Abstract
Vaccinia uses actin-based motility for virion movement in host cells, but the specific protein components have yet to be defined. A cardinal feature of Listeria and Shigella actin-based motility is the involvement of vasodilator-stimulated phosphoprotein (VASP). This essential adapter recognizes and binds to actin-based motility 1 (ABM-1) consensus sequences [(D/E)FPPPPX(D/E), X = P or T] contained in Listeria ActA and in the p90 host-cell vinculin fragment generated by Shigella infection. VASP, in turn, provides the ABM-2 sequences [XPPPPP, X = G, P, L, S, A] for binding profilin, an actin-regulatory protein that stimulates actin filament assembly. Immunolocalization using rabbit anti-VASP antibody revealed that VASP concentrates behind motile virions in HeLa cells. Profilin was also present in these actin-rich rocket tails, and microinjection of 10 μM (intracellular) ABM-2 peptide (GPPPPP)3 blocked vaccinia actin-based motility. Vinculin did not colocalize with VASP on motile virions and remained in focal adhesion contacts; however, another ABM-1-containing host protein, zyxin, was concentrated at the rear of motile virions. We also examined time-dependent changes in the location of these cytoskeletal proteins during vaccinia infection. VASP and zyxin were redistributed dramatically several hours before the formation of actin rocket tails, concentrating in the viral factories of the perinuclear cytoplasm. Our findings underscore the universal involvement of ABM-1 and ABM-2 docking sites in actin-based motility of Listeria, Shigella, and now vaccinia.
The association of actin filaments with intracellular vaccinia was first discovered over two decades ago (1). However, characterization of the mechanisms responsible for virion-induced host-cell actin filament assembly has proved to be a daunting task. One strategy for exploring how vaccinia induces actin assembly is to identify and assess the contribution of viral protein analogues of cytoskeleton components known to be involved in actin-based motility. For example, genetic deletion of vaccinia’s A42R open-reading frame for a profilin-like protein failed to impair actin assembly (2), and there was no effect on deletion of the F8L viral protein containing a 42 residue region with homology to the Listeria ivanovi surface protein iActA (3). On the other hand, two viral membrane proteins, B5R and A34R, do affect actin rocket tail formation (4–7). Viral maturation and subsequent locomotory competence require that the intracellular mature virion be wrapped by membranes derived from the trans-Golgi network or by early endosomes to form the intracellular enveloped virion (6). Subsequently, the outer membrane of the intracellular enveloped virion fuses with the plasma membrane, releasing an extracellular enveloped virion. B5R and A34R affect intracellular enveloped virion and extracellular enveloped virion formation, respectively (5, 7). The observation that mutants defective in intracellular enveloped virion and extracellular enveloped virion formation are defective in rocketing implies that induction of actin tail formation requires components of the outer Golgi-derived envelope.
The bacterial pathogens Listeria and Shigella stimulate explosive assembly of new actin filaments near their surface, thereby providing the force for their intracellular movement and filopod formation required for cell-to-cell spread (8, 9, 10). Cudmore et al. (11) have demonstrated that vaccinia also uses a host-cell, actin-based mechanism to move at speeds comparable to those observed with Listeria and Shigella. Recent success in defining the actin-based motors of Listeria (12, 13) and Shigella (14) provide important clues about vaccinia motility. Listeria ActA, a surface protein required for actin-based motility, contains four oligoproline sequences of the type FEFPPPPTDE. This sequence represents an actin-based motility 1 (ABM-1) binding motif important for attracting the actin regulatory protein vasodilator-stimulated phosphoprotein (VASP) (15, 16, 17). VASP, in turn, contains a tandem series of oligoproline repeats of the type GPPPPP, representing typical ABM-2 binding sequences that attract the actin-monomer binding protein profilin (13, 15, 18). Profilin stimulates actin filament growth by enhancing addition of actin monomers to barbed or fast growing ends of actin filaments (19). Shigella generates a similar actin-based motor by using its IcsA surface protein, which binds to a proteolytic fragment of vinculin containing an ABM-1 sequence that links VASP to the surface of motile Shigella (14). Another actin-based motile process is endosomal rocketing, which is generated by treating bone marrow macrophages with lanthanum/zinc. After lanthanum treatment, endosomes generate actin filament rocket tails and move through the cytoplasm at velocities similar to those observed with Listeria and Shigella. Endosomes require a different adapter protein, namely zyxin, a host-cell actin-regulatory protein containing two ABM-1 sequences (20). We now demonstrate (i) that vaccinia also assembles an ABM complex (consisting of zyxin, VASP, and profilin) and (ii) that during viral infection, VASP and zyxin are recruited from focal adhesion contacts to viral assembly factories. These observations underscore the universal use of ABM-1 and ABM-2 docking sites in actin-based motility.
