Abstract
The Cdc7p protein kinase is essential for the G1/S transition and initiation of DNA replication during the cell division cycle in Saccharomyces cerevisiae. Cdc7p appears to be an evolutionarily conserved protein, since a homolog Hsk1 has been isolated from Schizosaccharomyces pombe. Here, we report the isolation of a human cDNA, HsCdc7, whose product is closely related in sequence to Cdc7p and Hsk1. The HsCdc7 cDNA encodes a protein of 574 amino acids with predicted size of 64 kDa. HsCdc7 contains the conserved subdomains common to all protein-serine/threonine kinases and three “kinase inserts” that are characteristic of Cdc7p and Hsk1. Immune complexes of HsCdc7 from cell lysates were able to phosphorylate histone H1 in vitro. Indirect immunofluorescence staining demonstrated that HsCdc7 protein was predominantly localized in the nucleus. Although the expression levels of HsCdc7 appeared to be constant throughout the cell cycle, the protein kinase activity of HsCdc7 increased during S phase of the cell cycle at approximately the same time as that of Cdk2. These results, together with the functions of Cdc7p in yeast, suggest that HsCdc7 may phosphorylate critical substrate(s) that regulate the G1/S phase transition and/or DNA replication in mammalian cells.
The important events of the eukaryotic cell cycle are that cells must duplicate their genetic material precisely, and then subsequently segregate the sister chromatids into two daughter cells. The first event, DNA replication, is apparently regulated at multiple distinct levels. Genetic and biochemical studies indicate that the initiation of DNA replication occurs from discrete chromosomal locations (replication origins) in all eukaryotic cells. This event can occur asynchronously and over an extended period of many hours. When DNA replication is initiated, the cell must ensure that all its genome is replicated and that the replication of every DNA section occurs once and only once during the cell cycle. In the past several years, identification and isolation of factors that interact with DNA replication origins have greatly advanced our understanding of the mechanisms of DNA replication.
In the budding yeast Saccharomyces cerevisiae, replication origins, known as autonomously replicating sequences (ARSs), are bound to the origin recognition complex (ORC) and several other factors. ORC consists of six protein subunits, designated Orc1–6, that are all essential for cell division and for the initiation of DNA replication (1–5). Although the binding of ORC to ARSs is detected at all stages of the cell cycle (1), in vivo footprinting experiments have shown that different patterns of nucleotide contacts are detected before and after DNA replication (2, 6). Thus, it is proposed that ORC functions to recruit other replication factors to replication origins in a cell cycle dependent manner and thereby to establish a complex competent for initiation of DNA replication. Homologs of the ORC genes have been identified in a range of different eukaryotes including humans (7–10). Recent studies have demonstrated that both Orc1 and Orc2 are also essential for the initiation of DNA replication in higher eukaryotes, even though the characteristics of replication origins in higher eukaryotes are poorly defined (8, 9).
Other factors that interact with ORC and/or origins have been identified and isolated by various methods from budding yeast. They include proteins named for their minichromosome maintenance function (MCM proteins), Cdc6p, Cdc7p, and Dbf4p (11). There are six MCM proteins (MCM2, MCM3, CDC46/MCM5, CDC47, Cdc21, and Mis5) and they share sequence homology in their DNA-dependent ATPase domains. The MCM proteins were originally identified in a screen for mutants unable to initiate DNA replication efficiently at a particular origin (12, 13). Although the expression levels of MCM proteins remain constant, the subcellular localization of MCM proteins appears to be cell cycle dependent. The MCM proteins become bound to chromatin during late mitosis and remain there until they are gradually removed as S phase progresses (12–14). This observation suggests that elimination of MCM proteins from DNA after initiation of replication prevents DNA rereplication in the same cell cycle. MCM family members have also been identified from a wide range of organisms including mammals (15).
Cdc6p and its fission yeast Schizosaccharomyces pombe homolog Cdc18 play an unique role in regulating DNA replication. In contrast to the ORC and MCM proteins, Cdc6p and Cdc18 are expressed at specific stages of the cell cycle (16–19). The protein level of Cdc6p peaks at the end of mitosis, whereas Cdc18 is expressed just prior to the G1/S phase transition (17, 19). Cdc6p is essential for viability, and temperature sensitive cdc6 mutants cause growth arrest with partially unreplicated DNA at the restrictive temperature (5). Overexpression of Cdc18 leads to rereplication in the absence of mitosis (19–21). Reduced activity of Cdc6p and Cdc18 results in premature mitosis without finishing replication (5, 17, 18). Recently, proteins highly related to Cdc6p have been identified in Xenopus and humans (refs. 22 and 23; W.J. and T.H., unpublished results) and it has been shown that the Xenopus Cdc6-related protein is required for initiation of DNA replication in Xenopus egg extracts (22).
