Abstract
A locus in feline DNA, termed flvi-1, which may play an important role in the natural induction of lymphomas by feline leukemia virus (FeLV) was identified. Examination of a bank of 21 naturally occurring FeLV-positive feline lymphomas revealed that FeLV proviral integration occurs at flvi-1 in four independent tumors (19%). Independent integrations occurred within a 2.4-kilobase region of flvi-1, the probability of which by random chance can be estimated as 10(-16). Several lines of evidence, including sequence analysis of the long terminal repeat, demonstrated that proviruses integrated at flvi-1 are exogenously acquired and are oriented in the same transcriptional direction with respect to the locus. Molecularly cloned flvi-1 did not hybridize with probes representing several previously described proviral integration domains or with probes representing 10 oncogenes. The natural feline lymphomas examined in this study were heterogeneous with respect to tissue of origin, cell type, and number of monoclonal proviral integrations. The four tumors in which flvi-1 is interrupted were classified as members of a phenotypic subgroup containing seven lymphomas, i.e., at least four (57%) of seven lymphomas of this type contained FeLV proviral integration at flvi-1. Members of this phenotypic subgroup are non-T-cell lymphomas isolated from the spleen and contain an average of three proviruses, compared with an average of eight among all of the tumors examined. The small number of proviral integrations in tumors of this subgroup suggests that an early proviral integration event into flvi-1 can induce malignant change.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Berry B. T., Ghosh A. K., Kumar D. V., Spodick D. A., Roy-Burman P. Structure and function of endogenous feline leukemia virus long terminal repeats and adjoining regions. J Virol. 1988 Oct;62(10):3631–3641. doi: 10.1128/jvi.62.10.3631-3641.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordereaux D., Fichelson S., Sola B., Tambourin P. E., Gisselbrecht S. Frequent involvement of the fim-3 region in Friend murine leukemia virus-induced mouse myeloblastic leukemias. J Virol. 1987 Dec;61(12):4043–4045. doi: 10.1128/jvi.61.12.4043-4045.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Buchberg A. M., Bedigian H. G., Taylor B. A., Brownell E., Ihle J. N., Nagata S., Jenkins N. A., Copeland N. G. Localization of Evi-2 to chromosome 11: linkage to other proto-oncogene and growth factor loci using interspecific backcross mice. Oncogene Res. 1988;2(2):149–165. [PubMed] [Google Scholar]