MATERIALS AND METHODS
Cells and Viruses.
HeLa cells S3 (ATCC CCL 2.2) were grown in MEM (GIBCO/BRL) supplemented with 10% fetal bovine serum. Infected or mock-infected HeLa cells were grown on 35-mm tissue culture dishes to 80% confluency. Vaccinia virus strain WR was obtained from the American Type Culture Collection (ATCC VR-1354), and infectivity was determined by plaque assay. Vaccinia virus infections of cells were carried out in media containing 10% fetal bovine serum and were incubated at 37°C in a 5% CO2 atmosphere as described (21, 22). Cells were infected with vaccinia virus at a multiplicity of infection of 10. Shigella infection of HeLa cells has been described by Zeile et al. (10).
Antibodies.
Anti-vinculin 11–5 mouse mAb (Vin 11–5), anti-α-tubulin mouse mAb, and anti-α-actinin mouse mAb were obtained from Sigma. The rabbit anti-VASP and anti-profilin polyclonal antibodies were generated by immunizing with recombinant human VASP and recombinant human profilin coupled to keyhole limpet hemocyanin (Cocalico Biologicals, Reamstown, PA). Antiprofilin monospecific IgG was isolated by immunoaffinity chromatography on recombinant profilin coupled to cyanogen bromide-activated Sepharose 4B. Antizyxin B38 antibody was a kind gift from M. C. Beckerle (University of Utah, Salt Lake City).
Indirect Immunofluorescence.
At specified times, the cells were fixed in 3.7% formalin in PBS for 20 min at room temperature. The cells were permeabilized for 5 min with 0.2% Triton X-100 at room temperature. Antibody staining was performed as described (23). Secondary antibodies were prepared in blocking buffer at a dilution of 1:500. Secondary antibodies used were anti-mouse or anti-rabbit IgG-conjugated to fluorescein isothiocyanate (Sigma). F-actin was stained with rhodamine-conjugated phalloidin (Molecular Probes). In all experiments, F-actin staining was performed simultaneously with antibody staining and demonstrated colocalization of F-actin with profilin, VASP, and zyxin in vaccinia-infected HeLa cells (data not shown). For Hoechst staining of nucleic acids, a stock solution of Hoechst stain (Pierce) of 1 mg/ml in dimethyl sulfoxide was diluted to 1:500 in PBS.
Microscopy.
A Nikon Diaphot inverted microscope equipped with a cooled charge-coupled device camera (Hamamatsu Photonics, Hamamatsu City, Japan) was used to obtain digital images that were processed by using an Image-1 computer image analyzer (Universal Imaging, Media, PA).
Western Blot Analysis.
HeLa cells were grown on 60-mm tissue culture dishes to 100% confluency, were washed three times in PBS, and were harvested by scraping into 200 μl of lysis buffer (0.5 M Tris⋅HCl, pH 7.5/0.15 M NaCl/1% Triton X-100/0.1% SDS/2 μg/ml aprotinin/1 μg/ml leupeptin/ 1 μg/ml pepstatin/0.1 mM phenylmethylsulfonyl fluoride). Cells were disrupted by sonication with a microtip sonicator for ≈1 min on ice. Sucrose gradient purified vaccinia virus, according to Joklik (22), was solubilized in lysis buffer and was sonicated as above. The resulting viral preparations have been shown to consist primarily of intracellular mature virion (22). Protein concentration of cell lysates and vaccinia lysates was determined by the bicinchoninic acid assay (Pierce). HeLa cell total protein (20 μg) and vaccinia virus total protein (10 μg) were separated by SDS/PAGE, were transferred to PVDF membrane, and were treated as described (14). Primary antibodies antivinculin clone 11–5, antiprofilin, and anti-VASP were used at a dilution of 1:200. Secondary antibodies, including goat anti-rabbit IgG and rabbit anti-mouse IgG conjugated to horseradish peroxidase, were used at a dilution of 1:20,000.