The protein kinase Cdc7p is essential for the G1/S transition, with an execution point very similar to that of the MCM protein Cdc46/Mcm5 (24). Although the expression level of Cdc7p remains constant through the cell cycle, Cdc7p kinase activity increases at the G1/S boundary (25). This increase is due, at least in part, to its association with the regulatory subunit Dbf4p whose transcript level reaches the maximum at the G1/S boundary (25). Genetic evidence indicates that Cdc7p interacts with ORC and that Dbf4 interacts with the origin of DNA replication (26), suggesting that the Cdc7/Dbf4 protein complex is directly involved in initiation of DNA replication. The targets of the Cdc7/Dbf4 protein kinase are not known; nonetheless, MCMs have been implicated as candidates (24). An S. pombe Cdc7p homolog, Hsk1, has been isolated, indicating that this protein kinase is also evolutionarily conserved in eukaryotes (27). However, until now a homolog of Cdc7p has not been identified in higher eukaryotes.
In this communication we report identification of a human protein kinase, HsCdc7, whose structure is highly related to the yeast Cdc7p and Hsk1. HsCdc7 is predominantly localized in the nucleus. Although HsCdc7 protein levels remained constant throughout the cell cycle, its protein kinase activity increased in the S phase of the cell cycle.
MATERIALS AND METHODS
Cloning.
The expressed sequence tag (EST) databases of higher eukaryotic sequences were searched for potential reading frame sequences that might be involved in G1/S phase transition and DNA replication with budding yeast cell division control proteins. One of them (GenBank accession no. N40295) showed ≈50% homology to the kinase domain of budding yeast Cdc7p (amino acids 291–359) and fission yeast Cdc7p homolog Hsk1 (amino acids 261–249). cDNAs corresponding to the EST sequence were isolated from HeLa and human primary foreskin fibroblast (HSF8) cells using reverse transcription–PCR (RT-PCR). The cDNA fragment from HeLa cells was radiolabeled and used as a probe for screening a HeLa cDNA library constructed in λGEX5 (28). A screen of 106 phage plaques yielded nine positive clones. The 5′ and 3′ DNA sequences of these clones were determined using the dideoxy-sequencing method. Two clones contained an entire ORF designated as HsCdc7. Conceptual translation of full-length HsCdc7 cDNA reveals a 574 deduced amino acid ORF with predicted size of 64 kDa.
Antibodies.
Polyclonal antibodies were developed in rabbits against a synthetic peptide, CASRITAEEALLHPFFKDMSL, corresponding to the C-terminal residues 555–574 of HsCdc7 coupled to keyhole limpet hemocyanin via the added N-terminal cysteine. Polyclonal antibodies against HsCdc7 were purified from the serum of rabbit no. 6018 using a glutathione S-transferase-HsCdc7 C-terminus (amino acids 538–574) fusion protein affinity column. The 9E10 anti-Myc mAb was used for immunoblotting, immunostaining and protein kinase assay of the Myc-tagged proteins. Rabbit polyclonal antibodies against human Cdk2 were used for protein kinase assays, as described (29).
Mammalian Expression Constructs, Site-Directed Mutagenesis, and Transient Transfection.
For transient transfections, we used the cytomegalovirus-based pCS3 mammalian expression vector, which provides an N-terminal (Myc)6 epitope tag (a gift from J. Cooper). The entire coding sequence of HsCdc7 (amino acids 2–574) was generated by PCR amplification using Pfu polymerase (Stratagene) and subcloned into the XhoI site of pCS3. The pCS3HsCdc7 (K-R) mutant of HsCdc7, in which Lys90 in the kinase subdomain II was replaced by Arg, was generated by quick change mutagenesis method according to the manufacturer’s description (Stratagene) using oligonucleotides 5′-GAGAAAATTGCTGTAAGACACTTGATTCCAACA-3′ and 5′-TGTTGGAATCAAGTGTCTTACAGCAATTTTCTC-3′. The PCR and mutagenesis products were verified by DNA sequencing. Human 293 and HeLa cells (obtained from ATCC) were transfected with pCS3HsCdc7, pCS3HsCdc7 (K-R), and control vector, pCS3, using the calcium phosphate precipitation method, as described (30). Cells were lysed 36–48 hr posttransfection, and the resulting cell lysates were used for immunoprecipitation, immunoblotting, and protein kinase assays.
Cell Cycle Analysis.
Subconfluent 293 cells were fractionated at specific stages of the cell cycle using centrifugal elutriation, as previously described (30). Ninety percent of the cells from each elutriation point were lysed and then subjected to immunoblotting and protein kinase assays. The remaining 10% of the cells from each elutriation point were subjected to flow cytometric analysis. For flow cytometric analysis, cells were first fixed in 70% ethanol and then stained with propidium iodide (40 μg/ml final concentration) plus RNase A (50 μg/ml final concentration) for more than 30 min at room temperature. Samples of 104 cells were analyzed to determine DNA content in G1, S, G2/M phase on a Becton Dickinson FACScan, as described (30).
