Abstract
The v-jun oncogene encodes a nuclear DNA binding protein that functions as a transcription factor and is part of the activator protein 1 complex. Oncogenic transformation by v-jun is thought to be mediated by the aberrant expression of specific target genes. To identify such Jun-regulated genes and to explore the mechanisms by which Jun affects their expression, we have fused the full-length v-Jun and an amino-terminally truncated form of v-Jun to the hormone-binding domain of the human estrogen receptor. The two chimeric proteins function as ligand-inducible transactivators. Expression of the fusion proteins in chicken embryo fibroblasts causes estrogen-dependent transformation.
Keywords: activator protein 1, oncogenic transformation, target genes
The v-jun oncogene is the transforming gene of avian sarcoma virus 17 (1). It is a mutated form of the cellular jun (c-jun) gene and induces sarcomas in chickens and transforms chicken embryo fibroblasts (CEFs) in culture (2).
The jun gene codes for a “basic leucine zipper” protein characterized by a basic DNA binding domain adjacent to a carboxyl-terminal leucine zipper that serves as a dimerization region. Dimers of Jun with related basic leucine zipper proteins form activator protein 1 (AP-1) transcription factor complexes (3, 4). The amino-terminal half of the Jun protein functions as a transactivation domain. Dimerization of Jun is required for DNA binding, and DNA binding is a prerequisite for transactivation. All three—dimerization, DNA binding, and transactivation—are essential for oncogenic transformation (5). Transformation therefore may result from aberrant regulation of gene expression. However, no direct correlation has been seen between the transactivation potential as measured on the AP-1 consensus sequence and the oncogenicity of various jun mutants (6, 7). Presumably, the spectrum of target promoters responsive to oncogenic v-Jun is not identical to that of nononcogenic c-Jun. Little is known about the target promoters that are differentially regulated in Jun-transformed cells. Recently, several genes have been identified that are specifically up-regulated in such cells (8, 9). It is not known whether any of these genes have an essential function in the initiation and maintenance of transformation. To characterize genes that are controlled by v-Jun we have constructed a conditional v-Jun expression system by fusing the full-length v-Jun protein and a truncated v-Jun protein to the hormone binding domain of the human estrogen receptor (ER). Here we describe hormone-dependent transcriptional activation and oncogenic transformation induced by the Jun-ER chimeras.
MATERIALS AND METHODS
Cells and Viruses.
Primary CEF cultures were prepared from White Leghorn embryos as described (10). To grow virus stocks, secondary CEF cultures were transfected by the calcium phosphate method with DNA of the replication competent avian retroviral RCAS vector containing various inserts (11). The cultures were passaged two to three times, and culture supernatants containing infectious RCAS virus were harvested from confluent plates and stored at −80°C. Focus assays were performed as described (2); estrogen was added to the agar overlays at 2 μM and tamoxifen at 200 nM. The human choriocarcinoma cell line JEG-3 (12) was obtained from the American Type Culture Collection and maintained in MEM supplemented with 10% fetal bovine serum. For transfections, 10% donor calf serum was used instead of fetal bovine serum because of its lower estrogen content.
Plasmid Constructs.