RESULTS
Antibodies directed against known actin regulatory proteins were used to identify potential components of the ABM complex of vaccinia. This strategy requires that antibodies be specific and not cross-react with vaccinia proteins. Western blots of HeLa lysates and purified vaccinia lysate were probed with antibodies against human VASP, human profilin, and human vinculin. Each antibody specifically recognized a single polypeptide in HeLa lysate and did not recognize epitopes in the vaccinia lysate. Preimmune sera of anti-VASP and antiprofilin had no reactivity toward HeLa cell or vaccinia lysates (data not shown).
VASP Is Localized to the Actin Tail Interface of Vaccinia Virus.
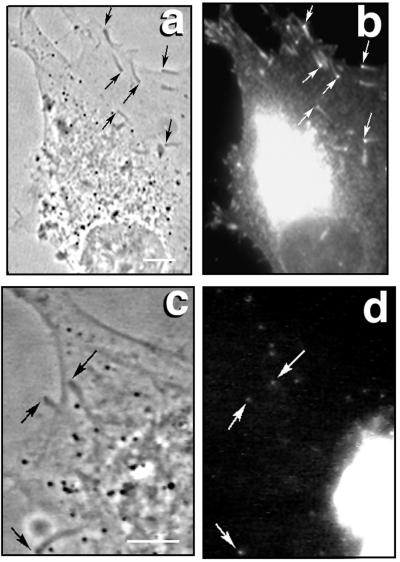
As noted earlier, VASP is an essential adapter protein used in Shigella, Listeria, and endosomal rocketing. We now demonstrate that VASP is localized at the junction of the actin rocket tail and the intracellular virion (Fig. 1). Fig. 1 a and b are the respective phase-contrast and fluorescent images of vaccinia-infected HeLa cells 7 hours after initiating the infection. Fig. 1a shows the phase-dense rocket tails behind the intracellular virions. As previously observed (24), simultaneous staining with rhodamine phalloidin confirmed that these rocket tails were rich in actin filaments (data not shown). In Fig. 1b, the anti-VASP antibody reacted intensely at the virion-rocket tail interface. Less intense staining was observed throughout the rocket tails. Hoechst staining proved that each rocket tail was preceded by a DNA containing virion (Fig. 1c, phase-contrast image; Fig. 1d, fluorescent image). Simultaneous staining with the Hoechst dye and anti-VASP antibody revealed that VASP was localized directly behind each viral particle (data not shown). As a control, we used Shigella-infected cells to demonstrate that the anti-VASP antibody was also highly concentrated behind each motile bacterium (data not shown). To assess the functional importance of VASP for viral actin-based locomotion, the ABM-2 oligoproline peptide GPPPPPGPPPPPGPPPPP (estimated intracellular concentration, 10 μM; needle concentration, 100 μM) was microinjected into vaccinia-infected HeLa cells at 7 h. This peptide can competitively inhibit profilin binding to VASP (13) and, when introduced into cells, caused the abrupt inhibition of virion movement (mean preinjection velocity, 0.06 ± 0.02 μm/sec, n = 24 vs. postinjection velocity of 0.01 ± 0.01 μm/sec, n = 24, P < 0.0001). Microinjection of an equivalent volume of PBS had no significant effect on the velocity of virion movement (mean preinjection velocity 0.03 ± .01 μm/sec, n = 18 vs. post-injection velocity of 0.04 ± 0.01 n = 18, P = 0.21). Identical concentrations of the (GPPPPP)3 peptide were shown to inhibit Listeria and Shigella intracellular movement (10, 18).
Figure 1.
VASP localization in vaccinia-infected cells. (a and c) Phase-contrast micrographs of HeLa cells infected with vaccinia for 7 h. (b) Immunofluorescent micrograph using anti-VASP antibody. Arrows point to the virus-actin tail interface. Viral factories also are intensely stained. (d) Hoechst dye staining for double-stranded DNA locates motile viral particles (arrows) and viral factories in the cytoplasm.
VASP would be expected to concentrate profilin in the same regions of the vaccinia rocket tail. As shown in Fig. 2 a and b, profilin was routinely detected in the vaccinia rocket tails. On occasion, profilin appeared to be more concentrated at the viral-actin tail interface (lower right-hand arrow in Fig. 2b) but more often was dispersed throughout the actin tail. Profilin also was seen throughout the peripheral cytoplasm.