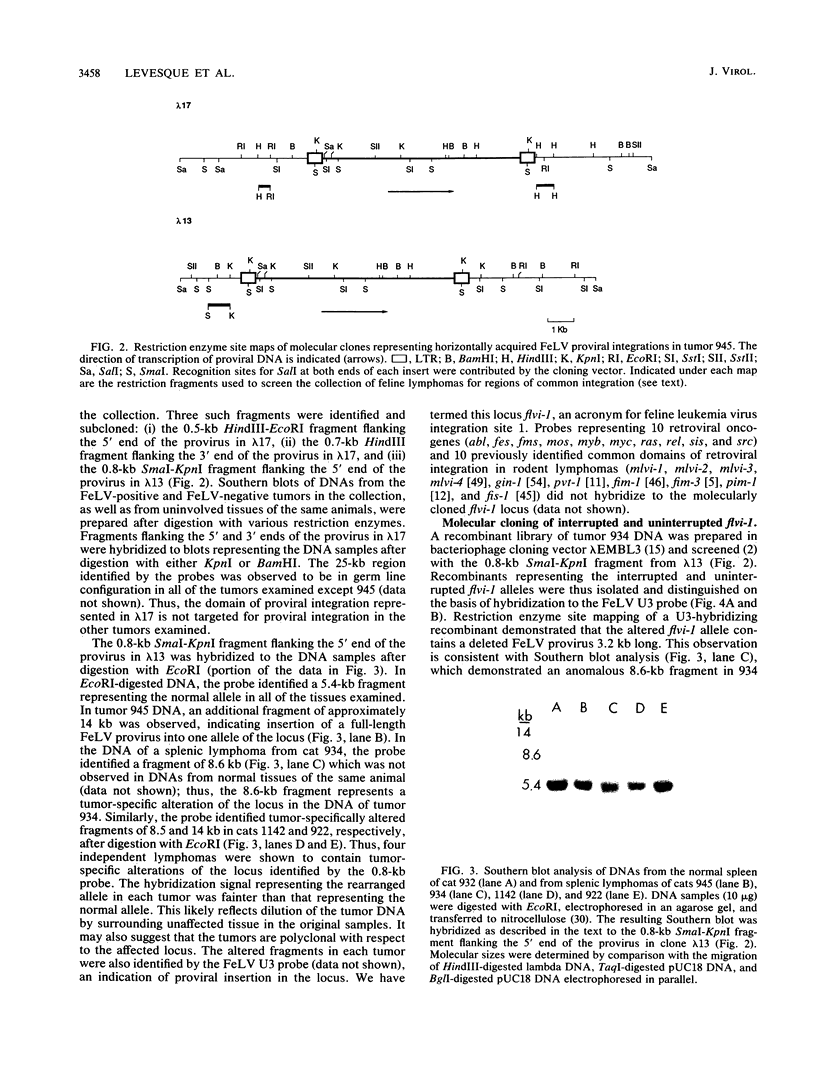
- Casey J. W., Roach A., Mullins J. I., Burck K. B., Nicolson M. O., Gardner M. B., Davidson N. The U3 portion of feline leukemia virus DNA identifies horizontally acquired proviruses in leukemic cats. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7778–7782. doi: 10.1073/pnas.78.12.7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran L. M., Adams J. M., Dunn A. R., Cory S. Murine T lymphomas in which the cellular myc oncogene has been activated by retroviral insertion. Cell. 1984 May;37(1):113–122. doi: 10.1016/0092-8674(84)90306-4. [DOI] [PubMed] [Google Scholar]
- Corden J., Wasylyk B., Buchwalder A., Sassone-Corsi P., Kedinger C., Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980 Sep 19;209(4463):1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- Cory S., Graham M., Webb E., Corcoran L., Adams J. M. Variant (6;15) translocations in murine plasmacytomas involve a chromosome 15 locus at least 72 kb from the c-myc oncogene. EMBO J. 1985 Mar;4(3):675–681. doi: 10.1002/j.1460-2075.1985.tb03682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuypers H. T., Selten G., Quint W., Zijlstra M., Maandag E. R., Boelens W., van Wezenbeek P., Melief C., Berns A. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell. 1984 May;37(1):141–150. doi: 10.1016/0092-8674(84)90309-x. [DOI] [PubMed] [Google Scholar]
- Dickson C., Smith R., Brookes S., Peters G. Tumorigenesis by mouse mammary tumor virus: proviral activation of a cellular gene in the common integration region int-2. Cell. 1984 Jun;37(2):529–536. doi: 10.1016/0092-8674(84)90383-0. [DOI] [PubMed] [Google Scholar]
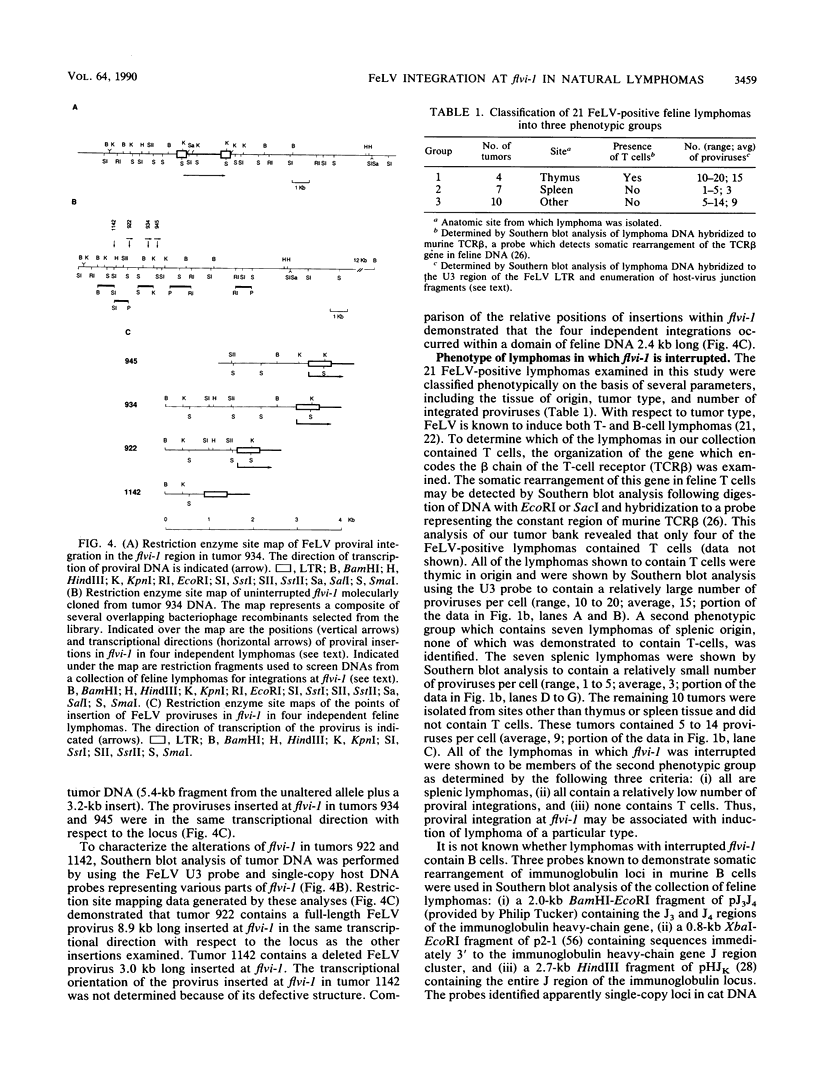
- Forrest D., Onions D., Lees G., Neil J. C. Altered structure and expression of c-myc in feline T-cell tumours. Virology. 1987 May;158(1):194–205. doi: 10.1016/0042-6822(87)90253-4. [DOI] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- Fung Y. K., Lewis W. G., Crittenden L. B., Kung H. J. Activation of the cellular oncogene c-erbB by LTR insertion: molecular basis for induction of erythroblastosis by avian leukosis virus. Cell. 1983 Jun;33(2):357–368. doi: 10.1016/0092-8674(83)90417-8. [DOI] [PubMed] [Google Scholar]
- Gallahan D., Callahan R. Mammary tumorigenesis in feral mice: identification of a new int locus in mouse mammary tumor virus (Czech II)-induced mammary tumors. J Virol. 1987 Jan;61(1):66–74. doi: 10.1128/jvi.61.1.66-74.