The v-jun clone VJ1 has been described (2). To construct fusion proteins between v-Jun and the carboxyl-terminal domain of the human ER, the cloning process was separated into two steps. In the first step, an adapter oligonucleotide containing a BglII restriction site was inserted into the MaeII site immediately before the v-jun stop codon. To this end, plasmid pG4–26-1 containing the v-jun ORF was digested with NcoI and MaeII. The 501-bp NcoI–MaeII fragment was gel-purified and ligated together with the annealed oligonucleotides ON236 and ON237 into the adapter plasmid CLA12Nco (11) cleaved with NcoI and EcoRI. This clone contains amino acid residues 128–296 of v-Jun plus five oligonucleotide-encoded amino acid residues and was designated ΔVJ(BglII). The full-length ORF of v-jun was restored by cloning the 366-bp NcoI fragment of pG4–26-1 encoding amino acids 6–127 of v-Jun into ΔVJ(BglII) cleaved with NcoI, resulting in VJ1(BglII). In the second step VJ1(BglII) and ΔVJ(BglII) were fused in-frame to the ER domain contained in plasmid HE14 (13). The 0.9-kb BamHI–EcoRI insert of pHE14 was ligated into VJ1(BglII) and ΔVJ(BglII) cleaved with BglII and EcoRI, resulting in VJ1-hER and ΔVJ-hER, respectively. The correct reading frame of the fusion was confirmed by sequencing. The Jun domain and the receptor domain are linked via three additional amino acid residues encoded by the oligonucleotide adapter that serves as flexible spacer (Gly-Gly-Ser) between the two protein parts. For a control construct containing the ER domain alone, the HE14 insert was cloned as a EcoRI fragment into the adapter plasmid. For transactivation and transformation experiments, the constructs VJ1-hER, ΔVJ-hER, ΔVJ, and hER were transferred as ClaI fragments into the retroviral vector RCAS (11). The correct orientation of inserts was determined by restriction analysis with SalI. The sequences of oligonucleotides were: ON236 (upper strand) 5′-CGTTTGGCGGTTCAGATCTCTAAG-3′; ON237 (lower strand) 5′-AATTCTTAGAGATCTGAACCGCCAAA-3′.
Hormone Treatment.
β-Estradiol (Sigma) was dissolved at 2 mM in ethanol and added to the medium at 2 μM. (Z)-4-hydroxytamoxifen (Research Biochemicals) was dissolved at 0.2 mM in ethanol and added at 200 nM. Control cultures received an equivalent amount of ethanol. Use of phenolred-free media or charcoal-stripped serum was not necessary, because we did not observe activation of the ER fusion proteins by phenolred or endogenous estrogen contained in sera. Cells were treated with cycloheximide (Sigma; 50 mg/ml stock solution in ethanol) at a final concentration of 50 μg/ml).
Transfection and Transactivation.
Transactivation by the recombinant proteins was measured by using luciferase assays. The HTLV-1 Lux reporter plasmid contains six copies of the AP-1 responsive element from the HTLV-1 long terminal repeat linked to the luciferase gene. JEG-3 cells (12) were cotransfected with 0.5 μg RCAS expression vector and 1 μg reporter plasmid and assayed for luciferase activity 48 hr after transfection.
Western Blotting.
To verify expression of the recombinant proteins, CEFs were transfected by the calcium phosphate method with the various constructs. Cells were passaged two to three times, and confluent dishes were lysed in 1 × SDS/PAGE sample buffer for Western blot analysis (14). Proteins transferred to nitrocellulose were detected with rabbit antiserum to chicken c-Jun (USC30–4) or rabbit antiserum to the human ER (Santa Cruz Biotechnology; ER HC-20) at a 1:2,500 dilution. After incubation with horseradish peroxidase-conjugated secondary antibody, bound proteins were detected by incubation with chemiluminescent substrate (Renaissance, DuPont/NEN) and exposed to Kodak X-Omat XAR5 film.
Northern Blotting.
Total RNA was isolated with the RNA STAT-60 reagent according to the protocol of the supplier (Tel-Test, Friendswood, TX). Northern blots were performed according to standard procedures and probed with 32P-labeled DNA probes (14).
RESULTS
Construction of v-Jun-ER Chimeras.