Immunoblotting, Immunoprecipitation, and Protein Kinase Assays.
Cells were lysed in Nonidet P-40 lysis buffer containing 50 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 1% Nonidet P-40, 1 mM PMSF, 1 mM DTT, 10 units/ml aprotinin, 20 μg/ml leupeptin, and 10% glycerol. Cell lysates were cleared by centrifugation, and protein concentrations were determined using the Bio-Rad protein assay. For protein kinase assays, endogenous HsCdc7 or the (Myc)6-tagged HsCdc7 and (Myc)6-tagged HsCdc7 (K-R) mutant were immunoprecipitated with anti HsCdc7 antibodies or the 9E10 anti-Myc mAb as described above. The immune complexes were washed twice with lysis buffer and twice with kinase buffer (50 mM Hepes, pH 7.4/10 mM MgCl2/1 mM DTT/10 mM NaF/10 mM β-glycerophosphate). After a 60 min incubation at 30°C in 25 μl kinase buffer containing 25 μM ATP, 5 μCi [γ-32P]ATP (1 Ci = 37GBq), and 1 μg histone H1, the kinase reaction was terminated by adding an equal volume of 2× sample buffer, and the reaction products separated on denaturing SDS/polyacrylamide gels prior to autoradiography. For immunoprecipitation and immunoblotting analyses, the immune complexes or proteins were separated on denaturing SDS/polyacrylamide gels and transferred to Immobilon-P membranes (Millipore) and then blotted with primary and secondary antibodies and visualized by enhanced chemiluminescence (Amersham), as described (31).
Immunofluorescence Staining.
HeLa cells were grown on glass coverslips and transiently transfected with the indicated plasmids. Thirty-six hours after the transfection, cells were fixed in PBS solution containing 3% formaldehyde and 2% sucrose at room temperature. After permeabilization in PBS containing 0.4% Triton X-100, cells were blocked with blocking solution (PBS containing 2% goat serum), and then incubated with 9E10 mAb (1:1, 500 dilution) for 1 hr at room temperature. After five washes in PBS, secondary antibody incubations were performed with 1:200 dilutions of Texas red-conjugated goat anti-mouse Ig. Five washes in PBS were performed prior to staining with Hoechst dye 33258 (1 μg/ml), and cells were then mounted and photographed with a fluorescence microscope (Nikon).
RESULTS
Cloning and Analysis of HsCdc7 cDNA.
To identify novel proteins that may be involved in DNA replication and cell cycle control in higher eukaryotes, the EST databases were screened with budding yeast cell division control proteins. A putative reading frame of a partial human cDNA was found with homology to the C-terminal kinase domain of the budding yeast protein kinase Cdc7p and fission yeast Cdc7p homolog Hsk1. A corresponding full-length human cDNA, HsCdc7, was isolated from a HeLa cDNA library (see Materials and Methods). Analysis of the full-length cDNA demonstrated that HsCdc7 contains the 11 conserved subdomains found in all protein-serine/threonine kinases (Fig. 1 A and B, and ref. 32). Sequence similarity analysis using the blast program on the National Center for Biotechnology Information database indicated that the kinase domain of HsCdc7 was most closely related to Cdc7p and Hsk1. As shown in Fig. 1B, the kinase domain of HsCdc7 has 44% and 42% identity in amino acid sequence with Cdc7p and Hsk1, respectively. This result was consistent with our initial EST screen.
Figure 1.
(A) Nucleotide and deduced amino acid (single letter amino acid code) sequences of HsCdc7. HsCdc7 contains an ORF of 574 amino acids and its kinase subdomains are indicated in the boxes. The asterisk represents the stop codon. (B) Multiple alignment of amino acid sequences in HsCdc7, Cdc7p and Hsk1 kinase subdomains (I to XI), which correspond to the boxes in A, using the blast program on the National Center for Biotechnology Information database. Amino acid residues that are identical are indicated by single letters between the sequence lines, and amino acid residues that are similar are indicated by +.
Besides the kinase subdomains, HsCdc7 has 39 amino acid residues at its N terminus and three additional sequences (kinase inserts) between the kinase subdomains I and II, VII and VIII, and X and XI (Fig. 1A). Cdc7p and Hsk1 also have kinase inserts at the same locations (27). However, the sequences and lengths of these kinase inserts are not conserved among HsCdc7, Cdc7p, and Hsk1. Both Cdc7p and Hsk1 contain a distinct C-terminal region after the kinase domain, while HsCdc7 lacks this region (Fig. 1A and ref. 27). It is noteworthy that the second and third kinase inserts in HsCdc7, Cdc7p, and Hsk1 are unusually large when compared with the insert sequences found in other protein-serine/threonine kinases. The second and third kinase inserts of HsCdc7, for instance, contain 158 and 98 amino acid residues, respectively (Fig. 1A). Thus far, the biological functions of these large kinase inserts have not been determined.