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M., Wellinger R., Vessaz A., Diggelmann H. A new site of integration for mouse mammary tumor virus proviral DNA common to BALB/cf(C3H) mammary and kidney adenocarcinomas. EMBO J. 1986 Jan;5(1):127–134. doi: 10.1002/j.1460-2075.1986.tb04186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray D. A., McGrath C. M., Jones R. F., Morris V. L. A common mouse mammary tumor virus integration site in chemically induced precancerous mammary hyperplasias. Virology. 1986 Jan 30;148(2):360–368. doi: 10.1016/0042-6822(86)90332-6. [DOI] [PubMed] [Google Scholar]
- Hayward W. S., Neel B. G., Astrin S. M. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981 Apr 9;290(5806):475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- Koehne C. F., Lazo P. A., Alves K., Lee J. S., Tsichlis P. N., O'Donnell P. V. The Mlvi-1 locus involved in the induction of rat T-cell lymphomas and the pvt-1/Mis-1 locus are identical. J Virol. 1989 May;63(5):2366–2369. doi: 10.1128/jvi.63.5.2366-2369.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy J. A. The multifaceted retrovirus. Cancer Res. 1986 Nov;46(11):5457–5468. [PubMed] [Google Scholar]
- Levy L. S., Fish R. E., Baskin G. B. Tumorigenic potential of a myc-containing strain of feline leukemia virus in vivo in domestic cats. J Virol. 1988 Dec;62(12):4770–4773. doi: 10.1128/jvi.62.12.4770-4773.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy L. S., Gardner M. B., Casey J. W. Isolation of a feline leukaemia provirus containing the oncogene myc from a feline lymphosarcoma. 1984 Apr 26-May 2Nature. 308(5962):853–856. doi: 10.1038/308853a0. [DOI] [PubMed] [Google Scholar]
- Lewis S., Rosenberg N., Alt F., Baltimore D. Continuing kappa-gene rearrangement in a cell line transformed by Abelson murine leukemia virus. Cell. 1982 Oct;30(3):807–816. doi: 10.1016/0092-8674(82)90285-9. [DOI] [PubMed] [Google Scholar]
- Li Y., Holland C. A., Hartley J. W., Hopkins N. Viral integration near c-myc in 10-20% of mcf 247-induced AKR lymphomas. Proc Natl Acad Sci U S A. 1984 Nov;81(21):6808–6811. doi: 10.1073/pnas.81.21.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Miura T., Shibuya M., Tsujimoto H., Fukasawa M., Hayami M. Molecular cloning of a feline leukemia provirus integrated adjacent to the c-myc gene in a feline T-cell leukemia cell line and the unique structure of its long terminal repeat. Virology. 1989 Apr;169(2):458–461. doi: 10.1016/0042-6822(89)90172-4. [DOI] [PubMed] [Google Scholar]
- Moreau-Gachelin F., Tavitian A., Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature. 1988 Jan 21;331(6153):277–280. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- Morishita K., Parker D. S., Mucenski M. L., Jenkins N. A., Copeland N. G., Ihle J. N. Retroviral activation of a novel gene encoding a zinc finger protein in IL-3-dependent myeloid leukemia cell lines. Cell. 1988 Sep 9;54(6):831–840. doi: 10.1016/s0092-8674(88)91175-0. [DOI] [PubMed] [Google Scholar]