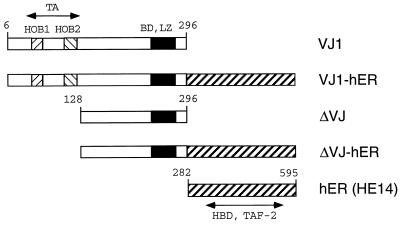
The structures of the v-Jun-ER fusion proteins analyzed in this study are shown in Fig. 1. The VJ1 cDNA, encoding the chicken v-Jun protein without the gag-region (2), was fused in-frame to a cDNA fragment (HE14) encoding the hormone binding domain of the ER. The HE14 ER-construct was chosen because it contains an amino acid substitution at position 400 (Gly → Val), which reduces the affinity for estrogen and thus reduces activation by trace amounts of hormone (15). The amino-terminal truncation construct ΔVJ was created by removing the 127 amino-terminal amino acid residues up to the internal NcoI restriction site. This construct was fused to the ER as described for VJ1-hER. ΔVJ lacks the major transactivation domain of v-Jun and was used to reveal the contribution of the transactivation function 2 (TAF-2) of the ER to the activity of chimeric proteins. A construct containing the ER part alone (amino acids 282–595) served as a control and demonstrated that the biological effects of the fusion proteins require the v-Jun part. The VJ1 construct served as standard for transactivation and transformation experiments. All constructs were cloned into the replication competent retroviral vector RCAS (11).
Figure 1.
Structure of v-Jun and v-Jun-ER fusion proteins. The basic DNA binding region (BD), the leucine zipper (LZ), and the homology boxes 1 and 2 (HOB1, HOB2) of Jun are shown as patterned boxes. Numbering of amino acid residues is according to c-Jun. The carboxyl-terminal half of the human ER HE14 (amino acids 282–595) comprising the hormone binding domain (HBD) and TAF-2 is depicted as a diagonally striped box. The v-Jun protein and the ER part are separated by a spacer consisting of the amino acids glycine-glycine-serine.
Expression of v-Jun-ER Chimeras in Chicken Cells.
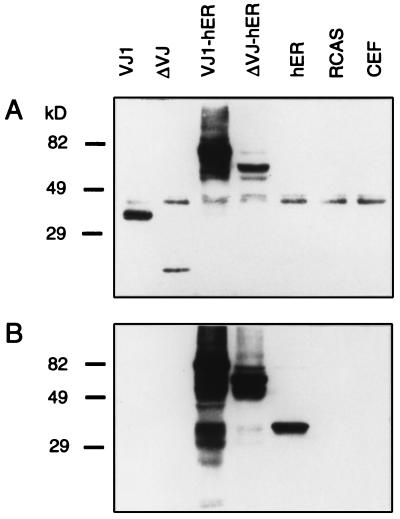
CEFs were infected with RCAS viral stocks expressing the chimeric constructs and control constructs VJ1, ΔVJ, and hER. The cells were passaged twice to allow spread of infectious virus. Expression of the chimeric proteins was examined by Western blot analysis by using rabbit antiserum to chicken v-Jun and to the human ER (Fig. 2). The Jun antiserum detected a band of 39 kDa in all samples, corresponding to endogenous c-Jun (Fig. 2A). The intensity of this band, however, was reduced in transformed cell cultures. This down-regulation of c-Jun by v-Jun has been documented in recent publications (16, 17). The VJ1 protein moved slightly faster than c-Jun because of the deletion of the delta region in v-Jun. The truncated protein ΔVJ appeared as a single band with an apparent molecular mass of 25 kDa. The expression level of ΔVJ was significantly lower than that of the full-length VJ1 protein. The fusion protein VJ1-hER gave a major band of 75 kDa and several faster migrating bands, presumably representing breakdown products. The deletion construct ΔVJ-hER generated a protein band of 60 kDa and a less intense smaller species. Similar to ΔVJ, the deletion construct was less well expressed than the full-length v-Jun when linked to the ER domain. A second blot prepared in parallel and probed with the ER antiserum (Fig. 2B) revealed multiple bands with higher mobility than the VJ1-hER protein and an additional strong band as well as several weak bands for the ΔVJ-hER chimera. The carboxyl-terminal part of the ER protein alone (HE14) migrated as a single band of 35 kDa. We consistently detected additional bands with higher mobility than the full-length fusion proteins. Presumably, the fusion proteins were more susceptible to proteolytic degradation than the parental v-Jun and ER proteins. In the case of VJ1-hER, the high expression level may make degradation products easier to detect.
Figure 2.