Figure 2.
Immunolocalization of profilin and α-actinin in vaccinia-infected cells. (a and c) Phase-contrast micrographs of HeLa cells infected for 7 h with vaccinia. (b) Immunolocalization with antiprofilin antibody. (d) Anti-α-actinin antibody staining.
The actin filaments of stress fibers and the actin filaments of bacterial actin rocket tails are stabilized by the actin bundling protein α-actinin. Hiller et al. (25) demonstrated that α-actinin was present in the actin tail of vaccinia virus. We also found that this important component was colocalized with the vaccinia-induced actin rocket tails (Fig. 2 c and d).
Identification of Zyxin as the ABM-1 Adapter in the Vaccinia Actin Motor.
Taken together with earlier evidence for VASP involvement in Listeria motility (17) and endosomal rocketing (20), the above experiments indicate that VASP is a universal component in actin-based motility. In order for VASP to be concentrated at the back of virions, a protein containing ABM-1 modules must be present. Searches of eukaryotic protein databases indicate that vinculin and zyxin are the only two host cell proteins that contain ABM-1 sequences (15). To identify the ABM-1 donor for vaccinia, we considered two possibilities: that vaccinia provides its own ABM-1 module for binding VASP or that the virion attracts vinculin or zyxin. We can exclude the first possibility because our database search of the vaccinia genome failed to identify any protein containing FPPPP ABM-1 sequences. Immunofluorescence staining with antivinculin antibody detected this protein in the focal contacts but not near the virions or in the virus-induced rocket tails (see Fig. 5a). The other candidate VASP docking protein, zyxin, contains two ABM-1 sequences and binds VASP in vitro (32) and in vivo (20). Zyxin could be detected readily at the vaccinia-actin tail interface as well as along the length of the rocket tail (Fig. 3). The intensity of VASP staining at the vaccinia-actin tail junction was somewhat greater than seen for zyxin; however, scans of the relative fluorescence intensity along the tails did demonstrate that zyxin, like VASP and filamentous actin, was concentrated more highly near the viral particle (data not shown). Finally, the upper right quadrant of the cell shown in Fig. 3 also contains several virally induced filopods. Such structures also are observed in Listeria and Shigella infection, and these pathogen-containing membrane projections are thought to play an important role in cell-to-cell spread.
Figure 5.
Comparisons of HeLa cells before and after a 7-h infection with vaccinia. (a) Antivinculin immunofluorescence staining of uninfected (upper right panel) and infected (lower right panel) cells. (b) Antitubulin immunofluorescence staining of uninfected (upper right panel) and infected (lower right panel) cells. Corresponding phase-contrast images are shown in the left-hand panels.
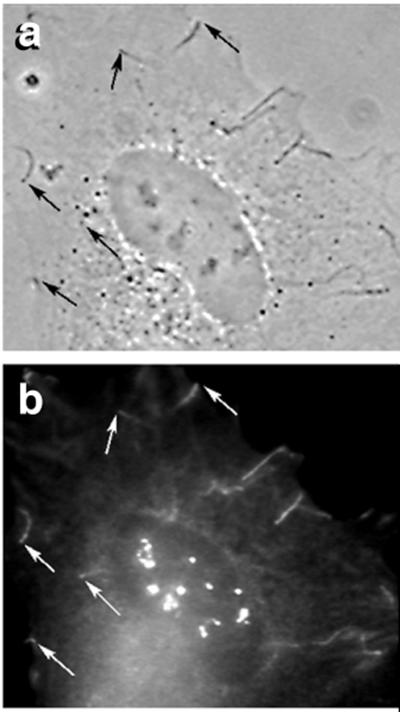
Figure 3.
Immunolocalization of zyxin in vaccinia-infected HeLa cells. (a) Phase-contrast image 7 h after initiation of vaccinia infection. (b) Immunofluorescence micrograph of same cell with anti-zyxin antibody. The interfaces between the viral particles and actin-rocket tails are indicated with arrows. The structures not marked by arrows at the upper right are viral-induced filopodia, resembling those observed in Listeria- and Shigella-infected cells. Note the nucleolar staining as well as the fainter perinuclear staining at the lower left.