- Mucenski M. L., Taylor B. A., Ihle J. N., Hartley J. W., Morse H. C., 3rd, Jenkins N. A., Copeland N. G. Identification of a common ecotropic viral integration site, Evi-1, in the DNA of AKXD murine myeloid tumors. Mol Cell Biol. 1988 Jan;8(1):301–308. doi: 10.1128/mcb.8.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R., Varmus H. E. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982 Nov;31(1):99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- Nusse R., van Ooyen A., Cox D., Fung Y. K., Varmus H. Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature. 1984 Jan 12;307(5947):131–136. doi: 10.1038/307131a0. [DOI] [PubMed] [Google Scholar]
- Payne G. S., Bishop J. M., Varmus H. E. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature. 1982 Jan 21;295(5846):209–214. doi: 10.1038/295209a0. [DOI] [PubMed] [Google Scholar]
- Peters G., Brookes S., Smith R., Dickson C. Tumorigenesis by mouse mammary tumor virus: evidence for a common region for provirus integration in mammary tumors. Cell. 1983 Jun;33(2):369–377. doi: 10.1016/0092-8674(83)90418-x. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. Sequence at the 3' end of globin mRNA shows homology with immunoglobulin light chain mRNA. Nature. 1974 Nov 29;252(5482):359–362. doi: 10.1038/252359a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selten G., Cuypers H. T., Berns A. Proviral activation of the putative oncogene Pim-1 in MuLV induced T-cell lymphomas. EMBO J. 1985 Jul;4(7):1793–1798. doi: 10.1002/j.1460-2075.1985.tb03852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selten G., Cuypers H. T., Zijlstra M., Melief C., Berns A. Involvement of c-myc in MuLV-induced T cell lymphomas in mice: frequency and mechanisms of activation. EMBO J. 1984 Dec 20;3(13):3215–3222. doi: 10.1002/j.1460-2075.1984.tb02281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Ong G. L., Potter M., Mushinski J. F., Lavu S., Reddy E. P. Activation of the c-myb locus by viral insertional mutagenesis in plasmacytoid lymphosarcomas. Science. 1984 Nov 30;226(4678):1077–1080. doi: 10.1126/science.6093260. [DOI] [PubMed] [Google Scholar]