Expression of the chimeric constructs in CEFs. Secondary CEFs infected with RCAS constructs bearing the chimeric inserts were lysed in sample buffer and then separated through a 10% SDS-polyacrylamide gel. The proteins were transferred to nitrocellulose and probed with rabbit anti-Jun antiserum (A) or rabbit anti-ER antiserum (B) followed by detection with anti-rabbit horseradish peroxidase conjugate. The position of molecular mass markers (in kDa) is shown on the left.
Transactivation by v-Jun-ER Chimeras.
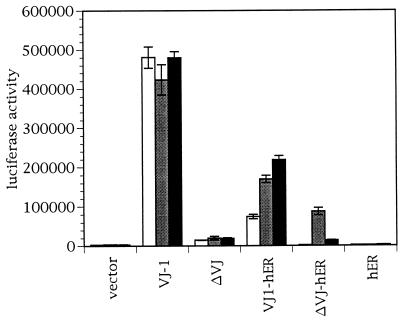
To test the hormone inducibility and the transactivation potential of the fusion proteins, we performed transient transfection experiments in JEG-3 cells, a human choriocarcinoma cell line with low endogenous AP-1 activity (Fig. 3). We chose the HTLV-lux reporter plasmid, containing a hexamer of the AP-1 responsive element in the HTLV-1 long terminal repeat, because this synthetic promoter was highly responsive to v-Jun (18). Cotransfection of the full-length VJ1 protein activated the HTLV promoter (100 × above background). Treatment of cells with estrogen or the partial agonist tamoxifen did not affect the level of VJ1 dependent transactivation. The ΔVJ truncated protein lacks the major transactivation domain of v-Jun and enhances transcription of the HTLV reporter only marginally (4% of VJ1 activity). Surprisingly, the VJ1-hER chimera showed some activity in the absence of inducer (15% of VJ1 activity), but could be further stimulated by the addition of estrogen or tamoxifen (50% of VJ1 activity). Tamoxifen was slightly more effective than estrogen presumably because the TAF-2 function of the hormone binding domain cannot interfere with the activation domain of Jun when stimulated by tamoxifen. A possible activation of VJ1-hER by traces of estrogen in the serum supplement of the tissue culture medium was ruled out by two observations. First, charcoal-stripped serum did not abolish transactivation by VJ1-hER (data not shown). Second, the ΔVJ-hER chimera was completely inactive in the absence of exogenous inducer (background level of activity), even when untreated serum was used. A probable explanation for the ligand-independent transactivation of VJ1-hER is proteolytic cleavage of the ER domain (see above). With the ΔVJ-hER construct, addition of estrogen to the cultures induced transactivation of the reporter (17% of VJ1 activity), but addition of tamoxifen was only minimally effective. These data show that in contrast to VJ1-hER, ΔVJ-hER represents a tightly regulatable chimeric transcription factor. The differential induction of ΔVJ-hER by estrogen suggested that the transactivation capacity of the fusion protein is due to the TAF-2 function localized in the hormone binding domain of the ER (19). It is known that estrogen, but not tamoxifen, activates the TAF-2 function (20).
Figure 3.
Transactivation by v-Jun-ER chimeras. Cells were cotransfected with a luciferase reporter plasmid containing the HTLV promoter together with RCAS expression vectors as indicated. Cells were treated with 2 μM estrogen (gray bars), 200 nM tamoxifen (black bars), or ethanol as solvent control (white bars). Luciferase activity was normalized to the protein content of the samples. The result of a typical experiment done in triplicate for each inducer is shown. The transfection was repeated three times with consistent outcomes.
Transformation of CEF by v-Jun-ER Chimeras.