Redistribution of VASP and Zyxin in the Cytoplasm of Vaccinia-Infected Cells.
We observed a dramatic redistribution of VASP (Fig. 4a) and zyxin (Fig. 4b) in the cytoplasm of vaccinia-infected cells. Before infection (Fig. 4 a and b, top), VASP and zyxin were concentrated at focal adhesion contacts in essentially the same pattern as vinculin (Fig. 5a, upper). One hour after infection, the cells began to round up and were less adherent; however, VASP and zyxin remained localized at sites of focal adhesion (Fig. 4, 1 h). Within 3–5 h after infection, VASP and zyxin were depleted from the focal contacts while vinculin remained (Fig. 4, 3- to 7-h time points vs. Fig. 5a, lower). The loss of zyxin and VASP from focal contacts was associated with flattening and spreading of the cells. In addition to being localized to the moving virions, VASP and zyxin were seen in the region adjacent to the nucleus known to contain viral factories, as determined by Hoechst staining for nucleic acids (data not shown). In order for vaccinia to move from the perinuclear region toward the peripheral membrane, actin-based motility must begin near the viral factories; therefore, we would expect VASP to be present in this region. Zyxin also became concentrated in the nucleus, particularly nucleoli. At 7 h, the number of rocketing virions reached its maximum, and VASP and zyxin were found at each actin tail–virus junction as well as being concentrated in the perinuclear and, in the case of zyxin, nuclear region. Over the next 3 h of infection, VASP and zyxin localization to the rocket tails and the perinuclear region remained unchanged (Fig. 4, 10h). Throughout the entire time course of infection, host cell vinculin remained localized to the focal adhesion contacts (Fig. 5a, lower). Moreover, vinculin never was found at the virus–actin tail interface, indicating that vinculin does not serve as an ABM-1 adapter for vaccinia motility. We also examined the microtubule cytoskeleton in infected and uninfected cells. It was evident that the microtubule structure was not affected to any major degree during infection by the vaccinia virus (Fig. 5b).
Figure 4.
Time-course of VASP and zyxin redistribution in vaccinia-infected cells. (a) Immunofluorescence images using anti-VASP antibody (right-hand panels) and corresponding phase-contrast images (left-hand panels). The time after initiating infection is shown for each pair of micrographs. (b) Immunofluorescence images using antizyxin antibody (right-hand panels). Note the intense nuclear staining at the later time points in addition to faint perinuclear staining. Phase-contrast images are in left-hand panels.
DISCUSSION
Over 20 years have elapsed since the discovery that polar actin filament structures assemble at one end of the intracellular vaccinia virions (1, 25). The complex life-cycle of this pathogen (2, 7, 26, 27) hampered identification of those viral and host proteins needed for actin-based motility and cell-to-cell spread. Rather than approaching this problem from the perspective of viral proteins, we sought to identify the host cell protein components of the ABM complex of vaccinia. Vaccinia, Listeria, and Shigella all achieve similar rates of intracellular locomotion and form actin filament rocket tails. These similarities raised the possibility that this virus might use the same host cell components as the bacteria. A cardinal feature of Listeria and Shigella motility is the polarized accumulation of VASP at the site of new actin filament assembly. The immunofluorescence experiments presented here now clearly demonstrate that VASP concentrates in an identical fashion behind motile virions. We also have found that VASP is released from the focal contacts of the host cell, becoming highly concentrated in the same perinuclear region in which viral factories are located. VASP relocation precedes actin rocket tail formation by several hours. This reorganization is specific for VASP, not being observed with tubulin or vinculin. These findings, combined with the inhibition of virion movement by microinjection of the ABM-2 peptide (GPPPPP)3, indicate that VASP plays a critical functional role in the assembly of the viral ABM complex. However, we cannot exclude the possibility that VASP also may be involved intimately in viral maturation.