HsCdc7 Is a Protein Kinase.
To determine whether HsCdc7 has protein kinase activity, (Myc)6-HsCdc7 and (Myc)6-HsCdc7 (K-R) in which the conserved Lys in the kinase subdomain II (K90) was mutated to Arg to reduce catalytic activity, were expressed using the cytomegalovirus-based eukaryotic expression vectors, pCS3HsCdc7 and pCS3HsCdc7 (K-R), respectively (see Materials and Methods). Lysates of 293 cells transiently transfected with pCS3HsCdc7, pCS3HsCdc7 (K-R), or the control vector (pCS3) were resolved by SDS/PAGE and then immunoblotted with the 9E10 anti-Myc-tag mAb. The 9E10 mAb specifically recognized an 87-kDa band in cells transfected with pCS3HsCdc7 and pCS3HsCdc7 (K-R) constructs and a 23 kDa band, which represents the (Myc)6 polypeptide, in cells transfected with the pCS3 control vector (Fig. 2). Since the predicted size of HsCdc7 is 64 kDa, this result indicates that the 87-kDa bands detected in cells transfected with pCS3HsCdc7 and pCS3HsCdc7 (K-R) are the (Myc)6-tagged HsCdc7 and (Myc)6-tagged HsCdc7 (K-R) mutant proteins.
Figure 2.
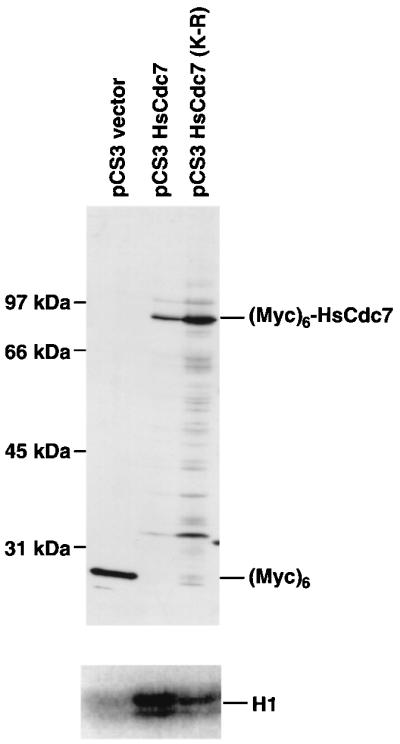
Immunoblot and protein kinase assays of HsCdc7 in 293 cells transfected with pCS3HsCdc7, pCS3HsCdc7 (K-R) and pCS3 control vector plasmids. Lysates from 293 cells transfected with pCS3HsCdc7, pCS3HsCdc7 (K-R) and pCS3 were separated by SDS/PAGE, transferred to Immobilon-P membrane, and then blotted with the 9E10 anti-Myc-tag mAb (Upper). The same cell lysates used for the immunoblotting experiment were immunoprecipitated with 9E10 mAb, and subjected to in vitro kinase reactions using histone H1 as substrate plus [γ-32P]ATP. The products of the kinase reactions were separated by SDS/PAGE prior to autoradiography (Lower) (for details see Materials and Methods).
The same cell lysates used in the immunoblotting experiment were immunoprecipitated with the 9E10 mAb. Protein kinase assays were performed on 9E10 immunoprecipitates using histone H1 as a substrate. Histone H1 was chosen as a potential HsCdc7 substrate, because previous studies have demonstrated that yeast Cdc7p phosphorylates histone H1 in vitro (33, 34). Histone H1 kinase activity was very weak in the 9E10 immunoprecipitate from cells transfected with the pCS3 control vector, whereas histone H1 kinase activity was clearly detected in the 9E10 immunoprecipitate from cells transfected with the pCS3HsCdc7 vector (Fig. 2). Consistent with HsCdc7 having intrinsic protein kinase activity, the (Myc)6-HsCdc7 (K-R) immunoprecipitate had much lower activity than the (Myc)6-HsCdc7 immunoprecipitate, even though the expression level of (Myc)6-HsCdc7 (K-R) was significantly higher than that of (Myc)6-HsCdc7 (Fig. 2). In the same assay, autophosphorylation of HsCdc7 was not observed, although it may occur at a level below the detection of this assay (data not shown). We conclude that HsCdc7 is a protein kinase.
Subcellular Localization of HsCdc7 by Indirect Immunofluorescence Staining.