The v-Jun-ER fusion proteins together with VJ1 and ΔVJ were tested for their ability to transform CEFs in vitro in focus and agar colony assays. The result of a typical focus assay is shown in Table 1. The viral Jun protein VJ1 induced characteristic foci of fusiform cells. Focus formation was not affected by incorporation of estrogen or tamoxifen in the overlay agar. The ΔVJ truncated protein that lacks the major Jun transactivation domain did not form foci. The VJ1-hER fusion protein induced foci even in the absence of estrogen, but focus numbers were higher after addition of estrogen. Interestingly, VJ1-hER induces typical Jun-like foci of elongated cells packed in parallel arrays in the absence of estrogen, whereas induction with estrogen resulted in morphologically different foci. They were not as clearly demarcated, spread more diffusely on the background of the normal monolayer than typical Jun-induced foci, and did not show the pronounced parallel orientation of the transformed cells. The ΔVJ-hER chimera induced foci only in the presence of estrogen, and focus-forming titers were comparable to those of VJ1-hER in the presence of ligand. Tamoxifen did not activate focus formation by the ΔVJ-hER construct. This observation suggests that the ability of ΔVJ-hER to transform cells depends on the TAF-2 transactivation function of the ER portion that is activated by estrogen but not by tamoxifen (cf. Fig. 3). The Jun-derived DNA binding portion of the chimeric construct also plays an essential role in transformation by the chimera as indicated by the lack of transforming activity of the ER hormone binding domain alone. Transfected CEFs also were tested for anchorage-independent growth by colony formation in cloning agar. Each of the constructs capable of inducing neoplastic transformation in monolayer cultures stimulated growth of agar colonies. The VJ1-hER expressing CEFs formed agar colonies without added estrogen, albeit at a lower efficiency, whereas ΔVJ-hER expressing CEFs were strictly dependent on estrogen for anchorage independent growth (Table 2). Additional information on the proliferative capacity of CEFs expressing the chimeric proteins was obtained by determining cell growth and saturation densities. The ΔVJ protein caused a decrease of cell growth as compared with CEF controls possibly by acting as a dominant negative mutant of endogenous Jun, which is essential for the progression through the cell cycle (21, 22). The ΔVJ-hER chimera also showed a decrease in cell growth in the absence of estrogen, but accelerated growth when estrogen was present. This effect was reversible, and withdrawal of estrogen from VJ1-hER transfected cells led to a retardation of cell growth (data not shown).
Table 1.
Focus formation on CEFs infected with VJ-hER constructs
| Infecting construct | Virus stock, FFU/ml
|
||
|---|---|---|---|
| No additive | Estrogen | Tamoxifen | |
| VJ1 | 1.3 × 106 | 1.3 × 106 | 1.3 × 106 |
| ΔVJ | 0 | 0 | 0 |
| VJ1-hER | 6.0 × 104 | 2.1 × 106 | 3.6 × 105 |
| ΔVJ-hER | 0 | 3.0 × 106 | 0 |
| hER | 0 | 0 | 0 |
| RCAS vector | 0 | 0 | 0 |
FFU, focus-forming unit.
Table 2.
Agar colony formation by CEFs infected with VJ-hER constructs
| Infecting construct | Colonies/104 Cells
|
|
|---|---|---|
| No additive | Estrogen | |
| VJ1 | 5 × 103 | 5 × 103 |
| ΔVJ | 0 | 0 |
| VJ1-hER | 4 × 102 | 2 × 103 |
| ΔVJ-hER | 0 | 5 × 103 |
| hER | 0 | 0 |
| RCAS vector | 0 | 0 |
Ligand-Dependent Up-Regulation of the bkj Gene.