How is VASP attracted to the virions? As noted in the introduction, Listeria uses its surface protein ActA, which contains a series of ABM-1 oligoproline repeats that bind VASP with high affinity, and Shigella uses its surface protein IcsA to recruit a 90-kDa proteolytic fragment of host cell vinculin containing an unmasked ABM-1 sequence (15). In the case of vaccinia, deletion of the viral genes with sequences homologous to profilin or iActA fails to impair vaccinia actin-based motility. Therefore, it is unlikely that a viral protein is involved directly in attracting VASP to the virion surface. Earlier studies demonstrated that the virus particles pass through the Golgi apparatus and become enveloped by host cell membranes during their maturation (7, 28). In light of our discovery that VASP becomes highly concentrated in this same perinuclear region, we suspect that the virions may form structures analogous to the rocketing endosomes in lanthanum-treated bone marrow macrophages. Within 30 to 60 minutes after exposure to lanthanum, these macrophages develop large endosomes with zyxin on their surfaces to recruit VASP to generate an ABM complex (20). Zyxin is an 88-kDa host cell protein normally concentrated in focal contacts, and this protein contains two ABM-1 sequences that interact with VASP (14). Our immunofluorescence studies with antizyxin support a similar role for zyxin as an adapter protein for the assembly by vaccinia of an ABM-complex. Like VASP, zyxin is concentrated in the actin rocket tails of moving virions. Zyxin does not appear to be concentrated as highly as VASP at the viral–actin rocket tail junction. This difference may reflect the lower affinity of the antizyxin antibody as compared with our anti-VASP antibody. Secondly, because VASP binds with high affinity to zyxin, VASP binding may mask zyxin epitopes and thereby reduce antizyxin antibody binding. The fainter staining of zyxin along the rocket tails may result from the well established binding of zyxin to α-actinin (29); this would explain why VASP is likewise found at a lower intensity along the rocket tail. A similar pattern of staining was seen with Shigella (14) and, in that case, vinculin also is known to possess a weak binding site for α-actinin (30). Finally, zyxin displays high-affinity binding to vaccinia, providing further evidence that zyxin is an integral component of the ABM-complex (W.L.Z., R.C.C., D.L.P. and F.S.S., unpublished work).
Beyond its role in focal adhesion contacts, zyxin contains zinc-finger motifs for DNA binding as well as a nuclear export sequence (31). During the course of viral infection, we discovered that zyxin became highly concentrated in the nucleus, suggesting that the nuclear export sequence in zyxin may be masked or deleted during viral infection. It is also possible that the virus disrupts nuclear-cytoplasmic transport.
In conclusion, the present investigation points to the universal involvement of ABM-1 and ABM-2 sequences in actin-based motility. Listeria, Shigella, endosomes, and now vaccinia all use ABM-1 sequences to link VASP to their surfaces. Each VASP molecule contains 20–24 ABM-2 sequences, and each binds a profilin molecule (see Fig. 6). The high concentrations of profilin generated by these binding interactions create a polymerization zone for explosive assembly of actin filaments. This same mechanism is likely to apply to other actin-based motile processes, such as axonal growth cone movement, platelet shape changes, and amoeboid motion.
Figure 6.
Schematic representation of the binding interactions leading to the formation of an ABM complex.
Acknowledgments
This project was supported by National Institutes of Health RO1 Grants AI23262, AI34276, and AI18094.
ABBREVIATION
- VASP
vasodilator-stimulated phosphoprotein
References
- 1.Stokes G V. J Virol. 1976;18:636–643. doi: 10.1128/jvi.18.2.636-643.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blasco R, Cole N B, Moss B. J Virol. 1991;65:4598–4608. doi: 10.1128/jvi.65.9.4598-4608.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higley S, Way M. J Gen Virol. 1997;78:2633–2637. doi: 10.1099/0022-1317-78-10-2633. [DOI] [PubMed] [Google Scholar]