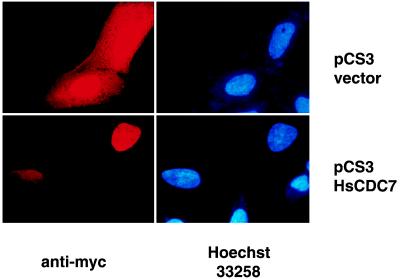
To investigate HsCdc7 function, we first determined its subcellular localization. This may also help us to narrow down the range of its potential in vivo substrates and proteins with which it interacts. Indirect immmunofluorescence staining was performed on HeLa cells, that were transiently transfected with the pCS3HsCdc7 vector and the pCS3 control vector using 9E10 antibody. To verify the position of the nuclei, cells were also co-stained with Hoechst 33258. Representative photomicrographs are shown in Fig. 3. Intense immunofluorescence staining was detected in both the cytoplasm and nuclei of the cells transfected with pCS3 that expressed the 23 kDa (Myc)6 tag polypeptide (see also Fig. 2). This result indicates that the (Myc)6 tag polypeptide alone was not localized in any specific cellular compartment. By contrast, intense immunofluorescence staining was only detected in nuclei of the cells transfected with pCS3HsCdc7 vector. Thus, the HsCdc7 protein appears to be predominantly localized in the nucleus. This result is consistent with the observation that overexpressed Cdc7p is localized in the nucleus in budding yeast (33, 34).
Figure 3.
Immunofluorescence staining in HeLa cells transfected with pCS3HsCdc7 and pCS3 control vector plasmids using 9E10 mAb. HeLa cells were grown on glass coverslips and transiently transfected with the indicated plasmids. After fixation, cells were incubated with the 9E10 mAb. Immunofluorescence staining was performed, and nuclei were visualized by staining with Hoechst dye 33258 as described in Materials and Methods.
Cell Cycle Regulation of HsCdc7 Kinase Activity.
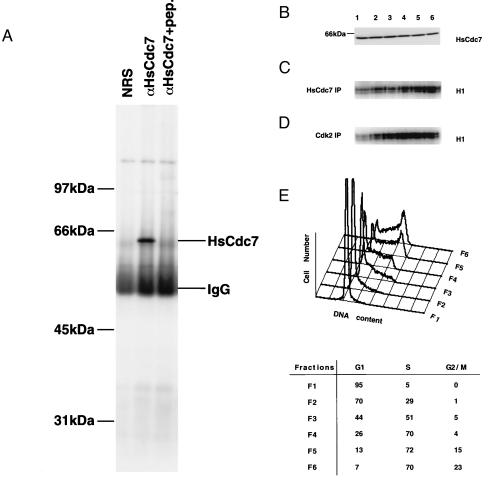
To determine the functions of endogenous HsCdc7 protein during the cell cycle, we generated rabbit polyclonal antibodies against an HsCdc7 C-terminal peptide (see Materials and Methods). Subconfluent 293 cells were lysed and the lysates were immunoprecipitated with nonimmune rabbit serum, HsCdc7 antiserum or HsCdc7 antiserum that had previously been incubated with the HsCdc7 C-terminal peptide. These immunoprecipitates were subjected to SDS/PAGE, transferred to Immobilon-P membranes and subsequently blotted with HsCdc7 antiserum. A single 64-kDa band was detected in the immunoprecipitates with HsCdc7 antiserum, but not in the immunoprecipitates with nonimmune rabbit serum or with HsCdc7 antiserum that was prebound to peptide antigen (Fig. 4A). Similar results were also obtained with HeLa and U2OS cells (data not shown). As mentioned above, the predicted size of HsCdc7 protein is 64 kDa, consistent with the 64 kDa band identified in 293, HeLa and U2OS cells. We conclude that the antibodies raised against the HsCdc7 C-terminal peptide are able to detect the endogenous 64 kDa HsCdc7 protein.
Figure 4.
The expression levels of HsCdc7 protein and its kinase activity during the cell cycle in 293 cells. (A) Lysates from 293 cells were immunoprecipitated with nonimmune rabbit serum (NRS), HsCdc7 antiserum or HsCdc7 antiserum that had been prebound to HsCdc7 C-terminal peptide. After washing, the immunoprecipitates were subjected to SDS/PAGE, transferred to Immobilon-P membrane and then blotted with HsCdc7 antiserum. (B) Subconfluent 293 cells were fractionated at specific stage of the cell cycle using centrifugal elutriation as described in Materials and Methods. Cell lysates from the indicated fractions (F1–F6) were subjected to SDS/PAGE, transferred to Immobilon-P membrane and then blotted with affinity-purified HsCdc7 antibodies. (C and D) The same cell lysates used in B were immunoprecipitated with HsCdc7 antibodies (C) or Cdk2 antibodies (D), and subjected to in vitro kinase reactions using histone H1 as substrate plus [γ-32P]ATP. The products of the kinase reactions were separated by SDS/PAGE prior to autoradiography. (E) 293 cells from each elutriated fraction as described in B were collected and analyzed for DNA content by flow cytometry. (Lower) The values represent the percentage of cells in the indicated phase(s) of the cell cycle in fractions (F1–F6), determined as described in Materials and Methods.