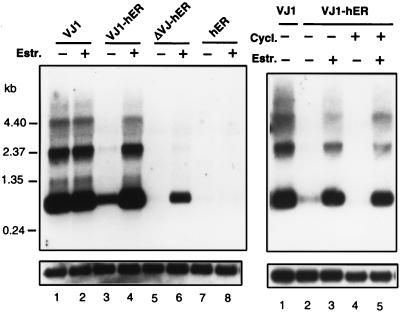
Recently, a quail gene termed bkj, a member of the β-keratin gene family, has been shown to be specifically up-regulated in v-Jun transformed cells (8). To test the effects of the Jun ER chimeras on an endogenous target, we analyzed expression of the bkj gene by Northern blot analysis (Fig. 4). The bkj mRNA is up-regulated in quail embryo fibroblasts transformed by the v-Jun protein VJ1 (Fig. 4, lane 1) as compared with control cultures expressing the hormone binding domain of the ER (Fig. 4, lanes 7 and 8). The VJ1-hER chimera induced expression of bkj only weakly in the absence of hormone. In the presence of estrogen, a stronger induction of bkj was observed. In contrast, induction of the bkj gene by ΔVJ-hER was strictly dependent on estrogen (Fig. 4, lanes 5 and 6), although induction by ΔVJ-hER was not as efficient as by VJ-hER. However, both chimeras were equally active in transformation of CEF in the focus formation assay. This difference may indicate that the bkj gene does not represent a typical transformation-mediating target gene of Jun. To test whether the bkj gene is a direct target of v-Jun, we analyzed the induction of the bkj gene in the presence of 50 μg/ml cycloheximide. Control experiments showed that this concentration of cycloheximide inhibited protein synthesis virtually completely (data not shown). Expression of the bkj gene was induced by estrogen even in cycloheximide-treated cells expressing VJ-hER (Fig. 4). No induction was observed in control cells expressing the hormone binding domain of the estrogen receptor. This result suggests that activation of the bkj gene was a direct effect of Jun.
Figure 4.
Estrogen-dependent activation of the bkj gene. Quail embryo fibroblasts transfected with various RCAS expression vectors were treated with (+) or without (−) estrogen as indicated for 48 hr (Left). Ten micrograms of total RNA per lane was analyzed by Northern blotting. The blot was hybridized with the 32P-labeled bkj cDNA sequence. Equal loading was confirmed by rehybridization with a glyceraldehyde-3-phosphate dehydrogenase probe. The sizes and positions of molecular mass markers are indicated on the left. Cells were grown in the presence (+) or absence (−) of 50 μg/ml cycloheximide either with (+) or without (−) 2 μM estrogen (Right). The hormone was added to the cultures 15 min later than cycloheximide. Cells were harvested 5 hr after addition of hormone. Fifteen micrograms of total RNA per lane were analyzed by Northern blotting.
DISCUSSION
The strategy followed in designing the conditional Jun is based on the observation that the hormone binding domain of steroid receptors can confer ligand-dependent activation to a fusion partner. In the absence of ligand, fusion proteins are inactive because of sequestration by heat-shock binding proteins. This strategy has been used to generate steroid receptor fusion proteins with the adenovirus EIA protein (23), c-Myc (24), c-Fos (25, 26), v-Myb (27), and JunD (28). Recently, two Jun-ER fusions also have been described (29, 30). They show hormone regulatable transcriptional activation, but do not induce oncogenic transformation in the cell systems studied. A crucial advantage of the ER system is the activation of the fusion protein by hormone in the absence of de novo protein synthesis. Activation of Jun while protein synthesis is inhibited allows a distinction between direct targets that will respond under these conditions and indirect targets that will remain unaffected because their regulation depends on the product of a primary target.
The principal result of the current study is that fusion of an amino-terminally truncated v-Jun protein to the hormone binding domain of the ER results in a chimeric protein whose transactivating and oncogenic potentials are tightly regulatable by estrogen. In contrast to ΔVJ-hER, the activity of the fusion product between full-length v-Jun and the ER is not tightly hormone dependent. A likely explanation of this leakiness is the proteolysis observed on Western blots. Even partial degradation of the ER portion would release the chimeric protein from hormone control and convert it into a constitutive transcriptional activator. The same kind of proteolytic cleavage also would act on ΔVJ-hER and would liberate ΔVJ-hER from ligand control. However, ΔVJ lacks a transactivation domain and is not oncogenic. The levels of ΔVJ generated by proteolysis also appear too low to reveal a potential transdominant negative effect that could arise from Jun-specific DNA binding in the absence of transactivation.
Because the transactivation function of ΔVJ-hER is induced only by estrogen but not by tamoxifen, we conclude that the transactivation potential of this fusion product depends on the TAF-2 domain localized within the hormone binding region of the ER. TAF-2 responds to estrogen but not to tamoxifen (20). The effectiveness of TAF-2 in ΔVJ-hER shows that the transactivation domain of Jun can be replaced by a heterologous one and that this domain can be fused to the carboxyl-terminus of the basic leucine zipper region of Jun. This observation is in accord with previous experiments that replaced the transactivation domain of Jun with that of the herpes simplex virus protein VP16. The fusion protein retained transcriptional regulatory as well as oncogenic potential (18).
