Madsen L, Radutoiu S, Frantescu M, Quistgaard E, Miwa H, Downie J, James E, Felle H, Haaning L, et al. A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc Natl Acad Sci U S A. 2006;103:359–364. doi: 10.1073/pnas.0508883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lee J, Hart S, Skalnik D. Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis. 2004;38:32–38. doi: 10.1002/gene.10250. [DOI] [PubMed] [Google Scholar]
- Loiodice I, Alves A, Rabut G, van Overbeek M, Ellenberg J, Sibarita J, Doye V. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. Npap60: a new player in nuclear protein import. Trends Cell Biol. 2003;13:61–64. doi: 10.1016/s0962-8924(02)00044-2. [DOI] [PubMed] [Google Scholar]
- Okita K, Kiyonari H, Nobuhisa N, Aizawa S, Taga T. Targeted disruption of the mouse ELYS gene results in embryonic death at peri-implantation development. Genes Cell. 2004;9:1083–1091. doi: 10.1111/j.1365-2443.2004.00791.x. [DOI] [PubMed] [Google Scholar]
- Orjalo A, Amaotov A, Shen Z, Boyarchuk Y, Zeitlin S, Fontura B, Briggs S, Dasso M, Forbes D. The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol Biol Cell. 2006;17:3806–3818. doi: 10.1091/mbc.E05-11-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani A, Davies J, Liu H, Nile A, Osmani S. Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol Biol Cell. 2006;17:4946–4961. doi: 10.1091/mbc.E06-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Qin B, Liu N, Scholer H, Pei D. Identification of a Nuclear Localization Signal in OCT4 and Generation of a Dominant Negative Mutant by Its Ablation. J Biol Chem. 2004;279:37013–37020. doi: 10.1074/jbc.M405117200. [DOI] [PubMed] [Google Scholar]
- Pevny L, Lovell-Badge R, Smith A. Generation of purified neural precursors from embryonic stem cells by lineage selection. Curr Biol. 1998;8:971–974. doi: 10.1016/s0960-9822(98)70399-9. [DOI] [PubMed] [Google Scholar]
- Pfister S, Steiner K, Tam P. Gene expression pattern and progression of embryogenesis in the immediate post-implantation period of mouse development. Gene Expr Patterns. 2007;7:558–573. doi: 10.1016/j.modgep.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Ralston A, Rossant J. Genetic regulation of stem cell origins in the mouse embryo. Clin Genet. 2005;68:106–112. doi: 10.1111/j.1399-0004.2005.00478.x. [DOI] [PubMed] [Google Scholar]
- Rhinn M, Dierich A, Shawlot W, Behringer R, Le Meur M, Ang S. Sequential roles for Otx2 in visceral endoderm and neuroectoderm for forebrain and midbrain induction and specification. Development. 1998;125:845–856. doi: 10.1242/dev.125.5.845. [DOI] [PubMed] [Google Scholar]
- Ruzinova M, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- Sabri N, Roth P, Xylourgidis F, Sadeghifar F, J A, Samakovlis C. Distinct functions of the Drosophila Nup153 and Nup214 FG domains in nuclear protein transport. J Cell Biol. 2007;178:557–565. doi: 10.1083/jcb.200612135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savatier P, Lapillonne H, van Grunsven L, Rudkin B, Samarut J. Withdrawal of differentiation inhibitory activity/leukemia inhibitory factor up-regulates D-type cyclins and cyclin-dependent kinase inhibitors in mouse embryonic stem cells. Oncogene. 1996;12:309–322. [PubMed] [Google Scholar]