Affinity-purified HsCdc7 antibodies were used to examine the expression and kinase activity of HsCdc7 during the cell cycle in 293 cells. 293 cells were chosen in these experiments since they expressed relatively higher levels of the endogenous HsCdc7 protein than other cell lines tested. Subconfluent 293 cells were subjected to centrifugal elutriation and then collected at different elutriation points as described in Materials and Methods. Ten percent of cells collected at indicated elutriation points were analyzed by flow cytometry to verify cell cycle stages (Fig. 4E). The remaining 90% of the cells were lysed, and the lysates were separated by SDS/PAGE and then immunoblotted with purified anti-HsCdc7 antibodies. The levels of the 64 kDa HsCdc7 protein were constant, irrespective of the proportion of cells in specific phases of the cell cycle (Fig. 4B). Comparable levels of HsCdc7 were also detected in asynchronous cells (data not shown).
In contrast, HsCdc7 kinase activity was clearly regulated during the cell cycle. HsCdc7 immunoprecipitates from the same cell lysates used in immunoblotting were analyzed for histone H1 kinase activity. As a control, Cdk2 kinase activity was examined in parallel using anti-Cdk2 polyclonal antibodies. HsCdc7 kinase activity was low in cells in the G1 phase of the cell cycle and clearly increased in S phase (Fig. 4 C and E). Since the amount of HsCdc7 protein did not fluctuate during the cell cycle, we conclude that increased HsCdc7 kinase activity at specific cell cycle stages is the result of posttranslational regulation. Increased Cdk2 kinase activity was also detected at those elutriation points where cells were at S phase of the cell cycle (Fig. 4 D and E). When a double thymidine block and release protocol was used as an alternative means to synchronize cells, similar results were obtained, with HsCdc7 and Cdk2 histone H1 kinase activities rising together in S phase and then falling as cells entered the next cycle (data not shown). These results are consistent with previous studies in which Cdk2 in association with cyclin E and cyclin A has been shown to be the important regulator in late G1 to S phase transition of the cell cycle (35). The observation that HsCdc7 kinase activity is concomitant with Cdk2 kinase activity in S phase of the cell cycle, taken together with Cdc7p function in the yeast cell cycle, implies that HsCdc7 may phosphorylate critical substrate(s) involved in the regulation of G1/S phase transition and/or DNA replication in mammalian cells.
DISCUSSION
The initiation of DNA replication is highly regulated in eukaryotic cells and a large body of evidence demonstrates that many proteins are involved in this complex process. Identification of replication origins (ARSs) and isolation of proteins (ORC, MCMs, Cdc6p, Cdc7p, and Dbf4p) that assemble as complexes at the replication origins in budding yeast have deepened our understanding of the biochemical mechanisms of DNA replication process (11). However, initiation of DNA replication in higher eukaryotes including humans is poorly understood, since the characteristic sequence(s) of replication origins have not been defined and identification of proteins involved in DNA replication is less complete.
In present studies, we have isolated a human cDNA encoding a protein kinase, HsCdc7, that is structurally related to S. cerevisiae Cdc7p and its S. pombe homolog Hsk1. The kinase domain of HsCdc7 is remarkably conserved by comparison to Cdc7p and Hsk1; HsCdc7 shares 44% and 42% identity in amino acid sequence with Cdc7p and Hsk1, respectively. HsCdc7 also contains three “kinase inserts” between kinase subdomains I and II, VII and VIII, and X and XI, which are the unique features of Cdc7p and Hsk1 even though the sequences and lengths of these kinase inserts are divergent among HsCdc7, Cdc7p, and Hsk1. The degree of conservation of structure and sequence between these proteins suggest that HsCdc7 is likely to be the human homolog of Cdc7p. While this paper was under review, the cloning and characterization of human and Xenopus homologs of Cdc7 was reported (36); huCdc7 is identical to HsCdc7.
We have demonstrated that HsCdc7 is predominantly localized in the nucleus and has a protein kinase activity. The expression level of HsCdc7 remains constant, but the kinase activity of HsCdc7 increases in S phase of the cell cycle. Previous studies have demonstrated that Cdc7p is a nuclear protein-serine/threonine kinase whose function is required at the G1/S phase transition for the initiation of DNA replication in the mitotic cell cycle in budding yeast S. cerevisiae (33, 34, 37). Cdc7p binds to its regulatory subunit Dbf4p, whose function is also required for the initiation of DNA replication and the Cdc7p/Dbf4p complex has maximal kinase activity at the G1/S phase boundary (25, 38, 39). The expression level of Cdc7p remains constant throughout the cell cycle, whereas DBF4 is transcriptionally regulated during the cell cycle with the highest levels of RNA in the G1/S phase, indicating that Dbf4p regulates, at least in part, Cdc7p/Dbf4p protein kinase complex activity (25). Therefore, it seems likely that HsCdc7 will require a regulatory subunit (e.g., a Dbf4-like protein) to regulate its kinase activity, although we cannot exclude the possibility that posttranslational modifications of HsCdc7 (e.g., phosphorylation or dephosphorylation) may also contribute to regulation of its kinase activity. Using a two-hybrid screen we have recently identified a human Dbf4p-related protein that interacts with HsCdc7 (unpublished observations), and we are currently investigating whether expression of this protein is cell cycle regulated and whether it modulates HsCdc7 kinase activity.