A recent study reports on a fusion protein between ER and an amino-terminally deleted c-Jun, TAM-67ER (29). This fusion protein is structurally similar to the ΔVJ-hER described here. In the absence of ligand, TAM-67ER exerts a transdominant negative effect on AP-1 activity. This inhibition is relieved by the estrogen-dependent activation of TAF-2. However, the transdominant effect of TAM-67ER on Ras-Jun cotransformation is not abrogated by ligand, in contrast to the hormone regulatable transformation seen with ΔVJ-hER. These observations suggest that the direct oncogenic transformation induced by v-Jun in CEF and the cooperative transformation seen with c-Jun and Ras in mammalian cells depend on different properties of the transactivation domains and may be characterized by divergent downstream mechanisms.
In the absence of estrogen, ΔVJ-hER exerted a growth inhibitory effect on CEF reminiscent of the transdominant actions of other amino-terminal deletions of c-Jun (29). However, this transdominant effect was not strong enough to prevent transformation by coexpressed v-Jun (data not shown), presumably because expression levels of the amino-terminally deleted v-Jun are always low. It may appear surprising that the ΔVJ-hER protein transforms CEFs with an efficiency comparable to v-Jun although its activation potential, relying largely on the TAF-2 function of the ER, amounts to only about 20% of that seen with v-Jun in transient transfection assays. This observation could be explained by proposing that transformation-relevant target genes respond differently to ΔVJ-hER than does the synthetic HTLV-derived reporter plasmid that may not be representative of transformation-specific promoters.
In this study we applied the regulatable Jun transformation system to the analysis of one previously reported Jun target, bkj (8). The activation of the bkj gene by VJ1-hER and ΔVJ-hER shows that the Jun-ER fusion proteins can regulate endogenous targets of Jun. Independence of this activation from de novo protein synthesis operationally classifies bkj as a direct target of Jun. Tightly regulatable transactivation and transformation will be essential in tracing the downstream effects of Jun via target genes to their combined manifestation in the oncogenic cellular phenotype.
Acknowledgments
We thank Susan Burke for the preparation of the manuscript. We thank Pierre Chambon for his generous gift of plasmid HE14 and Markus Hartl for the bkj cDNA clone. Quail eggs were generously provided by the laboratory of Barry W. Wilson, Division of Avian Sciences, University of California at Davis. This work was supported by U.S. Public Health Service Grant CA 42564.
ABBREVIATIONS
- AP-1
activator protein 1
- CEF
chicken embryo fibroblast
- ER
estrogen receptor
- v-Jun
viral Jun protein
- c-Jun
cellular Jun protein
- HTLV
human T cell leukemia virus
- TAF-2
transactivation function 2
Note Added in Proof
M. Hartl and K. Bister (personal communication) have obtained independent evidence indicating that the bkj gene that they isolated is a direct target of Jun.