- Silver J., Kozak C. Common proviral integration region on mouse chromosome 7 in lymphomas and myelogenous leukemias induced by Friend murine leukemia virus. J Virol. 1986 Feb;57(2):526–533. doi: 10.1128/jvi.57.2.526-533.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola B., Fichelson S., Bordereaux D., Tambourin P. E., Gisselbrecht S. fim-1 and fim-2: two new integration regions of Friend murine leukemia virus in myeloblastic leukemias. J Virol. 1986 Nov;60(2):718–725. doi: 10.1128/jvi.60.2.718-725.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen D. Proviruses are adjacent to c-myc in some murine leukemia virus-induced lymphomas. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2097–2101. doi: 10.1073/pnas.81.7.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. A., Warnock M., Wheeler A., Wilkie N., Mullins J. I., Onions D. E., Neil J. C. Nucleotide sequences of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinational origin of subgroup B viruses. J Virol. 1986 Jun;58(3):825–834. doi: 10.1128/jvi.58.3.825-834.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsichlis P. N., Lohse M. A., Szpirer C., Szpirer J., Levan G. Cellular DNA regions involved in the induction of rat thymic lymphomas (Mlvi-1, Mlvi-2, Mlvi-3, and c-myc) represent independent loci as determined by their chromosomal map location in the rat. J Virol. 1985 Dec;56(3):938–942. doi: 10.1128/jvi.56.3.938-942.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto A. S., Grosschedl R., Guzman R. C., Parslow T., Varmus H. E. Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell. 1988 Nov 18;55(4):619–625. doi: 10.1016/0092-8674(88)90220-6. [DOI] [PubMed] [Google Scholar]
- Varmus H. Retroviruses. Science. 1988 Jun 10;240(4858):1427–1435. doi: 10.1126/science.3287617. [DOI] [PubMed] [Google Scholar]
- Vijaya S., Steffen D. L., Kozak C., Robinson H. L. Dsi-1, a region with frequent proviral insertions in Moloney murine leukemia virus-induced rat thymomas. J Virol. 1987 Apr;61(4):1164–1170. doi: 10.1128/jvi.61.4.1164-1170.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemur R., Monczak Y., Rassart E., Kozak C., Jolicoeur P. Identification of a new common provirus integration site in gross passage A murine leukemia virus-induced mouse thymoma DNA. Mol Cell Biol. 1987 Jan;7(1):512–522. doi: 10.1128/mcb.7.1.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver D., Costantini F., Imanishi-Kari T., Baltimore D. A transgenic immunoglobulin mu gene prevents rearrangement of endogenous genes. Cell. 1985 Aug;42(1):117–127. doi: 10.1016/s0092-8674(85)80107-0. [DOI] [PubMed] [Google Scholar]
- van Lohuizen M., Verbeek S., Krimpenfort P., Domen J., Saris C., Radaszkiewicz T., Berns A. Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell. 1989 Feb 24;56(4):673–682. doi: 10.1016/0092-8674(89)90589-8. [DOI] [PubMed] [Google Scholar]