Taken together, the similarities between HsCdc7 and Cdc7p suggest that HsCdc7 plays a role in regulating cell cycle progression and DNA replication in human cells. In budding yeast, replication of DNA is blocked by overexpression of kinase-inactive mutants of Cdc7p (40), confirming the essential role for Cdc7p in the initiation of DNA replication. In contrast, we have not observed inhibition of DNA replication when kinase-inactive K90R HsCdc7 is transiently overexpressed in human cells. This might imply that HsCdc7 is not required for the initiation of DNA replication, but other methods of abrogating HsCdc7 function, such as anti-HsCdc7 antibody microinjection, need to be tried before one can determine whether or not HsCdc7 plays a crucial role in DNA replication.
The interaction between Cdc7p and Dbf4p requires the Cdc7p C-terminal region beyond the kinase domain. This region is absolutely required for Cdc7p function, since exchange of the Cdc7p C-terminal region with the corresponding region of Hsk1 (no sequence homology with Cdc7p) leads to loss of Cdc7p complementation ability in a cdc7 temperature-sensitive (ts) mutant strain at the nonpermissive temperature (35°C) (27). In addition, Hsk1 does not complement the Cdc7p mutation in the cdc7(ts) strain at 35°C, nor does Cdc7 complement an hsk1 deletion mutation in S. pombe (27). In contrast to Cdc7p and Hsk1, HsCdc7 does not contain a C-terminal region beyond its kinase domain. Therefore, it is not surprising that HsCdc7 does not complement the hsk1 deletion mutation in S. pombe (unpublished results). Correspondingly, if HsCdc7 interacts with a regulatory subunit, it presumably uses other region(s) of the protein to interact with this subunit. Since HsCdc7 contains unusually large “kinase inserts” II and III (see Fig. 1A), these kinase inserts could be the potential sites mediating the interaction.
Genetic evidence has demonstrated that the Cdc7p/Dbf4p complex is directly involved in the initiation of DNA replication in budding yeast (26). It has been shown that Dbf4p interacts with the origins of DNA replication, suggesting that the Cdc7p/Dbf4p complex may directly phosphorylate other components of DNA replication complexes assembled at the origins. Although the physiological substrates of Cdc7p/Dbf4p have not be identified biochemically, recent genetic studies demonstrate that a recessive loss-of-function mutant mcm5/cdc46 can bypass the requirement for Cdc7p (41). Thus, it has been proposed that Cdc7p phosphorylates MCM5/Cdc46 (or other MCMs) in late G1 phase of the cell cycle leading to removal of MCMs from origins, and thus promoting the initiation of DNA replication. We are currently testing whether MCMs are substrates of HsCdc7 in vitro and in vivo.
Acknowledgments
We thank Dr. Greg Plowman (Sugen, Redwood City, CA) for sharing EST database search data; Dr. Heinz Ruffner (The Salk Institute, La Jolla, CA) for helping with elutriation experiments; and Jill Meisenhelder, Helen Mondala, and Suzy Simon for laboratory support. W.J. is supported by a postdoctoral fellowship from the American Cancer Society (PF-2474). T.H. is an American Cancer Society Research Professor. This work was supported by U.S. Public Health Service Grants CA14195 and CA39780 to T.H.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: ARS, autonomously replicating sequences; MCM proteins; proteins with a minichromosome maintenance function; ORC, origin recognition complex; EST, expressed sequence tag.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF 005209).