References
- 1.Maki Y, Bos T J, Davis C, Starbuck M, Vogt P K. Proc Natl Acad Sci USA. 1987;84:2848–2852. doi: 10.1073/pnas.84.9.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bos T J, Monteclaro F S, Mitsunobu F, Ball A R, Jr, Chang C H W, Nishimura T, Vogt P K. Genes Dev. 1990;4:1677–1687. doi: 10.1101/gad.4.10.1677. [DOI] [PubMed] [Google Scholar]
- 3.Angel P, Karin M. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 4.Karin M, Liu Z-G, Zandi E. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 5.Morgan I M, Ransone L J, Bos T J, Verma I M, Vogt P K. Oncogene. 1992;7:1119–1125. [PubMed] [Google Scholar]
- 6.Håvarstein S L, Morgan I M, Wong W-Y, Vogt P K. Proc Natl Acad Sci USA. 1992;89:618–622. doi: 10.1073/pnas.89.2.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliviero S, Robinson G S, Struhl K, Spiegelman B M. Genes Dev. 1992;6:1799–1809. doi: 10.1101/gad.6.9.1799. [DOI] [PubMed] [Google Scholar]
- 8.Hartl M, Bister K. Proc Natl Acad Sci USA. 1995;92:11731–11735. doi: 10.1073/pnas.92.25.11731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadman M, Gabos L, Loo M, Sehgal A, Bos T J. Oncogene. 1996;12:135–142. [PubMed] [Google Scholar]
- 10.Vogt P K. In: Fundamental Techniques in Virology. Habel K, Salzmann N P, editors. New York: Academic; 1969. pp. 198–211. [Google Scholar]
- 11.Hughes S H, Greenhouse J J, Petropoulos C J, Sutrave P. J Virol. 1987;61:3004–3017. doi: 10.1128/jvi.61.10.3004-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohler P O, Bridson W E. J Clin Endocrinol. 1971;32:683–687. doi: 10.1210/jcem-32-5-683. [DOI] [PubMed] [Google Scholar]
- 13.Kumar V, Green S, Staub A, Chambon P. EMBO J. 1986;5:2231–2236. doi: 10.1002/j.1460-2075.1986.tb04489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 15.Tora L, Mullick A, Metzger D, Ponglikitmongkol M, Park I, Chambon P. EMBO J. 1989;8:1981–1986. doi: 10.1002/j.1460-2075.1989.tb03604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kilbey A, Black E J, Unlu M, Gillespie D A F. Oncogene. 1996;12:2409–2418. [PubMed] [Google Scholar]
- 17.Gao M, Morgan I, Vogt P K. Cancer Res. 1996;56:4229–4235. [PubMed] [Google Scholar]
- 18.Schuur E R, Parker E J, Vogt P K. Cell Growth Differ. 1993;4:761–768. [PubMed] [Google Scholar]
- 19.Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, Chambon P. Cell. 1989;59:477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- 20.Webster N J G, Green S, Jin J R, Chambon P. Cell. 1988;54:199–207. doi: 10.1016/0092-8674(88)90552-1. [DOI] [PubMed] [Google Scholar]
- 21.Brown P H, Alani R, Preis L H, Szabo E, Birrer M J. Oncogene. 1993;8:877–886. [PubMed] [Google Scholar]
- 22.Johnson R S, van Lingen B, Papaioannou V E, Spiegelman B M. Genes Dev. 1993;7:1309–1317. doi: 10.1101/gad.7.7b.1309. [DOI] [PubMed] [Google Scholar]
- 23.Picard D, Salser S J, Yamamoto K R. Cell. 1988;54:1073–1080. doi: 10.1016/0092-8674(88)90122-5. [DOI] [PubMed] [Google Scholar]
- 24.Eilers M, Picard D, Yamamoto K R, Bishop J M. Nature (London) 1989;340:66–68. doi: 10.1038/340066a0. [DOI] [PubMed] [Google Scholar]
- 25.Superti-Furga G, Bergers G, Picard D, Busslinger M. Proc Natl Acad Sci USA. 1991;88:5114–5118. doi: 10.1073/pnas.88.12.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuermann M, Hennig G, Müller R. Oncogene. 1993;8:2781–2790. [PubMed] [Google Scholar]
- 27.Burk O, Klempnauer K-H. EMBO J. 1991;10:3713–3719. doi: 10.1002/j.1460-2075.1991.tb04939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Francis M K, Phinney D G, Ryder K. J Biol Chem. 1995;270:11502–11513. doi: 10.1074/jbc.270.19.11502. [DOI] [PubMed] [Google Scholar]
- 29.Kim S, Brown P H, Birrer M J. Oncogene. 1996;12:1043–1053. [PubMed] [Google Scholar]
- 30.Fialka I, Schwarz H, Reichmann E, Oft M, Busslinger M, Beug H. J Cell Biol. 1996;132:1115–1132. doi: 10.1083/jcb.132.6.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]