- 4.Herrera E, Mar Lorenzo M D, Blasco R, Isaacs S N. J Virol. 1998;72:294–302. doi: 10.1128/jvi.72.1.294-302.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathew E, Sanderson C M, Hollinshead M, Smith G L. J Virol. 1998;72:2429–2438. doi: 10.1128/jvi.72.3.2429-2438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmelz M, Sodeik B, Ericsson M, Wolffe E J, Shida H, Hiller G, Griffiths G. J Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolffe E J, Katz E, Weisberg A, Moss B. J Virol. 1997;71:3904–3915. doi: 10.1128/jvi.71.5.3904-3915.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernardini M L, Mounier J, D’Hauteville H, Coquis-Rondon M, Sansonetti P. Proc Natl Acad Sci USA. 1989;86:3867–3870. doi: 10.1073/pnas.86.10.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dabiri G A, Sanger J M, Portnoy D A, Southwick F S. Proc Natl Acad Sci USA. 1990;87:6068–6072. doi: 10.1073/pnas.87.16.6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeile W, Purich D L, Southwick F S. J Cell Biol. 1996;133:49–59. doi: 10.1083/jcb.133.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cudmore S, Cossart P, Griffiths G, Way M. Nature (London) 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 12.Southwick F S, Purich D L. Proc Natl Acad Sci USA. 1994;91:5168–5172. doi: 10.1073/pnas.91.11.5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinhard M, Giehl K, Abel K, Haffner C, Jarchau T, Hoppe V, Jockusch B M, Walter U. EMBO J. 1995;14:1583–1589. doi: 10.1002/j.1460-2075.1995.tb07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laine R O, Zeile W L, Kang F, Purich D L, Southwick F S. J Cell Biol. 1997;138:1255–1264. doi: 10.1083/jcb.138.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purich D L, Southwick F S. Biochem Biophys Res Communs. 1997;231:686–691. doi: 10.1006/bbrc.1997.6158. [DOI] [PubMed] [Google Scholar]
- 16.Pistor S, Chakroborty T, Walter U, Wheland J. Curr Biol. 1995;5:517–525. doi: 10.1016/s0960-9822(95)00104-7. [DOI] [PubMed] [Google Scholar]
- 17.Chakraborty T, Ebel F, Domann E, Niebuhr K, Gerstel B, Pistor S, Temm-Grove J, Jockusch B M, Reinhard M, Walter U, et al. EMBO J. 1995;14:1314–1321. doi: 10.1002/j.1460-2075.1995.tb07117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang F, Laine R O, Bubb M R, Southwick F S, Purich D L. Biochemistry. 1997;36:8384–8392. doi: 10.1021/bi970065n. [DOI] [PubMed] [Google Scholar]
- 19.Pantaloni D, Carlier M F. Cell. 1993;75:1007–1014. doi: 10.1016/0092-8674(93)90544-z. [DOI] [PubMed] [Google Scholar]
- 20.Southwick F S, Heuser J E, Beckerle M C, Shen P, Purich D L. Mol Biol Cell. 1997;8:169a. (abstr.). [Google Scholar]
- 21.Condit R C, Motyczka A. Virology. 1981;113:224–241. doi: 10.1016/0042-6822(81)90150-1. [DOI] [PubMed] [Google Scholar]
- 22.Joklik W K. Virology. 1962;18:9–18. doi: 10.1016/0042-6822(62)90172-1. [DOI] [PubMed] [Google Scholar]
- 23.Sanger J W, Sanger J M, Jockusch B M. J Cell Biol. 1983;96:961–969. doi: 10.1083/jcb.96.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiller G, Weber K. J Virol. 1982;44:647–657. doi: 10.1128/jvi.44.2.647-657.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiller G, Weber K, Schneider L, Parajsz C, Jungwirth C. Virology. 1979;98:142–153. doi: 10.1016/0042-6822(79)90533-6. [DOI] [PubMed] [Google Scholar]
- 26.Ramsey-Ewing A, Moss B. Virology. 1998;242:138–149. doi: 10.1006/viro.1997.8985. [DOI] [PubMed] [Google Scholar]
- 27.Hu X, Carroll L J, Wolffe E J, Moss B. J Virol. 1996;70:7669–7677. doi: 10.1128/jvi.70.11.7669-7677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sodeik B, Doms R W, Ericsson M, Hiller G, Machamer C E, van’t Hof W, van Meer G, Moss B, Griffiths G. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crawford A W, Michelsen J W, Beckerle M. J Cell Biol. 1992;116:1381–1393. doi: 10.1083/jcb.116.6.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wachsstock D H, Wilkins J A, Lin S. Biochem Biophys Res Commun. 1987;146:554–560. doi: 10.1016/0006-291x(87)90564-x. [DOI] [PubMed] [Google Scholar]
- 31.Beckerle M. BioEssays. 1997;19:949–957. doi: 10.1002/bies.950191104. [DOI] [PubMed] [Google Scholar]
- 32.Reinhard M, Jourenal K, Tripier D, Walter U. Proc Natl Acad Sci USA. 1995;92:7956–7960. doi: 10.1073/pnas.92.17.7956. [DOI] [PMC free article] [PubMed] [Google Scholar]