- Scholer H, Dressler G, Balling R, Rohdewohld H, Gruss P. Oct-4: a germline-specific transcription factor mapping to the mouse t-complex. EMBO J. 1990;9:2185–2195. doi: 10.1002/j.1460-2075.1990.tb07388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöblom T, Jones S, Wood L, Parsons D, Lin J, Barber T, Mandelker D, Leary R, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Smitherman M, Lee K, Swanger J, Kapur R, Clurman B. Characterization and targeted disruption of murine Nup50, a p27(Kip1)-interacting component of the nuclear pore complex. Mol Cell Biol. 2000;20:5631–5642. doi: 10.1128/mcb.20.15.5631-5642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern C. Neural induction: 10 years on since the 'default model'. Curr Opin Cell Biol. 2006;18:692–697. doi: 10.1016/j.ceb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Taddei A. Active genes at the nuclear pore complex. Curr Opin Cell Biol. 2007;19:305–310. doi: 10.1016/j.ceb.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Tam P, Loebel D. Gene function in mouse embryogenesis: get set for gastrulation. Nature Rev Genet. 2007;8:368–381. doi: 10.1038/nrg2084. [DOI] [PubMed] [Google Scholar]
- Tam P, Loebel D, Tanaka S. Building the mouse gastrula: signals, asymmetry and lineages. Curr Opin Genet Dev. 2006;16:419–425. doi: 10.1016/j.gde.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Tam P, Rossant J. Mouse embryonic chimeras: tools for studying mammalian development. Development. 2003;130:6155–6163. doi: 10.1242/dev.00893. [DOI] [PubMed] [Google Scholar]
- Terry L, Wente S. Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J Cell Biol. 2007;178:1121–1132. doi: 10.1083/jcb.200704174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E, Wente S. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- van Deursen J, J B, Kasper L, G G. G2 arrest and impaired nucleocytoplasmic transport in mouse embryos lacking the proto-oncogene CAN/Nup214. EMBO J. 1996;15:5574–5583. [PMC free article] [PubMed] [Google Scholar]
- Vasu S, Shah S, Orjalo A, Park M, Fischer W, Forbes D. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J Cell Biol. 2001;155:339–354. doi: 10.1083/jcb.200108007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, Hulsmann B, Kocher T, Wilm M, Allen T, et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- Wianny F, Real F, Mummery C, Van Rooijen M, Lahti J, Samarut J, Savatier P. G1-phase regulators, cyclin D1, cyclin D2, and cyclin D3: up-regulation at gastrulation and dynamic expression during neurulation. Dev Dyn. 1998;212:49–62. doi: 10.1002/(SICI)1097-0177(199805)212:1<49::AID-AJA5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Lieberam I, Porter J, Jessell T. Directed Differentiation of Embryonic Stem Cells into Motor Neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- Williams R, Azuara V, Perry P, Sauer S, Dvorkina M, Jørgensen H, Roix J, McQueen P, Misteli T, Merkenschlager M, et al. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J Cell Sci. 2006;119:132–140. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- Winnier G, Blessing M, Labosky P, Hogan B. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The Genomic Landscapes of Human Breast and Colorectal Cancers. Science. 2007:1145720. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Wu X, Kasper L, Mantcheva R, Mantchev G, Springett M, van Deursen J. Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proc Natl Acad Sci U S A. 2001;98:3191–3196. doi: 10.1073/pnas.051631598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Massague J. Nucleocytoplasmic shuttling of signal transducers. Nat Rev Mol Cell Biol. 2004;5:209–219. doi: 10.1038/nrm1331. [DOI] [PubMed] [Google Scholar]
- Yasuhara N, Shibazaki N, Tanaka S, Nagai M, Kamikawa Y, Oe S, Asally M, Kamachi Y, Kondoh H, Yoneda Y. Triggering neural differentiation of ES cells by subtype switching of importin-alpha. Nat Cell Biol. 2007;9:72–79. doi: 10.1038/ncb1521. [DOI] [PubMed] [Google Scholar]
- Ying Q, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Zuccolo M, Alves A, Galy V, Bolhy S, Formstecher E, Racine V, Sibarita J-B, Fukagawa T, Shiekhattar R, Yen T, et al. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007;26:1853–1864. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