References
- 1.Bell S P, Stillman B. Nature (London) 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 2.Diffley J F, Cocker J H. Nature (London) 1992;357:169–172. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- 3.Li J J, Herskowitz I. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- 4.Micklem G, Rowley A, Harwood J, Nasmyth K, Diffley J F. Nature (London) 1993;366:87–89. doi: 10.1038/366087a0. [DOI] [PubMed] [Google Scholar]
- 5.Liang C, Weinreich M, Stillman B. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- 6.Diffley J F, Cocker J H, Dowell S J, Rowley A. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- 7.Gavin K A, Hidaka M, Stillman B. Science. 1995;270:1667–1671. doi: 10.1126/science.270.5242.1667. [DOI] [PubMed] [Google Scholar]
- 8.Carpenter P B, Mueller P R, Dunphy W G. Nature (London) 1996;379:357–360. doi: 10.1038/379357a0. [DOI] [PubMed] [Google Scholar]
- 9.Rowles A, Chong J P, Brown L, Howell M, Evan G I, Blow J J. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- 10.Takahara K, Bong M, Brevard R, Eddy R L, Haley L L, Sait S J, Shows T B, Hoffman G G, Greenspan D S. Genomics. 1996;31:119–122. doi: 10.1006/geno.1996.0018. [DOI] [PubMed] [Google Scholar]
- 11.Stillman B. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- 12.Hennessy K M, Lee A, Chen E, Botstein D. Genes Dev. 1991;5:958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- 13.Yan H, Merchant A M, Tye B K. Genes Dev. 1993;7:2149–2160. doi: 10.1101/gad.7.11.2149. [DOI] [PubMed] [Google Scholar]
- 14.Dalton S, Whitbread L. Proc Natl Acad Sci USA. 1995;92:2514–2518. doi: 10.1073/pnas.92.7.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura H, Ohtomo T, Yamaguchi M, Ishii A, Sugimoto K. Genes Cells. 1996;1:977–993. doi: 10.1046/j.1365-2443.1996.840284.x. [DOI] [PubMed] [Google Scholar]
- 16.Zwerschke W, Rottjakob H W, Kuntzel H. J Biol Chem. 1994;269:23351–23356. [PubMed] [Google Scholar]
- 17.Piatti S, Lengauer C, Nasmyth K. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly T J, Martin G S, Forsburg S L, Stephen R J, Russo A, Nurse P. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- 19.Muzi Falconi M, Brown G W, Kelly T J. Proc Natl Acad Sci USA. 1996;93:1566–1570. doi: 10.1073/pnas.93.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishitani H, Nurse P. Cell. 1995;83:397–405. doi: 10.1016/0092-8674(95)90117-5. [DOI] [PubMed] [Google Scholar]
- 21.Jallepalli P V, Kelly T J. Genes Dev. 1996;10:541–552. doi: 10.1101/gad.10.5.541. [DOI] [PubMed] [Google Scholar]
- 22.Coleman T R, Carpenter P B, Dunphy W G. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- 23.Williams R S, Shohet R V, Stillman B. Proc Natl Acad Sci USA. 1997;94:142–147. doi: 10.1073/pnas.94.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Botchan M. Proc Natl Acad Sci USA. 1996;93:9997–10000. doi: 10.1073/pnas.93.19.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson A L, Pahl P M, Harrison K, Rosamond J, Sclafani R A. Mol Cell Biol. 1993;13:2899–2908. doi: 10.1128/mcb.13.5.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dowell S J, Romanowski P, Diffley J F. Science. 1994;265:1243–1246. doi: 10.1126/science.8066465. [DOI] [PubMed] [Google Scholar]
- 27.Masai H, Miyake T, Arai K. EMBO J. 1995;14:3094–3104. doi: 10.1002/j.1460-2075.1995.tb07312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukunaga R, Hunter T. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe N, Broome M, Hunter T. EMBO J. 1995;14:1878–1891. doi: 10.1002/j.1460-2075.1995.tb07180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruffner H, Verma I M. Proc Natl Acad Sci USA. 1997;94:7138–7143. doi: 10.1073/pnas.94.14.7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang W, Kahn S M, Zhou P, Zhang Y J, Cacace A M, Infante A S, Doi S, Santella R M, Weinstein I B. Oncogene. 1993;8:3447–3457. [PubMed] [Google Scholar]
- 32.Hanks S K, Hunter T. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 33.Hollingsworth R E, Jr, Sclafani R A. Proc Natl Acad Sci USA. 1990;87:6272–6276. doi: 10.1073/pnas.87.16.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoon H J, Campbell J L. Proc Natl Acad Sci USA. 1991;88:3574–3578. doi: 10.1073/pnas.88.9.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hunter T, Pines J. Cell. 1994;79:573–582. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- 36.Sato N, Arai K, Masai H. EMBO J. 1997;16:4340–4351. doi: 10.1093/emboj/16.14.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartwell L H. Bacteriol Rev. 1974;38:164–198. doi: 10.1128/br.38.2.164-198.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chapman J W, Johnston L H. Exp Cell Res. 1989;180:419–428. doi: 10.1016/0014-4827(89)90068-2. [DOI] [PubMed] [Google Scholar]
- 39.Kitada K, Johnston L H, Sugino T, Sugino A. Genetics. 1992;131:21–29. doi: 10.1093/genetics/131.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohtoshi A, Miyake T, Arai K, Masai H. Mol Gen Genet. 1997;254:562–570. doi: 10.1007/s004380050452. [DOI] [PubMed] [Google Scholar]
- 41.Hardy C F, Dryga O, Seematter S, Pahl P M, Sclafani R A. Proc Natl Acad Sci USA. 1997;94:3151–3155. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]